Introduction
Cutaneous malignant melanoma (CMM) is the most
life-threatening primary skin malignancy, with a high global
incidence rate amongst the Caucasian population. In Bulgaria,
>470 new cases of melanoma are diagnosed each year (1). Once diagnosed, CMM can remain latent
for a long period of time or can rapidly metastasize. Following
distant metastasis, patient prognosis is poor, with an average
survival time of 6–8 months, with only 11% of patients surviving
beyond 2 years (2–4).
Numerous studies have investigated the genetic
factors involved in the development of sporadic melanoma, including
genes involved in the regulation of skin pigmentation, the cell
cycle, DNA repair, the oxidative stress defense system and the
production of immune modulatory mediators (5–12).
Previous evidence also suggested that patients with CMM mounted an
efficient immune response towards the tumor leading in some cases
to spontaneous regression, although in most cases these responses
did not prevent tumor progression (13,14). The
pro-inflammatory cytokines interleukin (IL)-6 and tumor necrosis
factor-α (TNF-α) are among the factors involved in this response
(13,15). IL-6 is a major pro-inflammatory
mediator produced by various cell types, including melanoma cells,
which exerts different biological activities towards a variety of
target cells (16,17). IL-6 is reportedly involved in the
differentiation of myeloid-derived suppressor cells and the
reinforcement of their suppressive function; it is also associated
with increased production of immunosuppressive cytokines by tumor
cells, and increased metastasis in melanoma (18,19).
Furthermore, the expression of IL-6 has been shown to promote the
progression of CMM. Elevated pre-treatment levels of serum IL-6
have been determined as an independent prognostic biomarker of
reduced overall survival (20).
Moreover, the pro-tumorigenic effects of IL-6 have been attributed
to its regulatory role in tumor angiogenesis, proliferation,
survival and tumor cell motility (15,21,22). The
effects of IL-6 are mediated by the stimulation of signal
transducer and activator of transcription 3, which profoundly
influences melanoma angiogenesis and cellular proliferation by
transcriptionally regulating basic fibroblast growth factor,
vascular endothelial growth factor, matrix metalloproteinase-2, Wnt
family member 5A, Twist and N-cadherin (22,23).
TNF-α also plays a role in the development of CMM.
Notably, increased TNF-α expression, stimulated by exposure to UV
radiation, has been reported to contribute to antitumor immune
escape (13). Together with IL-6,
TNF-α has been suggested as one of the key modulators of melanoma
cell aggressiveness, which these cells secrete in large volumes to
initiate a cascade of effects, such as upregulation of matrix
metalloproteinases (24).
Conflictingly, TNF-α has been reported to both inhibit and promote
tumor growth (25,26).
The genes encoding TNF-α and IL-6 are highly
polymorphic due to a variety of single nucleotide polymorphisms
(SNPs) in their regulatory and coding sequences. rs1800795, a SNP
in the promoter region of the IL6 gene (−174 G>C) is
reportedly associated with constitutive IL-6 expression, which
results in higher IL-6 expression in carriers of the GG/GC
genotype, compared with those of the CC genotype (27).
A G>A substitution at position −308 in the
promoter region of the TNFA gene has also been identified,
and the −308A allele has been associated with enhanced TNF-α
expression in vivo and in vitro, and increased plasma
levels of TNF-α compared with the −308G allele (28–30).
To date, only a limited number of studies have
investigated the role of polymorphisms in the IL6 and
TNFA genes in melanoma (13,31–34).
Thus, the current study aimed to clarify the possible effects of
the IL6 −174G>C and TNFA −308G>A SNPs on the
susceptibility and prognosis of CMM in a Bulgarian population. The
present study is the first, to the best of our knowledge, to
describe possible effects of these polymorphisms on the progression
of CMM in Bulgarian patients.
Materials and methods
Patients
In total, 76 patients with CMM treated at The
Oncology Center of Stara Zagora (Stara Zagora, Bulgaria) were
enrolled in the present study. All patients with melanoma diagnosed
for the first time between January 2011 and December 2015,
regardless of disease stage, were invited to participate in the
study. Demographic data and information on working conditions were
extracted from patient files. According to their occupation, the
patients were divided into two groups: i) Those working in
conditions less harmful for the skin (e.g., offices and schools);
and ii) those working in more harmful conditions (such as
agricultural workers, construction workers and those in open mine
shafts). The demographic and clinical data obtained from the
patients are presented in Table I.
The control group comprised 200 individuals without CMM and
included 94 (47%) men and 106 (53%) women aged between 19 and 85
years (median age, 58 years) from the same ethnic group and area of
Bulgaria. The recruited controls were volunteers or individuals
participating in prophylactic examinations who were reported not to
have cancer. The patients were treated and followed-up at the
Dermatology Unit of The Oncology Center of Stara Zagora. Informed
consent was obtained from all participants, and the protocol was
approved by The Ethics Committee of The Medical Faculty of Trakia
University (Stara Zagora, Bulgaria). The study was also performed
in accordance with the ethical standards of the 1964 Declaration of
Helsinki and its later amendments, or comparable ethical
standards.
 | Table I.Demographic and clinical data of the
patients with cutaneous malignant melanoma. |
Table I.
Demographic and clinical data of the
patients with cutaneous malignant melanoma.
| Parameter | N (%) |
|---|
| Sexa | 76 |
|
Male | 31 (41) |
|
Female | 45 (59) |
| Localization of the
tumora | 60 |
|
Extremities (legs/arms) | 24 (40) |
|
Trunk | 30 (50) |
|
Head | 6 (10) |
| pT
categorya | 61 |
|
pT1-2 | 32 (52) |
|
pT3-4 | 29 (48) |
| pN
categorya | 61 |
|
pN0 | 55 (90) |
|
pN1-3 | 6 (10) |
|
Metastasisa | 60 |
| No | 23 (38) |
|
Yes | 37 (62) |
| pTNM clinical
stagea | 60 |
| I | 22 (36) |
| II | 29 (48) |
|
III | 3 (5) |
| IV | 6 (10) |
| Clark's
levela | 50 |
| II | 17 (34) |
|
III | 20 (40) |
| IV | 9 (18) |
| V | 4 (8) |
| Outcome after
follow-up perioda | 75 |
|
Alive | 41 (55) |
|
Dead | 34 (45) |
| Breslow's
thickness, n=18, mmb | 2.00
(0.20–4.20) |
| Survival after
diagnosis, n=75, monthsb | 81.78
(55.90–301.45) |
| Overall survival,
n=75, monthsb | 19.42
(0.49–237.52) |
| Age at diagnosis,
n=76, yearsb | 59.74
(15.49–83.76) |
DNA isolation
Genomic DNA was isolated from 0.2 ml whole blood
using the GenElute™ Mammalian Genomic DNA Miniprep kit
(Sigma-Aldrich; Merck KGaA). The isolated DNA was stored at −80°C.
DNA concentration was determined using the NanoVue™
Spectrophotometer (GE Healthcare), and purity was assessed by
calculating the ratio of the optical density at 260 and 280 nm. The
purity and quality of the DNA samples was also confirmed by
electrophoresis, using a 1% agarose gel.
Genotyping for TNFA −308G>A
(rs1800629) and IL6 −174G>C (rs1800795) SNPs
Genotyping was performed using a PCR-restriction
fragment length polymorphism (RFLP)-based method, as previously
described (35). The amplification
reactions were performed in a final volume of 12 µl using the
Mastercycler® instrument (Eppendorf). The amplification
mix contained 30–50 ng genomic DNA, 0.8 pmol/µl each primer, 200 µM
dNTPs, 1.2 µl 10X buffer with 15 mM MgCl2
(Sigma-Aldrich; Merck KGaA), 0.5 U Taq Polymerase (Sigma-Aldrich;
Merck KGaA) and double-distilled H2O to a final volume
of 12 µl. The primers used were as follows: TNF-α, forward
5′-AGGCAATAGGTTTTGAGGGCCAT-3′, reverse 5′-TCCTCCCTGCTCCGATTCCG-3′;
IL-6, forward 5′-TTGTCAAGACATGCCAAGTGCT-3′ and reverse
5′-GCCTGAGAGACATCTCCAGTCC-3′.
Thermocycling conditions for rs1800629 were:
i) Pre-amplification denaturation at 95°C for 3 min; ii) 5 cycles
of denaturation for 30 sec at 94°C, annealing for 30 sec at 58°C,
and polymerization for 30 sec at 72°C; iii) 30 cycles of
denaturation for 30 sec at 94°C, annealing for 30 sec at 56°C and
polymerization for 30 sec at 72°C; and iv) final extension at 72°C
for 7 min.
For rs1800795, the thermocycling conditions
were: i) Pre-amplification denaturation at 95°C for 3 min; ii) 35
cycles of denaturation for 30 sec at 95°C, annealing for 30 sec at
62°C and polymerization for 30 sec at 72°C; and iii) final
extension at 72°C for 5 min.
Restriction digestion for the rs1800629 SNP
was performed in a final volume of 16 µl with 12 µl PCR product and
4.8 U NcoI in 4 µl 1X ONE Buffer (EUREX Sp. z o.o.) for 16 h
at 37°C. The restriction digestion reaction for the
rs1800795 SNP was performed in a final volume of 17 µl with
12 µl PCR product and 3 U HinI in 5 µl 1X Tango buffer
(Thermo Fisher Scientific, Inc.) for 16 h at 37°C. The obtained
restriction fragments were analyzed using electrophoresis with a
3.5% agarose gel stained with ethidium bromide (Sigma-Aldrich;
Merck KGaA), and detected using a UV transilluminator (Cleaver
Scientific Ltd.). The results were assessed using the Gel
documentation system EC3 Imaging system (Ultra-Violet Products
Ltd.).
Measurement of IL-6 serum
concentration
Serum IL-6 levels of 20 control individuals and 59
patients with CMM were determined using a commercial ELISA kit
(cat. no. D6050; R&D Systems, Inc.) according to the
manufacturer's protocol. The IL-6 concentrations were recorded in
comparison to the standards included in the kit and are presented
in pg/ml serum. Statistical analysis. Statistical analysis
was performed using SPSS v16.0 (SPSS, Inc.). The descriptive data,
including the mean, SEM and median, were assessed.
Kolmogorov-Smirnov's test and Shapiro-Wilks' W-test were used to
analyze the normality of the continuous variables. Continuous
variables with normal distribution were compared between ≥2
independent groups using one-way ANOVA followed by a least
significant difference post hoc test. Variables with non-normal
distribution were compared using a Mann-Whitney U test. The
manifestation frequencies of the qualitative (categorical)
variables were determined in 2×3 and 2×2 cross-tables and were
evaluated using the χ2 test. Fisher's exact test was
used as appropriate (when the expected numbers of ≥1 of the cells
of the 2×2 cross-tables were <5). The correlations between the
quantitative variables were evaluated using Pearson or Spearman's
test according to the distribution (normal or skewed,
respectively). The odds ratios and 95% CI values were calculated by
binary logistic regression analysis for evaluation of the risk of
outcome occurrence (development of melanoma). Hardy-Weinberg
equilibrium (HWE) was tested among the controls and patients using
the χ2 test.
Cumulative survival curves were constructed using
the Kaplan-Meier method, and the differences in survival were
calculated using the log rank test. The prognostic significance of
various factors regarding patient survival after surgery was
determined by univariate and multivariate Cox regression analyses.
P<0.05 was considered to indicate a statistically significant
difference.
Results
IL6 −174G>C SNP
Genotyping of the −174G>C polymorphism in the
promoter region of IL6 was performed by PCR-RFLP. The
resulting PCR product was 299 bp in length, and the restriction
reaction resulted in three fragments for the wild-type G allele
(227, 50 and 13 bp). For the variant C allele (CC genotype), the
restriction reaction resulted in four fragments of 118, 109, 50 and
13 bp (Fig. 1).
For this SNP, 59 patients with CMM and 173 control
individuals were successfully genotyped. The distribution of the
genotypes did not deviate from HWE in either group (P=0.997 and
P=0.799, respectively; χ2 test). The genotype
distributions in the patient group were 30 (50.8%) GG carriers, 22
(37.3%) GC carriers and 7 (11.9%) CC carriers. The control group
comprised 74 (42.8%) carriers of the GG genotype, 83 (48.0%) with
the GC genotype and 16 (9.2%) with the CC genotype. Both genotype
and allelic distributions did not differ between the patients and
controls (P=0.358 and P=0.878; χ2 test; Table II).
 | Table II.Genotype and allele frequencies of
the IL6 −174G>C gene polymorphism in patients with
cutaneous melanoma (n=59 and n=118, respectively) and controls
(n=173 and n=346, respectively) (binary logistic regression
analysis). |
Table II.
Genotype and allele frequencies of
the IL6 −174G>C gene polymorphism in patients with
cutaneous melanoma (n=59 and n=118, respectively) and controls
(n=173 and n=346, respectively) (binary logistic regression
analysis).
| A, Genotype
frequency (P=0.358; χ2 test) |
|---|
|
|---|
| Variable | Patients, n
(frequency) | Controls, n
(frequency) | OR (95% CI) | P-value |
|---|
| GG | 30 (0.508) | 74 (0.428) | 1.0
(reference) |
|
| GC | 22 (0.373) | 83 (0.480) | 0.654
(0.347–1.231) | 0.188 |
| CC | 7 (0.119) | 16 (0.092) | 1.079
(0.403–2.888) | 0.879 |
| GC+CC | 29 (0.492) | 99 (0.572) | 0.723
(0.399–1.307) | 0.283 |
|
| B, Allele
frequency (P=0.878; χ2 test) |
|
|
Variable | Patients, n
(frequency) | Controls, n
(frequency) | OR (95%
CI) | P-value |
|
| −174 G | 82 (0.695) | 231 (0.668) | 1.0
(reference) |
|
| −174 C | 36 (0.305) | 115 (0.332) | 0.882
(0.563–1.382) | 0.649 |
In patients with CMM, there were no associations
between different genotypes and biochemical/blood parameters such
as total protein, albumin, glucose, bilirubin, creatinine, enzymes
[aspartate aminotransferase (AsAT), alanine aminotransferase
(AlAT), γ-glutamyl transferase (GGT) and lactate dehydrogenase
(LDH)], red blood cell count, white blood cell (WBC) count and the
percentages of WBC subpopulations (Table III). According to the Clark scale
(36), carriers of the GG genotype
predominantly exhibited more advanced melanoma (Clark 3, 4 and 5)
than those with C allele genotypes (GC+CC) (P=0.037; χ2
test; Fig. 2). Similarly, GG
carriers more frequently possessed thicker tumors (≥2 mm; 75%) than
patients with other genotypes (52.4%; P=0.114; χ2 test;
Fig. 3).
 | Table III.Biochemical/blood parameters in
patients with cutaneous melanoma and different IL6 −174G>C
genotypes. |
Table III.
Biochemical/blood parameters in
patients with cutaneous melanoma and different IL6 −174G>C
genotypes.
| Biochemical/blood
parameters | IL6
−174GG | IL6
−174GC | IL6
−174CC |
P-valuea |
|---|
| AsAT, U/l | 21.34±2.03 | 24.77±5.97 | 18.00±2.51 | 0.621 |
| AlAT, U/l | 22.21±3.38 | 20.82±7.40 | 18.40±3.11 | 0.915 |
| GGT, U/l | 51.17±18.03 | 21.50±4.50 | 54.00±17.62 | 0.607 |
| LDH, U/l | 215.20±33.26 | 203.00±25.7 | 246.33±44.54 | 0.820 |
| RBC,
1012 cells/l | 4.82±0.14 | 4.71±0.11 | 4.62±0.30 | 0.777 |
| WBC, 109
cells/l | 6.94±0.53 | 7.59±0.46 | 9.16±2.09 | 0.239 |
| Lymphocytes, % | 36.27±3.01 | 32.80±3.65 | 31.70±4.93 | 0.654 |
| Granulocytes,
% | 62.88±1.75 | 62.65±4.45 | 71.35±1.35 | 0.080 |
On patient follow-up, the survival time after
diagnosis of IL6 −174GG genotype carriers was shorter,
although not significantly, than those with a genotype with ≥1
variant IL6 −174C allele (GC+CC) (mean, 132.58 vs. 166.55
months; P=0.299; log rank test; Fig.
4). When patients were categorized according to their working
conditions, those carrying the GG genotype and in occupational
conditions with longer periods of sunlight exposure (so called
‘high risk conditions’) had significantly shorter survival times
(24.09 months) compared with carriers of С allele genotype (104.33
months; P=0.016; log rank test; Fig.
5A). Similar results, although not significant, were obtained
for the subgroup of patients working indoors, i.e., in conditions
with rare sunlight exposure (so called ‘low risk conditions’): The
mean survival period of patients with the GG genotype who worked
indoors was 185.73 months, while that of the patients with C allele
genotypes (GC+CC group) was 256.00 months (P=0.121; log rank test;
Fig. 5B).
Serum IL-6 levels between patients with CMM (median,
5.68 pg/ml; mean ± SEM), 20.57±5.89 pg/ml) and the control subjects
(median, 4.17 pg/ml; mean ± SEM, 5.02±0.74 pg/ml) differed with
marginal significance (P=0.033; Fig.
6). No significant difference was observed when comparing the
three −174G>C genotypes of both the patient and control groups
(P=0.323 and 0.104, respectively; data not shown). However, the
IL-6 level was significantly lower in control subjects with the GG
genotype (median, 3.02 pg/ml), compared with those with variant C
allele genotypes (GC+CC; 5.01 pg/ml; P=0.039; Fig. 7). Although not statistically
significant, a similar trend was observed in the patient group
(median, 4.87 pg/ml vs. 8.57 pg/ml; P=0.161; Fig. 7).
Serum IL-6 levels were not associated with clinical
or histological tumor parameters such as the Clark scale (1+2 vs.
3+4+5; P=0.404), Breslow's thickness (<2 mm vs. ≥2 mm; P=0.808),
TNM staging (37) (1 vs. 2 vs. 3 vs.
4; P=0.187) and the presence of distant metastases at the time of
diagnosis (P=0.440; data not shown). However, the IL-6 serum levels
between the patients with disease progression and development of
new distant metastases (median, 22.81 pg/ml), and those without new
distant metastases (median, 5.10 pg/ml), were significantly
different (P=0.004; Fig. 8A).
Moreover, the serum levels differed significantly between patients
whose occupation was associated with increased exposure to sunlight
(presence of occupational hazard; median, 9.43 pg/ml) and those who
worked indoors (median, 4.56 pg/ml; P<0.0001; Fig. 8B).
IL-6 serum levels were positively correlated with
liver-specific enzyme levels, including AsAT (ρ=0.408; P=0.015)
AlAT (ρ=0.328; P=0.050) and GGT (ρ=0.758; P=0.007). IL-6 was also
positively correlated with blood glucose (Rho=0.621; P<0.0001)
and creatinine levels (ρ=0.434; P=0.015), as well as WBC count
(ρ=0.384; P=0.019; Table IV).
 | Table IV.Correlation between serum IL-6 and
serum biochemical and blood parameters in patients with cutaneous
melanoma. |
Table IV.
Correlation between serum IL-6 and
serum biochemical and blood parameters in patients with cutaneous
melanoma.
| Biochemical/ blood
parameters | AsAT | AlAT | GGT | Serum glucose | Serum
creatinine | WBC |
|---|
| ρ | 0.408 | 0.328 | 0.758 | 0.621 | 0.434 | 0.384 |
| P-value | 0.015 | 0.050 | 0.007 | <0.0001 | 0.015 | 0.019 |
Subsequently, the median patient IL-6 serum level
(5.68 pg/ml) was selected as the cut-off value for survival
analysis. Patients with higher serum IL-6 levels exhibited
significantly shorter survival times after diagnosis, compared with
those with IL-6 levels below the cut-off (median, 58.63 vs. 237.52
months; P=0.042; Fig. 9).
Cox univariate analysis demonstrated that several
demographic, clinical and blood parameters had significant adverse
effects on the survival of patients with CMM. These included male
sex (P<0.0001), advanced age (P=0.010), occupational conditions
with increased exposure to sunlight (P=0.022, low hemoglobin levels
(P=0.001), greater Clark scale depth (3+4+5; P=0.024) and the
presence of lymph node metastases (P<0.0001); higher serum IL-6
levels were also adversely associated with survival (P=0.048;
Table V).
 | Table V.Univariate and multivariate Cox
proportional analysis of the survival of patients with cutaneous
melanoma. (pTNM staging system is not included in multivariate
analysis because it depends on another variable in the analysis,
the presence of lymph node metastases, LN metastases) |
Table V.
Univariate and multivariate Cox
proportional analysis of the survival of patients with cutaneous
melanoma. (pTNM staging system is not included in multivariate
analysis because it depends on another variable in the analysis,
the presence of lymph node metastases, LN metastases)
|
| Univariate
analysis | Multivariate
analysis model 1, n=30 | Multivariate
analysis model 2, n=32 |
|---|
|
|
|
|
|
|---|
| Variable | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| Sex, category
(n) |
|
|
|
|
|
|
| Male
(32) vs. female (44) | <0.0001 | 4.51
(2.19–9.28) | 0.015 | 44.29
(2.08–942.27) | 0.004 | 14.09
(2.34–84.74) |
| Age | 0.010 | 1.03 | 0.217 | 1.06
(0.97–1.15) | 0.105 | 1.06 (0.99
−1.15) |
| Clark score,
category (n) |
|
|
|
|
|
|
| 3+4+5
(34) vs. 1+2 (18) | 0.024 | 4.41
(1.21–16.04) | 0.263 | 3.52
(0.39–31.90) | 0.719 | 1.41
(0.22–9.21) |
| LN metastasis,
category (n) |
|
|
|
|
|
|
| Yes
(6) vs. no (55) | <0.0001 | 9.54
(3.48–26.26) | 0.098 | 42.27
(0.50–358.20) | 0.211 | 16.43
(0.20–132.40) |
| pTNM staging,
category (n) |
|
|
|
|
|
|
| III–IV
(10) vs. I–II (54) | 0.002 | 4.04
(1.67–9.76) |
|
|
|
|
| Occupational
hazard, category (n) |
|
|
|
|
|
|
| Present
(36) vs. absent (30) | 0.022 | 3.03
(1.18–7.81) | 0.188 | 0.11
(0.01–2.93) | 0.047 | 6.83
(1.03–45.45) |
| Hemoglobin, g/l
(n) |
|
|
|
|
|
|
| <120
(10) vs. ≥120 (39) | 0.001 | 5.09
(2.01–12.88) | 0.588 | 2.29
(0.11–46.29) | 0.248 | 13.60
(0.16–113.70) |
| Serum IL-6, pg/ml
(n) |
|
|
|
|
|
|
| ≥5.68
(30) vs. <5.68 (28) vs. | 0.048 | 2.25
(1.01–5.02) | 0.299 | 5.67
(0.21–149.70) |
|
|
| ‒174G>C
IL6, SNP (n) |
|
|
|
|
|
|
| GG
(29) vs. GC+CC (29) | 0.303 | 1.51
(0.69–3.29) |
|
| 0.030 | 9.61
(1.24–74.28) |
Factors identified as significant using univariate
analysis were then assessed using the multivariate Cox's
proportional hazard model (Model 1). IL-6 serum level was no longer
a significant factor (P=0.299), and only the male sex remained an
independent risk factor for shorter survival time (P=0.015;
Table V). A second Cox's
proportional hazard model (Model 2) was also used, which included
genotypes with the IL6 −174G>C SNP together with all
significant factors identified during univariate analysis of
routine demographic, clinical and blood parameters. Male sex
(P=0.004), occupational conditions with increased exposure to
sunlight (P=0.047 and the GG genotype (P=0.030) remained
independent prognostic factors for shorter survival time (Table V).
TNFA −308G>A SNP
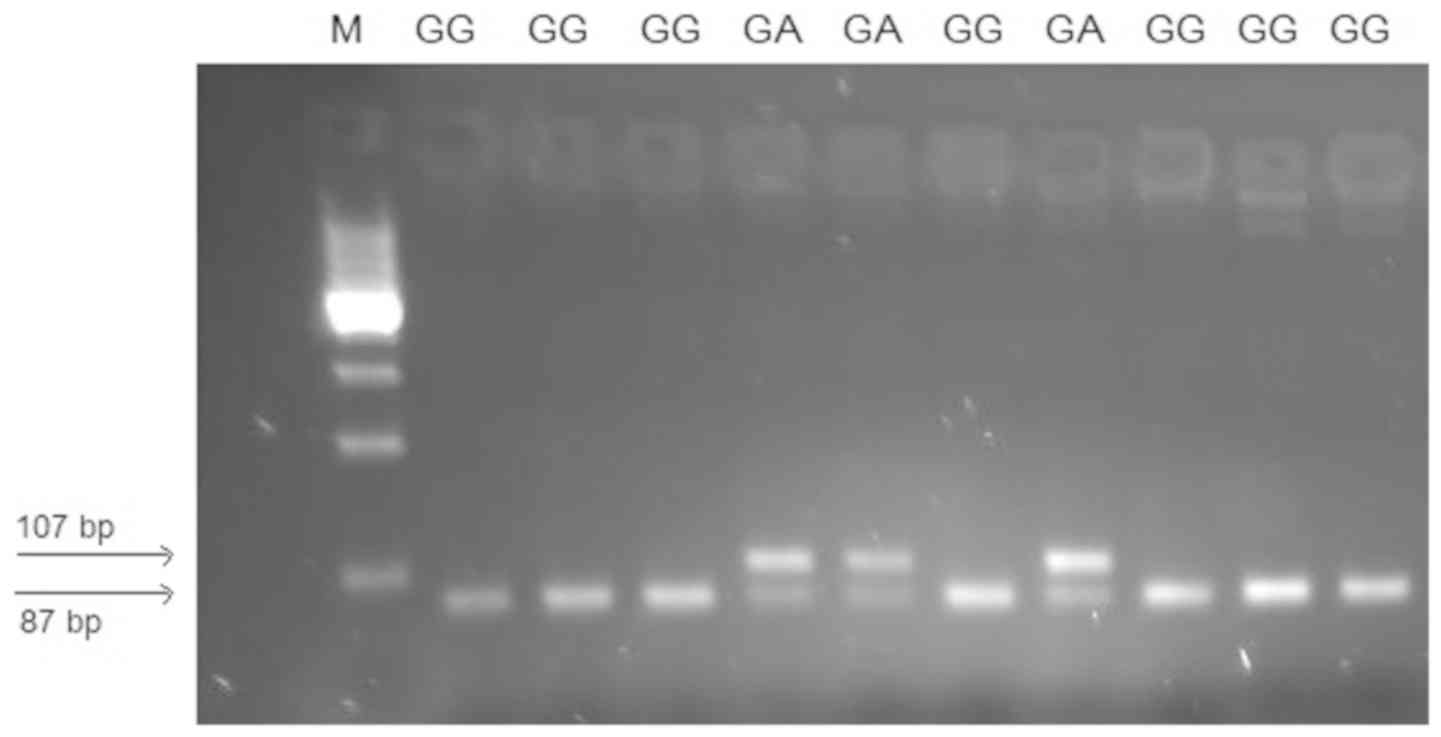
Genotyping for the −308G>A polymorphism in the
TNFA promoter was performed by PCR-RFLP. The resulting PCR
product was 107 bp in length, and the subsequent restriction
reaction for the G allele (GG genotype) resulted in 2 fragments of
87 and 20 bp. The variant А allele remained unchanged (107 bp;
Fig. 10).
Genotyping of the TNFA −308G>А SNP was
successfully performed in 76 patients with CMM and 198 control
individuals. The genotype distribution in both the control and
patient group did not deviate from HWE (P=0.148 and P=0.889,
respectively; χ2 test). In the patient group, 63 (82.9%)
carried the GG genotype, 12 (15.8%) were GA genotype-positive, and
only one patient (1.3%) carried the АА genotype. The genotype
distribution among the controls was 160 (80.8%) with the GG
genotype, 33 (16.7%) with GА genotype and 5 (2.5%) with the АА
genotype. No significant differences in genotype and allelic
distribution between the patients and controls were observed
(P=0.810 and P=0.982; χ2 test; Table VI). Furthermore, no associations
were found between genotypes with the TNFA −308G>A SNP
and biochemical or clinical parameters in patients with CMM (data
not shown).
 | Table VI.Genotype and allele frequencies of
the TNFA −308G>A gene polymorphism in patients with
cutaneous malignant melanoma (n=76 and n=152, respectively) and
controls (n=198 and n=396, respectively). |
Table VI.
Genotype and allele frequencies of
the TNFA −308G>A gene polymorphism in patients with
cutaneous malignant melanoma (n=76 and n=152, respectively) and
controls (n=198 and n=396, respectively).
| A, Genotype
frequency (P=0.810; χ2 test) |
|---|
|
|---|
| Variable | Patients, n
(frequency) | Controls, n
(frequency) | OR (95% CI) | P-value |
|---|
| GG | 63 (0.829) | 160 (0.808) | 1.0
(reference) |
|
| GA | 12 (0.158) | 33 (0.167) | 0.924
(0.449–1.901) | 0.829 |
| AA | 1 (0.013) | 5 (0.025) | 0.508
(0.058–4.434) | 0.540 |
| GA+AA | 13 (0.171) | 38 (0.192) | 0.869
(0.434–1.739) | 0.691 |
|
| B, Allele
frequency (P=0.878, χ2 test) |
|
|
Variable | Patients, n
(frequency) | Controls, n
(frequency) | OR (95%
CI) | P-value |
| −308
G | 138 (0.908) | 353 (0.891) | 1.0
(reference) |
|
| −308
A | 14 (0.092) | 43 (0.109) | 0.833
(0.445–1.558) | 0.641 |
Discussion
Melanoma cells are derived from normal melanocytes
transformed due to various extrinsic and intrinsic factors. The
immune response strongly influences the development and progression
of melanoma (14,34). Previous studies identified IL-6 as
one of the most important regulatory cytokines in tumor biology,
which is involved in key stages of tumor development, such as
proliferation, apoptosis, angiogenesis and differentiation
(38,39). However, IL-6 has pleiotropic effects
in carcinogenesis, including strong growth-stimulating, as well as
antitumor effects (39). During
tumor progression, IL-6 changes from a paracrine inhibitor of
normal melanocytes in the early stages, to an autocrine mitogen in
the advanced stages of disease (15). Additionally, it exerts a paracrine
effect on tumor angiogenesis and cells of the immune system
(15). Thus, IL-6 acts as an
inhibitor of tumor growth in early melanoma, but appears to be an
important growth factor in advances stages of the disease (33,40).
The expression of IL-6 depends on a variety of
factors, including polymorphisms in the IL6 gene. The
IL6 −174G>C (rs1800795) polymorphism is localized at the
promoter region of the gene, and is associated with altered
promoter activity and resulting protein expression levels (41). A previous study reported 2-fold lower
expression in HeLa cells transfected with a vector containing the C
allele construct, compared with cells transfected with the G allele
construct (41). Another in
vitro study also demonstrated that the G allele of the
IL6 −174G>C SNP was associated with an increased
transcriptional response to various stimuli (42). However, the results of studies
investigating the levels of circulating IL-6 are conflicting. In
healthy subjects, the G allele genotypes (GG and GC) were
associated with higher IL-6 plasma levels compared with the CC
genotype (41), while Jones et
al (43) detected high plasma
IL-6 levels in individuals with the C allele and CC genotypes. The
results of the present study are in a line with the latter study
(43), although carriers of C allele
genotypes (CG+CC) exhibited significantly higher serum IL-6 levels
in the control group only, not in patients with CCM.
The effects of the IL6 −174G>C SNP on the
progression of different cancer types have been widely explored,
with quite controversial results (27). Zhai et al (27) performed a comprehensive meta-analysis
of 17 studies including 4,304 patients with various cancer types,
including breast, colorectal, lung, ovarian cancer and lymphoma.
This previous study assessed the association between the IL6
−174 polymorphism and cancer prognosis. No association with overall
survival was observed from pooled analysis of the three genotypes
with this SNP. However, the GG genotype affected patient survival
compared with the C allele genotypes (GC+CC) in bladder, ovarian,
gastric and peritoneal cancer, as well as in neuroblastoma and
osteosarcoma.
To date, there are only a limited number of studies
concerning the possible role of the IL6 −174G>C SNP in
CMM (31,44). The present study is the first, to the
best of our knowledge, describing the possible effect of this
polymorphism on Bulgarian patients with CMM. The results of the
current study concur with those aforementioned (27), which also demonstrated no significant
differences in genotype or allelic frequency of the IL6
−174G>C SNP between patients with CMM and controls. Thus, these
results suggested that IL6 −174G>C is unlikely to be
heavily involved in patient susceptibility to CMM (31,44).
Investigating the associations between IL6
−174G>C SNP genotype frequencies and different clinical
characteristics of patients with CMM, the present results suggested
that the IL6 −174 GG genotype was associated with a more
advanced stage, thicker tumors and reduced overall survival.
Similarly, a previous study on the role of the IL6
−174G>C SNP demonstrated that the C allele decreased the risk of
developing urinary bladder cancer, and that the GG genotype was
associated with more advanced disease stages (45). By contrast, Leibovici et al
(46) reported that the CC genotype
was more frequently observed in the advanced stages of bladder
cancer.
The association between IL6 −174G>C and
disease progression may be due to the potential effects on IL-6
expression. The results of the present study indicated that higher
serum IL-6 levels were associated with unfavorable blood/serum
characteristics (poor WBC count, liver enzyme levels, blood glucose
and creatinine), tumor progression (development of new distant
metastases) and shorter survival time. These results concur with
those of Tobin et al (19),
which suggested an association between increased plasma IL-6 levels
in patients with stage IV melanoma and tumor burden, as well as
shorter survival.
A notable finding of the present study was the
relationship between serum IL-6 and patient working conditions.
Those working outdoors with supposedly increased exposure to
sunlight had significantly higher IL-6 levels compared with
patients working indoors. This finding suggested that sunlight may
stimulate IL-6 expression, which is in line with previous
observations of increased serum levels of the melanoma tumor
markers (IL-1α, IL-4, IL-6, IL-10, TNF-α and interferon-γ),
so-called ‘melanoma inhibitory activity’, following phototherapy
with UV light (47).
TNF-α is one of the most important pro-inflammatory
cytokines involved in cellular proliferation, differentiation and
apoptosis, and has been reported to play a critical role in
carcinogenesis (48). A previous
study demonstrated that protein expression and transcriptional
levels of TNF-α were related to several promoter polymorphisms in
cytokine-encoding genes, including the TNFA −308G>А SNP
(49). In previous studies, the A
allele of this SNP was associated with elevated TNF-α transcription
in vitro (50–52), and with increased serum TNF-α in
patients with acute myocardial infarction (53). By contrast, Sharma et al
(54) reported that the A allele of
the TNFA −863C>A SNP was associated with reduced serum
TNF-α levels in patients with asthma (54). Similarly, the wild-type G-allele of
TNFA −308G>A was associated with significantly higher
TNFA mRNA expression in human blood leucocytes, compared with the A
allele (55).
In the current study, no differences were found in
the genotype or allelic distributions between patients with CMM and
the controls. This result confirms the reported lack of association
between the TNFA −863C>A SNP and increased risk of
melanoma (13,33,56). By
contrast, previous meta-analyses suggested that the TNFA
−308G>A polymorphism was a risk factor for a range of other
malignancies, such as gastric, breast and hepatocellular cancer
(57–59), whilst other studies did not
demonstrate any significant association between TNFA
−308G>A and the risk of cancer (60).
In conclusion, the results of the present study
suggested that the IL6 −174G>C and the TNFA
−308G>A promoter polymorphisms were not predisposing factors for
CMM. However, the IL6 −174G>C SNP and IL-6 serum
concentrations are likely to influence the progression of CMM. The
GG genotype and higher serum levels may be associated with tumor
progression and shorter survival. Although the GG genotype was
associated with lower IL-6 levels, higher IL-6 levels may be
influenced by other factors, including sun light, and not only by
the genotype.
Acknowledgements
The authors would like to thank Dr Petya Peeva of
the Dermatology Unit of The Oncology Center (Stara Zagora,
Bulgaria) for providing patient clinical data and biological
materials. The present study was previously presented at the 2nd
International Multicenter European Cooperation in Science and
Technology Action (no. CA15129) on Diagnosis, Monitoring and
Prevention of Exposure Conference, Bentivoglio, near Bologna,
Italy, 30–31 October 2017 and the Abstract was published in a
supplement of the Journal of Health and Pollution.
Funding
The present study was supported by The Medical
Faculty of Trakia University (grant no. 1/2018), The National
Scientific Program for Support of Young Scientists and
Post-doctoral Scientists administered by The Ministry of Education
and Science (grant no. 577/17.08.2018), and by COST Action (grant
no. CA15129; DiMoPEX).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TV, MK, MD and DV conceived and designed the study.
TV, DD, DV and NO wrote and revised the manuscript. TV, AA and MK
performed the genotyping. TV, TT, DD, DV and AM collected the
biological material and clinical data, and analyzed the data. DD
and AM acquired the informed consent and approval by the Ethics
Committee, and were involved in drafting the manuscript. All
authors contributed to the writing of the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
participants included in the study. The study protocol was approved
by The Ethics Committee at The Medical Faculty of Trakia
University. The present study was performed in accordance with the
ethical standards laid down in the 1964 Declaration of Helsinki and
its later amendments, or comparable ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dimitrova N, Vukov M and Valerianova Z:
Cancer incidence in Bulgaria. 2013 National Oncological Hospital,
Bulgarian National Cancer Registry, Publisher AVIS-24 Ltd, .
24:1–171. 2015.
|
|
2
|
Balch CM, Soong SJ, Murad TM, Smith JW,
Maddox WA and Durant JR: A multifactorial analysis of melanoma. IV.
Prognostic factors in 200 melanoma patients with distant metastases
(stage III). J Clin Oncol. 1:126–134. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barth A, Wanek LA and Morton DL:
Prognostic factors in 1,521 melanoma patients with distant
metastases. J Am Coll Surg. 181:193–201. 1995.[see comments].
PubMed/NCBI
|
|
4
|
Brand CU, Ellwanger U, Stroebel W, Meier
F, Schlagenhauff B, Rassner G and Garbe C: Prolonged survival of 2
years or longer for patients with disseminated melanoma. An
analysis of related prognostic factors. Cancer. 79:2345–2353. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mössner R, Anders N, König IR, Krüger U,
Schmidt D, Berking C, Ziegler A, Brockmöller J, Kaiser R,
Volkenandt M, et al: Variations of the melanocortin-1 receptor and
the glutathione-S transferase T1 and M1 genes in cutaneous
malignant melanoma. Arch Dermatol Res. 298:371–379. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schoof N, von Bonin F, König IR, Mössner
R, Krüger U, Reich K, Berking C, Volkenandt M, Ziegler A, Böckmann
L, et al: Distal and proximal interleukin (IL)-10 promoter
polymorphisms associated with risk of cutaneous melanoma
development: A case--control study. Genes Immun. 10:586–590. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bishop DT, Demenais F, Iles MM, Harland M,
Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E,
et al: Genome-wide association study identifies three loci
associated with melanoma risk. Nat Genet. 41:920–925. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Landi MT, Kanetsky PA, Tsang S, Gold B,
Munroe D, Rebbeck T, Swoyer J, Ter-Minassian M, Hedayati M,
Grossman L, et al: MC1R, ASIP, and DNA repair in sporadic and
familial melanoma in a Mediterranean population. J Natl Cancer
Inst. 97:998–1007. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dolzan V, Rudolf Z and Breskvar K: Genetic
susceptibility to environmental carcinogenesis in Slovenian
melanoma patients. Acta Dermatovenerol Alp Pannonica Adriat.
15:69–78. 2006.PubMed/NCBI
|
|
10
|
Wei Q, Lee JE, Gershenwald JE, Ross MI,
Mansfield PF, Strom SS, Wang LE, Guo Z, Qiao Y, Amos CI, et al:
Repair of UV light-induced DNA damage and risk of cutaneous
malignant melanoma. J Natl Cancer Inst. 95:308–315. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lear JT, Smith AG, Strange RC and Fryer
AA: Detoxifying enzyme genotypes and susceptibility to cutaneous
malignancy. Br J Dermatol. 142:8–15. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fargnoli MC, Argenziano G, Zalaudek I and
Peris K: High- and low-penetrance cutaneous melanoma susceptibility
genes. Expert Rev Anticancer Ther. 6:657–670. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Howell WM, Turner SJ, Collins A, Bateman
AC and Theaker JM: Influence of TNFalpha and LTalpha single
nucleotide polymorphisms on susceptibility to and prognosis in
cutaneous malignant melanoma in the British population. Eur J
Immunogenet. 29:17–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Passarelli A, Mannavola F, Stucci LS,
Tucci M and Silvestris F: Immune system and melanoma biology: A
balance between immunosurveillance and immune escape. Oncotarget.
8:106132–106142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lázár-Molnár E, Hegyesi H, Tóth S and
Falus A: Autocrine and paracrine regulation by cytokines and growth
factors in melanoma. Cytokine. 12:547–554. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hirano T: Interleukin 6 and its receptor:
Ten years later. Int Rev Immunol. 16:249–284. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ohl K and Tenbrock K: Inflammatory
cytokines in systemic lupus erythematosus. J Biomed Biotechnol.
2011:4325952011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang Q, Bournazou E, Sansone P, Berishaj
M, Gao SP, Daly L, Wels J, Theilen T, Granitto S, Zhang X, et al:
The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and
metastasis. Neoplasia. 15:848–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tobin RP, Jordan KR, Kapoor P, Spongberg
E, Davis D, Vorwald VM, Couts KL, Gao D, Smith DE, Borgers JSW, et
al: IL-6 and IL-8 are linked with myeloid-derived suppressor cell
accumulation and correlate with poor clinical outcomes in melanoma
patients. Front Oncol. 9:12232019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoejberg L, Bastholt L, Johansen JS,
Christensen IJ, Gehl J and Schmidt H: Serum interleukin-6 as a
prognostic biomarker in patients with metastatic melanoma. Melanoma
Res. 22:287–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vlaykova T: Prognostic factors in
metastatic melanoma: special reference to tumor vascularity,
proliferation and Bcl-2 expression. Turku University. (Turku,
Finland). 2002.
|
|
22
|
Linnskog R, Jönsson G, Axelsson L, Prasad
CP and Andersson T: Interleukin-6 drives melanoma cell motility
through p38α-MAPK-dependent up-regulation of WNT5A expression. Mol
Oncol. 8:1365–1378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Na YR, Lee JS, Lee SJ and Seok SH:
Interleukin-6-induced Twist and N-cadherin enhance melanoma cell
metastasis. Melanoma Res. 23:434–443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rossi S, Cordella M, Tabolacci C, Nassa G,
D'Arcangelo D, Senatore C, Pagnotto P, Magliozzi R, Salvati A,
Weisz A, et al: TNF-alpha and metalloproteases as key players in
melanoma cells aggressiveness. J Exp Clin Cancer Res. 37:3262018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Donia M, Kjeldsen JW and Svane IM: The
controversial role of TNF in melanoma. OncoImmunology.
5:e11076992018. View Article : Google Scholar
|
|
26
|
Bertrand F, Rochotte J, Colacios C,
Montfort A, Andrieu-Abadie N, Levade T, Benoist H and Ségui B:
Targeting TNF alpha as a novel strategy to enhance CD8+ T
cell-dependent immune response in melanoma? OncoImmunology.
5:e10684952015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhai K, Yang Y, Gao ZG and Ding J:
Interleukin-6-174G>C gene promoter polymorphism and prognosis in
patients with cancer. Oncotarget. 8:44490–44497. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Banday MZ, Balkhi HM, Hamid Z, Sameer AS,
Chowdri NA and Haq E: Tumor necrosis factor-α (TNF-α)-308G/A
promoter polymorphism in colorectal cancer in ethnic Kashmiri
population - A case control study in a detailed perspective. Meta
Gene. 9:128–136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kroeger KM, Steer JH, Joyce DA and Abraham
LJ: Effects of stimulus and cell type on the expression of the −308
tumour necrosis factor promoter polymorphism. Cytokine. 12:110–119.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhuang L, Ma W, Cai D, Zhong H and Sun Q:
Associations between tumor necrosis factor-α polymorphisms and risk
of psoriasis: A meta-analysis. PLoS One. 8:e688272013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Howell WM, Turner SJ, Theaker JM and
Bateman AC: Cytokine gene single nucleotide polymorphisms and
susceptibility to and prognosis in cutaneous malignant melanoma.
Int J Immunogenet. 30:409–414. 2003. View Article : Google Scholar
|
|
32
|
Nikolova PN, Pawelec GP, Mihailova SM,
Ivanova MI, Myhailova AP, Baltadjieva DN, Marinova DI, Ivanova SS
and Naumova EJ: Association of cytokine gene polymorphisms with
malignant melanoma in Caucasian population. Cancer Immunol
Immunother. 56:371–379. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu F, Qureshi AA, Niu T, Kraft P, Guo Q,
Hunter DJ and Han J: Interleukin and interleukin receptor gene
polymorphisms and susceptibility to melanoma. Melanoma Res.
18:330–335. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu F, Qureshi AA, Kraft P, Guo Q, Hunter
DJ and Han J: Polymorphisms in genes involved in DNA repair, cell
growth, oxidative stress and inflammatory response, and melanoma
risk. Br J Dermatol. 161:209–212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kurzawski M, Pawlik A, Czerny B, Domański
L, Rózański J and Droździk M: Frequencies of the common promoter
polymorphisms in cytokine genes in a Polish population. Int J
Immunogenet. 32:285–291. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Garbe C, Ellwanger U, Tronnier M, Brocker
EB and Orfanos CE: The New American Joint Committee on Cancer
staging system for cutaneous melanoma: A critical analysis based on
data of the German Central Malignant Melanoma Registry. Cancer.
94:2305–2307. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dickson PV and Gershenwald JE: Staging and
prognosis of cutaneous melanoma. Surg Oncol Clin N Am. 20:1–17.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zarogoulidis P, Yarmus L, Darwiche K,
Walter R, Huang H, Li Z, Zaric B, Tsakiridis K and Zarogoulidis K:
Interleukin-6 cytokine: A multifunctional glycoprotein for cancer.
Immunome Res. 9:165352013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gomes M, Coelho A, Araújo A, Azevedo A,
Teixeira AL, Catarino R and Medeiros R: IL-6 polymorphism in
non-small cell lung cancer: A prognostic value? Tumour Biol.
36:3679–3684. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lu C, Vickers MF and Kerbel RS:
Interleukin 6: A fibroblast-derived growth inhibitor of human
melanoma cells from early but not advanced stages of tumor
progression. Proc Natl Acad Sci USA. 89:9215–9219. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fishman D, Faulds G, Jeffery R,
Mohamed-Ali V, Yudkin JS, Humphries S and Woo P: The effect of
novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6
transcription and plasma IL-6 levels, and an association with
systemic-onset juvenile chronic arthritis. J Clin Invest.
102:1369–1376. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Terry CF, Loukaci V and Green FR:
Cooperative influence of genetic polymorphisms on interleukin 6
transcriptional regulation. J Biol Chem. 275:18138–18144. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jones KG, Brull DJ, Brown LC, Sian M,
Greenhalgh RM, Humphries SE and Powell JT: Interleukin-6 (IL-6) and
the prognosis of abdominal aortic aneurysms. Circulation.
103:2260–2265. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Martínez-Escribano JA, Moya-Quiles MR,
Muro M, Montes-Ares O, Hernández-Caselles T, Frías JF and
Alvarez-López MR: Interleukin-10, interleukin-6 and
interferon-gamma gene polymorphisms in melanoma patients. Melanoma
Res. 12:465–469. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gautam KA, Muktanand T, Sankhwar SN, Goel
A, Sankhwar PL and Rajender S: Functional polymorphisms in the IL6
gene promoter and the risk of urinary bladder cancer in India.
Cytokine. 77:52–156. 2016. View Article : Google Scholar
|
|
46
|
Leibovici D, Grossman HB, Dinney CP,
Millikan RE, Lerner S, Wang Y, Gu J, Dong Q and Wu X: Polymorphisms
in inflammation genes and bladder cancer: From initiation to
recurrence, progression, and survival. J Clin Oncol. 23:5746–5756.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Datz E, Zeman F, Koller M, Szeimies RM,
Berneburg M, Landthaler M, Bosserhoff AK and Karrer S:
Phototherapy-induced elevation of serum level of melanoma
inhibitory activity. Photodermatol Photoimmunol Photomed.
35:255–260. 2019.PubMed/NCBI
|
|
48
|
Aggarwal BB, Gupta SC and Kim JH:
Historical perspectives on tumor necrosis factor and its
superfamily: 25 years later, a golden journey. Blood. 119:651–665.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Messer G, Spengler U, Jung MC, Honold G,
Blömer K, Pape GR, Riethmüller G and Weiss EH: Polymorphic
structure of the tumor necrosis factor (TNF) locus: An NcoI
polymorphism in the first intron of the human TNF-beta gene
correlates with a variant amino acid in position 26 and a reduced
level of TNF-beta production. J Exp Med. 173:209–219. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wilson AG, di Giovine FS, Blakemore AI and
Duff GW: Single base polymorphism in the human tumour necrosis
factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR
product. Hum Mol Genet. 1:3531992. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kroeger KM, Carville KS and Abraham LJ:
The −308 tumor necrosis factor-alpha promoter polymorphism effects
transcription. Mol Immunol. 34:391–399. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang S, Wang C, Xi B and Li X:
Association between the tumour necrosis factor-α −308G/A
polymorphism and chronic obstructive pulmonary disease: An update.
Respirology. 16:107–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ghaderian SM, Akbarzadeh Najar R and
Tabatabaei Panah AS: Tumor necrosis factor-α: Investigation of gene
polymorphism and regulation of TACE-TNF-α system in patients with
acute myocardial infarction. Mol Biol Rep. 38:4971–4977. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sharma S, Sharma A, Kumar S, Sharma SK and
Ghosh B: Association of TNF haplotypes with asthma, serum IgE
levels, and correlation with serum TNF-alpha levels. Am J Respir
Cell Mol Biol. 35:488–495. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Helmig S, Aliahmadi N, Stephan P, Döhrel J
and Schneider J: TNF-α −308 genotypes are associated with TNF-α and
TGF-β1 mRNA expression in blood leucocytes of humans.
Cytokine. 53:306–310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu N, Liu Gj and Liu J: Genetic
association between TNF-α promoter polymorphism and susceptibility
to squamous cell carcinoma, basal cell carcinoma, and melanoma: A
meta-analysis. Oncotarget. 8((32)): 53873–53885. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang J, Cao C, Luo H, Xiong S, Xu Y and
Xiong W: Tumour necrosis factor-α −308G/A polymorphism and risk of
the four most frequent cancers: a meta-analysis. Int J
Immunogenetics. 38:311–320. 2011. View Article : Google Scholar
|
|
58
|
Gorouhi F, Islami F, Bahrami H and
Kamangar F: Tumour-necrosis factor-A polymorphisms and gastric
cancer risk: A meta-analysis. Br J Cancer. 98:1443–1451. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang Y, Feng R, Bi S and Xu Y: TNF-alpha
polymorphisms and breast cancer. Breast Cancer Res Treat.
129:513–519. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Min L, Chen D, Qu L and Shou C: Tumor
necrosis factor-a polymorphisms and colorectal cancer risk: A
meta-analysis. PLoS One. 9:e851872014. View Article : Google Scholar : PubMed/NCBI
|