Introduction
Osteosarcoma (OS) is generally a high-grade tumor
that exhibits locally aggressive behavior and causes early systemic
metastases (1). OS primarily occurs
in the long bones of young adults and is the most common primary
bone sarcoma worldwide (1). The 5
year relative survival rate of early stage OS in the US was 20–30%
in children and adolescents diagnosed in the early 1970s
[1970-1973; (2)]. This increased to
64% in those diagnosed during 1986–1993, a figure which may be
attributed to the combined use of multiagent or neoadjuvant
chemotherapy and advanced surgery (3). However, in the past >20 years, no
significant improvements have been recorded in the 5 year survival
rate of OS: The 5-year relative survival rate of OS was ~65-70% in
children and adolescents diagnosed between 2008–2014 (4). Metastasis is primarily responsible for
an impediment in treatment development and can be held accountable
for therapeutic failure in patients with OS (5). There is a lack of effective strategies
to treat advanced OS and the 5-year survival rate of patients with
distant metastases was ~20-30% in the US and Italy during 1980–2000
(6,7). Thus, recent studies have focused on
identifying molecular markers that could be used for early
metastasis detection and prognosis assessment, serving as potential
therapeutic targets for advanced OS (3,5,8–10).
Differentially expressed in FDCP 6 homolog (DEF6)
and associated homologous genes (Def2, Def3, Def8) were first
described as novel mouse genes expressed in the hemopoietic system
(11). DEF6, also known as IFN
regulatory factor 4-binding protein (IBP), encodes an interferon
regulatory factor 4 binding protein and is diffusely expressed in
the human immune system (12). DEF6
is predominantly expressed in T lymphocytes and regulates several
T-cell processes, including T-helper-1/2/17 differentiation and the
coordination of actin cytoskeleton remodeling (13–15).
DEF6 serves a critical and unique role in regulating systemic
autoimmunity as a SWEF family, which is a small and unique protein
family of Rho guanine nucleotide exchange factors (GEF), only
comprised of DEF6 and SWAP-70, controlling both cytoskeletal
dynamics and the activity of IFN regulatory factor 4 (IRF4)
(13). Additionally, DEF6 reportedly
regulates bone remodeling by restraining osteoclastogenesis and
inflammatory bone resorption (16).
Although numerous studies have focused on the role of DEF6 in the
immune system, abnormal DEF6 expression levels in non-immune cells,
particularly tumor cells, indicate a potential function of DEF6 in
tumorigenesis regulation (12,17,18).
DEF6 has been reported to be highly expressed in breast carcinoma,
ovarian cancer, colorectal cancer, oral squamous cell carcinoma and
extraskeletal myxoid chondrosarcoma (17–21),
exhibiting various malignant behaviors and cancer cell biology,
including proliferation (19),
autophagy (22), invasion (20) and epithelial-mesenchymal transition
(12). Additionally, Rho-GTPase and
mammalian target of rapamycin (mTOR) signaling pathways, which
serve a key role in OS tumorigenesis and metastasis (3,8), are
targets of DEF6 that facilitate tumor cell metastasis and enhance
tumorigenic potential (12,22). However, the expression and biological
functions of DEF6 in OS are yet to be elucidated.
In the present study, the role of DEF6 with regards
to clinicopathological features of patients and its prognostic
value in human OS was investigated. DEF6 expression levels were
determined in OS and normal osteoblast cell lines. Furthermore,
DEF6 was demonstrated to act as an oncogenic driver that promoted
the metastatic potential of OS cells. To the best of our knowledge,
this is the first study to demonstrate the role of DEF6 in OS.
Materials and methods
Human specimens
Specimens of human osteosarcoma tissues were
collected from 58 patients with pathologically confirmed OS at the
Southwest Hospital and Xinqiao Hospital of the Army Medical
University from February 2011 to November 2015. The inclusion
criteria were as follows: No patients received pre-operative
anticancer treatment before pathological confirmation of OS and all
patients received standard treatment (chemotherapy and surgery)
following pathologically confirmation of OS. The exclusion criteria
were as follows: Patients who had other fatal diseases or cancer
and non-cancer associated mortality. Given that the paired normal
OS tissues were not completely collected during surgery, human
benign bone tumor specimens were used as the control group and
compared with the OS specimens in the present study. The specimens
were obtained from 12 patients with histopathologically-confirmed
osteoblastoma from Xinqiao Hospital from March 2015 to November
2017. The clinical staging and corresponding treatment for OS were
based on the Enneking system (23).
Patients were classified as with or without developed distant
metastasis at diagnosis or following treatment. The
clinicopathological features of all patients with osteosarcoma are
listed in Table I. Among the 58
patients with OS, 24 were women and 34 were men. The median age of
these patients was 22 years (age range, 8–59 years). A total of 22
patients with primary osteosarcoma had recurrence following surgery
and received chemotherapy, 12 patients had metastasis at first
diagnosis, and eight more patients developed distant metastasis
following treatment. Written informed consent was obtained from all
patients or their guardians. All experiments were approved by the
Ethics Committees of Southwest Hospital (approval no. 27–2011) and
Xinqiao Hospital (approval no. 2018-069-01).
 | Table I.Correlation between DEF6 expression
and clinicopathological features in patients with osteosarcoma. |
Table I.
Correlation between DEF6 expression
and clinicopathological features in patients with osteosarcoma.
|
| DEF6
expression |
|
|---|
|
|
|
|
|---|
| Characteristic | High | Low | χ2 |
|---|
| Age, years |
|
| 0.808 |
|
≤30 | 26 | 19 |
|
|
>30 | 8 | 5 |
|
| Sex |
|
|
0.097 |
|
Male | 23 | 11 |
|
|
Female | 11 | 13 |
|
| Clinical stage |
|
| 0.033a |
|
IIA | 7 | 7 |
|
|
IIB | 16 | 16 |
|
|
III | 11 | 1 |
|
| Local
recurrence |
|
| 0.544 |
|
Yes | 14 | 8 |
|
| No | 20 | 16 |
|
| Distant
metastasis |
|
| 0.003a |
|
Yes | 17 | 3 |
|
| No | 17 | 21 |
|
| Histological
type |
|
| 0.462 |
|
Osteoblastic | 17 | 17 |
|
|
Chondroblastic | 13 | 5 |
|
|
Fibroblastic | 2 | 1 |
|
|
Others | 2 | 1 |
|
| Tumor size |
|
| 0.233 |
| <8
cm | 22 | 19 |
|
| ≥8
cm | 12 | 5 |
|
| Tumor location |
|
| 0.085 |
|
Limbs | 32 | 19 |
|
|
Others | 2 | 5 |
|
Cell culture
MNNG/HOS, SaOS2, MG63 and U2OS OS cell lines were
purchased from Cellcook Biological Technology Co., Ltd (www.cellcook.com). The human osteoblast cell line
hFOB1.19 was obtained from the Cell Type Culture Collection of the
Chinese Academy of Sciences. A malignant transformed hFOB1.19 cell
line (MTF cells) was produced in the laboratory (24). MNNG/HOS, SaOS2, MG63, U2OS and MTF
cells were cultured in high-glucose DMEM (Hyclone; Cytiva)
supplemented with 10% FBS (Lonsera Science SRL) and 1%
penicillin-streptomycin (Hyclone; Cytiva), and maintained at 37°C
and 5% CO2. hFOB1.19 cells were cultured in DMEM/F12
(Hyclone; Cytiva) with 15% FBS (Lonsera Science SRL) and maintained
at 35°C and 5% CO2. Both cells were cultured to the
logarithmic growth phase of the third generation prior to use for
subsequent experimentation, and the medium was changed every 48–72
h.
Antibodies
Primary mouse anti-human DEF6 antibody (cat. no.
FAB-KY015-IBP) were provided by Professor Chuanmin Hu (Army Medical
University), which were produced and verified in their laboratory
(21). Primary rabbit anti-human
matrix metallopeptidase (MMP9) antibody (cat. no. 13667) was
purchased from Cell Signaling Technology, Inc. (CST) and primary
rabbit anti-human GAPDH antibody (cat. no. AB-P-R001) was purchased
from Hangzhou Goodhere Biotech Co., Ltd.
Immunohistochemistry (IHC)
IHC was performed using a kit (Beijing Zhongshan
Golden Bridge Biotechnology Co. Ltd., ZSGB-BIO, Beijing, China), as
previously described (8). Briefly,
human OS or osteoblastoma tissue sections were deparaffinized in
two replicate bottles of xylene for 10 min each at room
temperature. Tissue section were subsequently rehydrated with a
graded series of ethanol and then antigens were retrieved in 10 mM
citrate buffer by boiling for 10 min, and cooling to room
temperature. Sections were blocked with a ready to use goat serum
solution in the UltraSensitive™ SP (mouse/rabbit) IHC kit (cat. no.
KIT-9720; Fuzhou Maixin Biotech Co., Ltd.) for ~30 min at 37°C and
incubated with 3% hydrogen peroxide for ~15 min at 37°C to inhibit
endogenous peroxidase activity. Tissue sections were subsequently
incubated with primary antibodies against DEF6 (cat. no.
FAB-KY015-IBP) and MMP9 (cat. no. 13667, CST), dilution by primary
antibody dilution buffer (cat. no. ZLI-9028; ZSGB-BIO) overnight at
4°C. Subsequently, sections were incubated with ready-to-use
undiluted secondary antibodies conjugated with biotin in the
UltraSensitiveTM SP (mouse/rabbit) IHC kit (cat. no. KIT-9720;
Fuzhou Maixin Biotech Co., Ltd.) for 30 min at 37°C. Subsequently,
DAB staining (DAB kit; cat. no. DAB-0031; Fuzhou Maixin Biotech
Co., Ltd.) was used for 5 min at room temperature and the nuclei
were stained with hematoxylin for 2.5 min at room temperature. The
stained sections were observed under a light microscope (Olympus
Corporation, magnification, ×400). For quantitative assessment of
DEF6 (1:500; cat. no. FAB-KY015-IBP) protein expression in primary
site tissues or metastatic site tissues of OS, the percentage of
positive cells was determined in 5 randomly selected fields of view
using a light microscope and higher-magnification objectives
(magnification, ×400), including ≥50 cells. The final IHC score was
a product of the positive cell ratio score (0, no immunoreactivity;
1, ≤25% cells stained; 2, 26–50% cells stained; 3, 51–75% cells
stained; 4, ≥76% cells stained) and relative expression score (0,
negative; 1, light yellow staining; 2, yellow staining; 3, brown or
dark brown staining). Final scores of ≤1 indicated negative
expression of DEF6, scores between >1 and ≤3 indicated weak
positive expression, scores between >3 and <8 indicated
positive expression and scores ≥8 indicated overexpression.
Negative and weak positive expression of DEF6 represented low
protein levels, while positive and overexpression of DEF6
represented high protein levels.
The semiquantitative assessment of MMP9 (1:200; cat.
no. 13667; Cell Signaling Technology, Inc.) protein expression was
then performed. Positive MMP9 staining was observed under at least
5 fields of view, with >10 OS cells around tumor angiosomes,
using a light microscope (magnification, ×400).
Transfection of short interfering RNAs
(siRNAs)
Small interfering (si)RNAs targeting DEF6 were
purchased from Guangzhou RiboBio Co., Ltd. For transient knockdown
experiments, MTF and MNNG/HOS cells (exhibit significantly higher
DEF6 expression compared with other OS cells) were transfected with
30 nM targeting or scrambled DEF6 siRNAs using the RiboBio-FECT™ CP
kit (Guangzhou RiboBio Co., Ltd.), according to manufacturer's
protocol. Knockdown efficiency was assessed 48 or 72 h
post-transfection via RT-qPCR and western blotting analyses. The
siRNA target sequences used were DEF6-siRNA-1
(5′-ACAGTATGCTCTCCAATCA-3′) and DEF6-siRNA-2
(5′-CTGCTACTTTGGGAGTGAA-3′).
Reverse transcription quantitative PCR
(RT-qPCR)
Following appropriate treatments [transfection with
short interfering (si)RNAs], total RNA was extracted from
osteosarcoma cells using RNAiso™ Plus (Takara Biotechnology Co.,
Ltd.). RNA samples were then reverse transcribed into cDNA using a
QuantScript RT kit (Takara), according to manufacturer's protocol.
RT-qPCR was performed using a SYBR Premix kit (Takara), according
to the manufacturer's protocol. The thermocycling conditions were
used as previously described (25),
as follows: 95°C for 30 sec, followed by 40 cycles at 95°C for 15
sec and 60°C for 60 sec. The following primer sequences were used
for qPCR: DEF6 forward, 5′-GAAAGCTCGGCGAGATGAAG-3′ and reverse,
5′-GATGTAGCGCTCCTGCTCCT-3′; MMP9 forward,
5′-AAACCGAGTTGGAACCACGAC-3′ and reverse,
5′-AGACGGGTATCCCTTCGACG-3′; and GAPDH forward,
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse,
5′-GTAGAGGCAGGGATGATGTTCT-3′. Relative expression levels were
calculated using the 2−∆∆Cq method (26).
Cell proliferation and colony
formation assays
Cell proliferation assays were performed using a
Cell Counting Kit-8 (CCK-8; cat. no. C0038; Beyotime Institute of
Biotechnology), as previously reported (27). Cells (MNNG/HOS and MTF) were digested
using Trypsin (Hyclone; Cytiva) and seeded into four 96-well plates
(1.5×103 cells/well) following siRNA transfection.
Following incubation for 24, 48, 72 and 96 h, at 37°C in 5%
CO2, CCK-8 reagent mixed with serum-free DMEM (1:9) 100
µl was added and five wells with no cell were selected as blank
control also added 100 µl CCK-8 reagent mixed with serum-free DMEM.
Both of them were followed by incubation for 1.5 h at 37°C in 5%
CO2. A plate reader (Thermo Fisher Scientific, Inc.) was
used to measure the optical density (OD) at 450 nm. Final OD values
were obtained by subtracting the OD values of cell-containing wells
from blank wells, using he following formula: ODFinal =
ODmeasured - ODblank (ODblank
refers to the OD values of wells maintained in 100 µl CCK-8 reagent
and serum-free DMEM).
Colony formation assays were performed as previously
described (28). MTF and MNNG/HOS
cells, which were transfected with either scrambled siRNAs or DEF6
siRNA, were plated in six-well plates (100 cells/well) and
incubated at 37°C. On day 11 for MTF and on day 13 for MNNG/HOS
cells, cells were fixed with 4% paraformaldehyde for 15 min at room
temperature and then stained with 0.1% crystal violet (Beyotime
Institute of Biotechnology) for 5 min at room temperature. After
rinsing with water, colony numbers (>50 cells) were
determined.
Wound healing assay
The wound healing assay was performed as previously
described (8). Transfected MNNG/HOS
and MTF cells (~6×105 cells/well) were seeded in
six-well plates and cultured until they reached 80% confluence. A
100 µl micropipette tip was used to create a wound. Cells were
monitored at 0 and 24 h (MNNG/HOS) or 48 h (MTF) following wounding
and images of wound healing were captured using an inverted
phase-contrast light microscope (Olympus Corporation;
magnification, ×100) equipped with DP Controller software (version
3.1.1.267; Olympus Life Science; Olympus Corporation).
Transwell invasion assay
Transfected cells were resuspended in serum-free
DMEM, after which 200 µl of the cell suspension was seeded
(2×105 cells/ml) into the upper chambers of an 8 µm
Transwell filter (Merck KGaA), that were precoated with 1:3 diluted
Matrigel (BD Biosciences). DMEM supplemented with 10% FBS (600 µl)
was added to the lower chamber and incubated for 24 h at 37°C in 5%
CO2. Subsequently, invaded cells were fixed with 4%
paraformaldehyde for 15 min at room temperature and stained with
0.1% crystal violet for 5 min at room temperature. Cell counting
was performed in at least 5 randomly selected fields of view under
an inverted phase-contrast light microscope (Olympus Corporation;
magnification, ×200).
Western blotting
As previously reported (29), cells (MG63, hFOB1.19, SaOS2, U2OS,
MNNG/HOS and MTF) were collected following transfection and treated
with M-PER™ Mammalian Protein Extraction Reagent (cat. no. 78501;
Thermo Fisher Scientific, Inc.) containing a protease inhibitor
(Roche Diagnostics, Co. Ltd.). Protein concentration was determined
using the BCA Protein Assay kit (cat. no. P0010S; Beyotime
Institute of Biotechnology). Proteins (40–50 µg/lane) were
subsequently separated using 10% SDS Tris-glycine gels and
transferred onto PVDF membranes [cat. no. 3010040001; Roche
Diagnostics (Shanghai) Co., Ltd.]. The membranes were blocked with
5% fat-free milk and incubated at 4°C with the following primary
antibodies overnight: GAPDH (1:1,000), DEF6 (1:1,000) and MMP9
(1:400) were used. All primary antibodies were diluted to the
aforementioned concentration using Western Primary Antibody
Dilution buffer (cat. no. P0023A; Beyotime Institute of
Biotechnology). After rinsing with PBST. Secondary antibodies (goat
anti-rabbit or goat anti-mouse IgG conjugated with HRP; 1:5,000;
cat. nos. bs-0295G-HRP and bs-0368G-HRP; BIOSS) were subsequently
applied and incubated for 1 h at room temperature. Immunoreactivity
was detected using an ECL kit (BeyoECL Moon; Beyotime Institute of
Biotechnology).
Analysis using the R2 database
The R2 database (hgserver1.amc.nl) and subset data
named ‘mixed osteosarcoma-Kuijjer-127-vst-ilmnhwg6v2’ were used for
gene ontology (GO) analyses to investigate the relationship between
DEF6 and other cancer related genes in patients with OS. The
database was further used to analyze the correlation between DEF6,
common MMP genes and tissue inhibitor of metalloproteinase (TIMP)
genes associated with OS cell invasion (5).
Statistical analysis
Categorical data were analyzed using the
χ2 test and Spearman's or Pearson's rank correlation
coefficient. Quantitative data are presented as mean ± standard
deviation and analyzed using unpaired two-tailed Student's t-test
or Mann-Whitney test (nonparametric test when P-value and F test
<0.05) for between-group comparisons. ANOVA with Bonferroni's
correction was used for comparing data in >2 groups. Spearman
survival analysis was performed using the Kaplan-Meier method and
log-rank test. P<0.05 was considered to indicate a statistically
significant difference. All analyses were performed using SPSS
(version 20.0; IBM Corp.) or GraphPad Prism software (version 7.00;
GraphPad Software, Inc.). All in vitro experiments were
performed at least in triplicate.
Results
DEF6 is widely expressed in patients
with OS
Previous studies have reported an abnormal
expression of DEF6 in multiple malignant tumors, indicating that
DEF6 functions as an oncogene (17–21). IHC
staining was performed to examine the expression of DEF6 in 58
human OS tissues and to determine the role of DEF6 in OS
development. The results demonstrated that DEF6 was widely
expressed in these tissues. A total of 0.05% cases exhibited
negative staining for DEF6 (3/58; Fig.
1A), while 94.83% (55/58) cases exhibited positive staining for
DEF6. Furthermore, 36.20% cases exhibited weak positive expression
(21/58; Fig. 1B), 39.66% exhibited
positive expression (23/58; Fig. 1C)
and 18.97% exhibited overexpression (11/58; Fig. 1D). As evidenced, DEF6 was expressed
at high levels (positive expression/overexpression) in the majority
of cases (34/58).
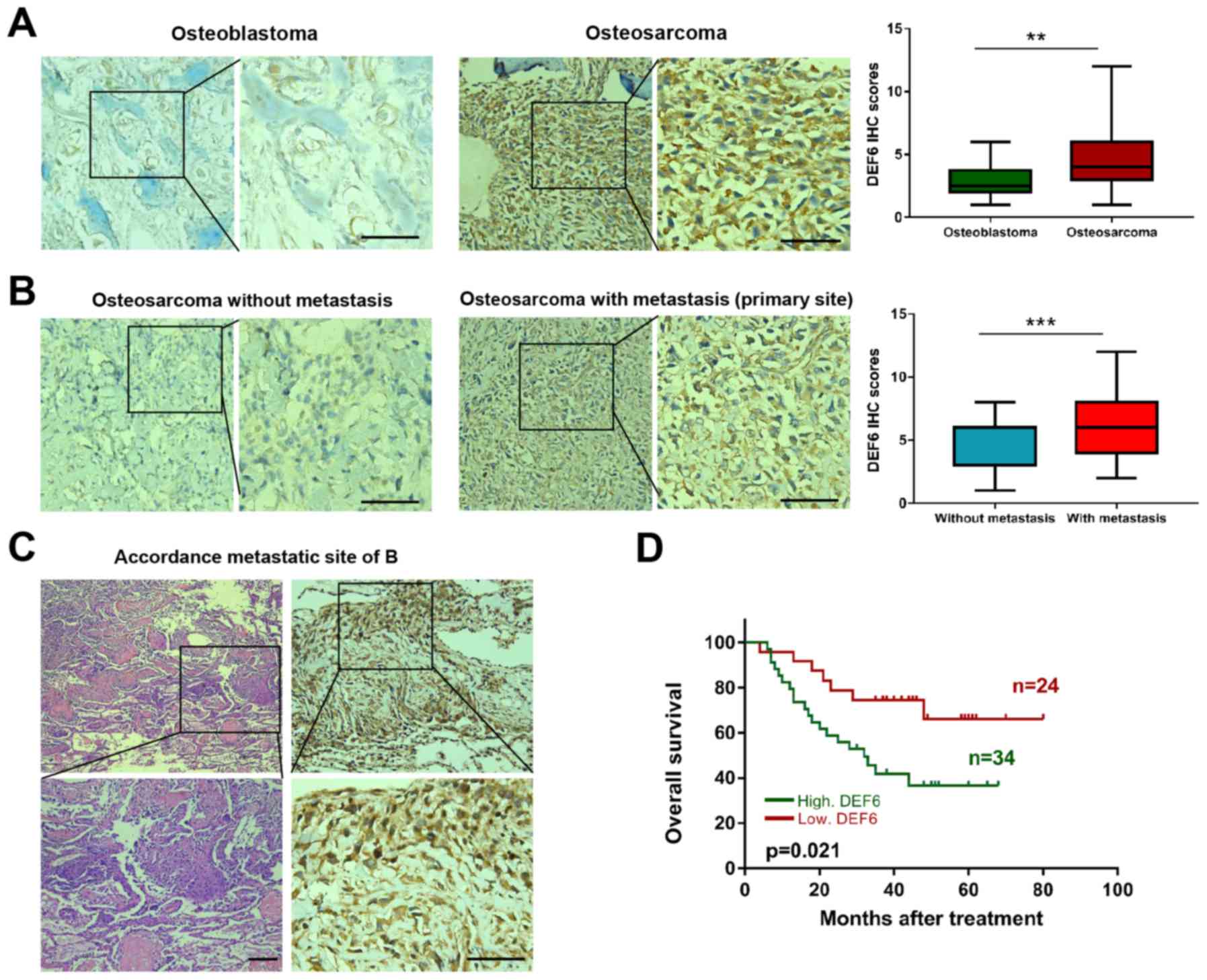
DEF6 is highly expressed, is
positively associated with distant metastasis and predicts poor
prognosis in patients with OS
IHC staining was utilized to examine DEF6 expression
in 12 human osteoblastomas (benign bone tumors) and 58 OS tissues.
As expected, DEF6 was observed to exhibit significantly higher
expression (P<0.05) in OS compared with osteoblastoma tissues
(Fig. 2A). Furthermore, the
correlation between DEF6 expression and clinicopathological
features of patients with OS via immunohistochemical staining was
analyzed. The results demonstrated high DEF6 expression in 44.74%
(17/38) of patients with OS and without metastasis, and 85.00%
(17/20) in patients with OS and metastasis. This difference was
statistically significant (P=0.003; Table I). Accordingly, a high IHC score for
DEF6 was observed more frequently in patients with distant
metastatic disease (Fig. 2B). DEF6
was highly expressed at lung metastatic sites in samples from the
case presented in Fig. 2B (Fig. 2C). Furthermore, a high DEF6
expression was significantly associated with worse clinical stage
(P<0.05), but not with age, sex, location, size, local
recurrence or histological subtype. Additionally, Kaplan-Meier
curves indicated that the overall survival of patients with OS and
high DEF6 expression levels was significantly shorter compared with
OS and low DEF6 levels (P=0.021; Fig.
2D). The results demonstrated that DEF6 was widely expressed in
OS tissues and that high DEF6 levels were positively associated
with metastasis and poor prognosis.
DEF6 is overexpressed in OS cell lines
and contributes to metastatic potential in vitro
Western blotting was utilized to determine the
expression of DEF6 in a series of cell lines. Excluding MG63 cells,
DEF6 protein levels were significantly higher in OS cell lines
compared with the osteoblast cell line hFOB1.19 (Fig. 3A). This validated that DEF6 that is
widely expressed in OS tissues, with upregulated levels in OS cell
lines. Furthermore, a significantly higher expression of DEF6 was
exhibited in MTF cells compared with hFOB1.19 osteoblasts
(P<0.001, Fig. 3A), indicating
that DEF6 upregulation is involved in the malignant transformation
of osteoblasts. To determine whether increased levels of DEF6
contributed to the malignant proliferation of OS cells, DEF6
expression was knocked down using two siRNA sequences of DEF6 in
MTF and MNNG/HOS cell lines (as they exhibit significantly higher
DEF6 expression compared with other OS cells). Both target
sequences demonstrated lower DEF6 protein levels, as demonstrated
via western blotting (Fig. 3B).
Furthermore, CCK-8 and colony formation assays were performed to
investigate the proliferation of cells with or without DEF6
knockdown. The results demonstrated that DEF6 knockdown had no
significant effect on the malignant proliferation of MTF or
MNNG/HOS cells (Fig. 3C and D).
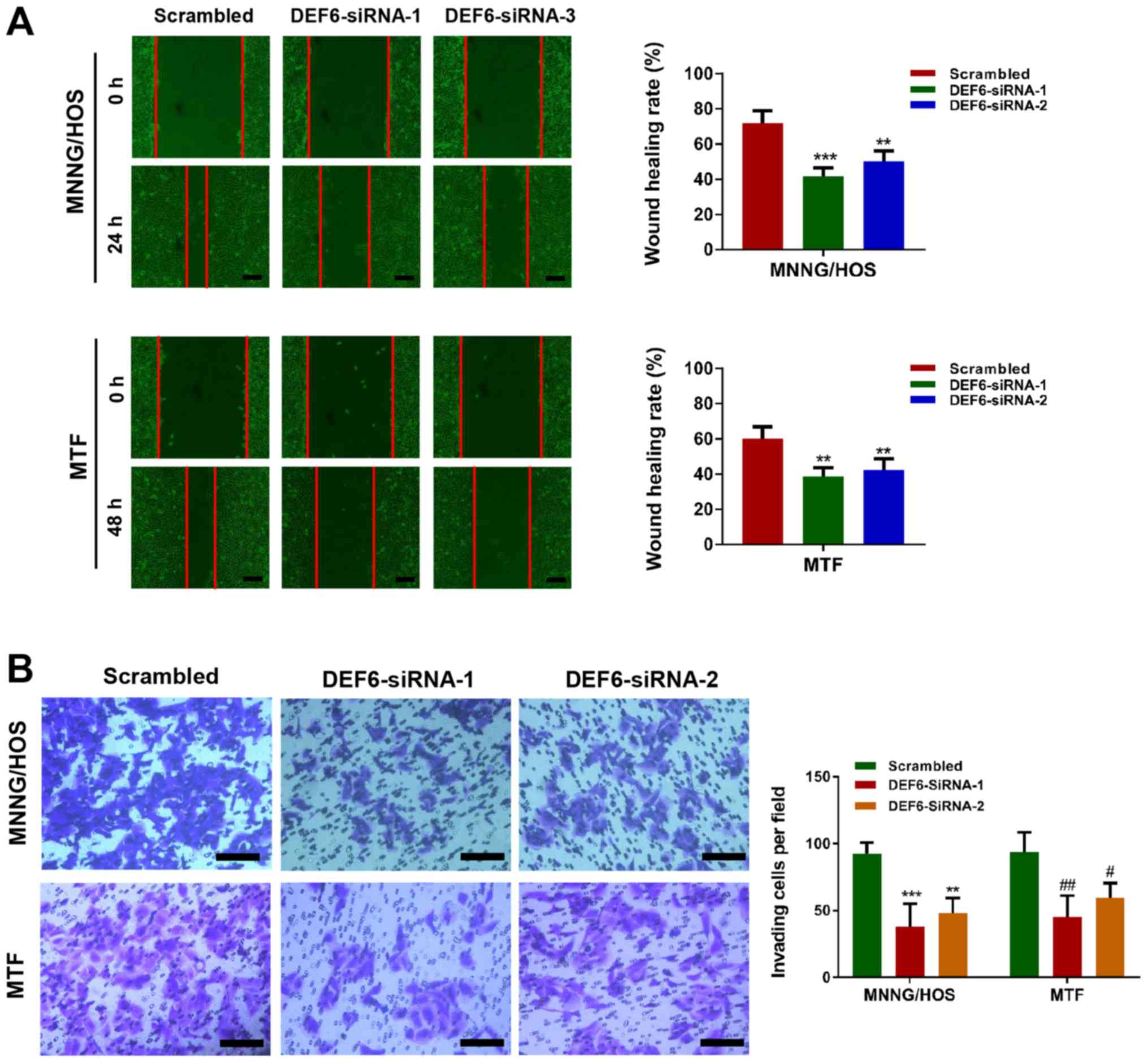
Considering the positive association between DEF6 expression and
metastasis in human OS tissues, wound healing and Transwell assays
were performed to determine whether DEF6 knockdown suppressed cell
motility. The results indicated that decreased levels of DEF6 in
MTF and MNNG/HOS cells caused a significant reduction in cell
migration and invasion, as evident from the decreased wound healing
rates (Fig. 4A) and numbers of
invading cells (Fig. 4B). In
summary, the results indicated that although DEF6 may not be a
major driver of OS cell proliferation, it significantly contributed
to metastatic potential in vitro.
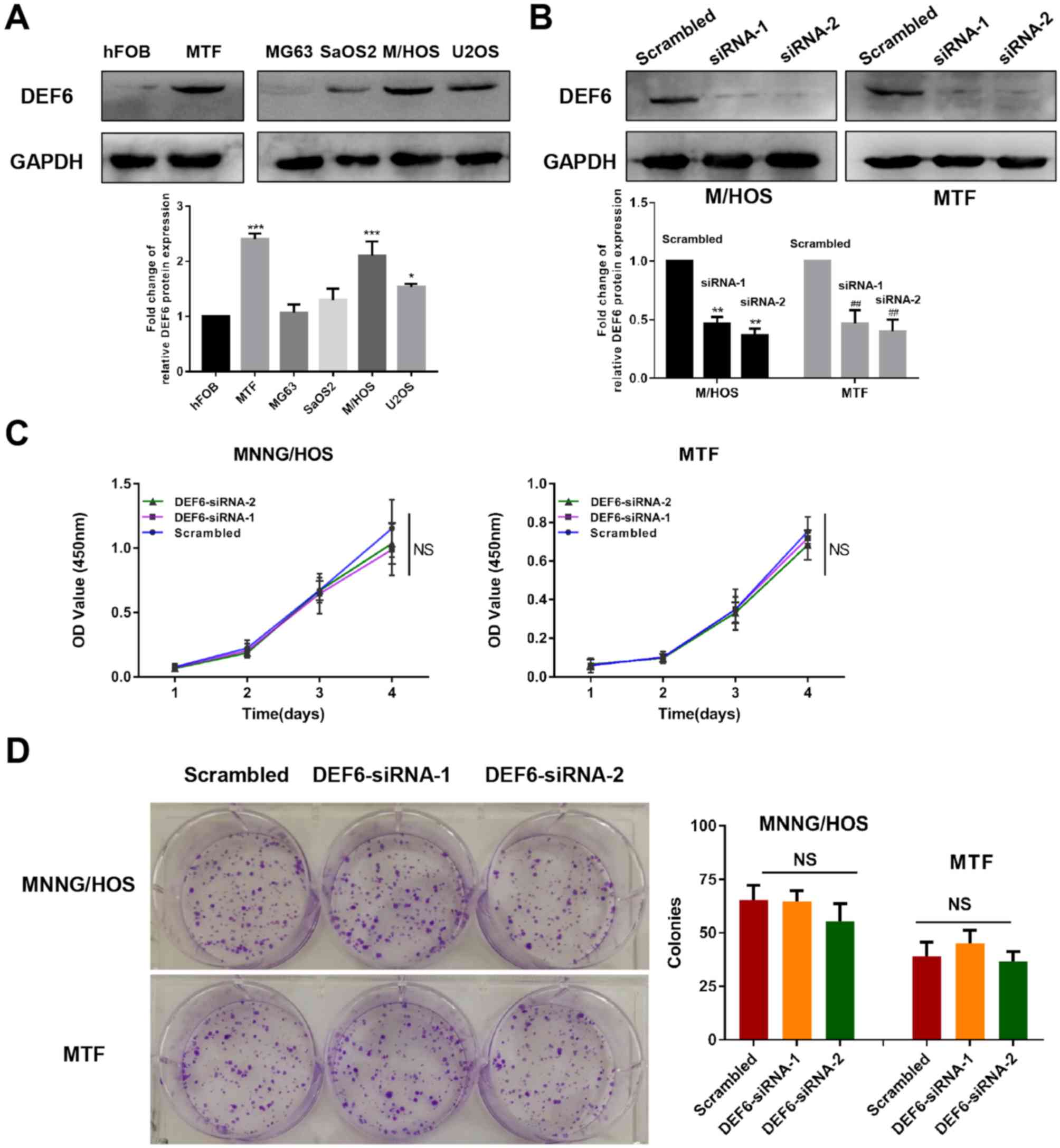 | Figure 3.DEF6 is highly expressed in OS cells;
however, high DEF6 expression may not contribute to high
proliferative ability. (A) DEF6 expression levels in OS and normal
osteoblast cell lines. DEF6 was highly expressed in OS cell lines,
particularly in MTF and MNNG/HOS cells compared with hFOB1.19
cells. (B) DEF6 expression in MTF and MNNG/HOS cells was
successfully knocked down by siRNAs. (C) DEF6 expression knockdown
by siRNAs in MTF and MNNG/HOS cells did not significantly affect
cell proliferation. (D) DEF6 knockdown in MTF and MNNG/HOS cells
did not significantly affect the ability of cell colony formation.
*P<0.05, ***P<0.001 vs. the hFOB group, **P<0.01 vs. the
M/HOS scrambled group, ##P<0.01 vs. the MTF scrambled
group. DEF6, differentially expressed in FDCP 6 homolog; OS,
osteosarcoma; MTF, malignant transformed hFOB1.19 cell line; siRNA,
short interfering RNA; NS, no significant difference. |
Knockdown of DEF6 expression
significantly decreases MMP9 expression in OS cells in vitro
To further investigate how DEF6 influenced OS cell
motility, the R2 database was used to identify a potential target
of DEF6 in OS. As predicted, the results of GO analysis supported
the aforementioned results of the current study and demonstrated
DEF6 is more likely to facilitate OS cell migration and invasion
rather than proliferation and survival (Fig. 5A). The results demonstrated that DEF6
was closely associated OS cell adhesion and motility. TIMPs
reportedly serve a key role in regulating extracellular matrix
remodeling, thereby affecting cell adhesion and motility (30). Additionally, MMPs and TIMPs
reportedly have considerable influence on the invasive ability of
cancer cells (31). Thus, the R2
database was used to investigate the correlation between DEF6 and
MMPs or their inhibitor-associated genes, both of which have been
widely reported to be correlated with OS metastases (10,30–36). The
results revealed that there may be a positive correlation between
DEF6 and MMP9 (P<0.0001; r=0.4278; Fig. 5B). Furthermore, the results
demonstrated that the mRNA and protein expression of MMP9 were
significantly downregulated in association with a decrease in DEF6
expression in OS cell lines (Fig.
5C-D). Collectively, the results indicated that the knockdown
of DEF6 expression had an obvious direct effect on restricting the
motility of OS cells in vitro.
DEF6 expression is positively
correlated with MMP9 expression in human OS tissues
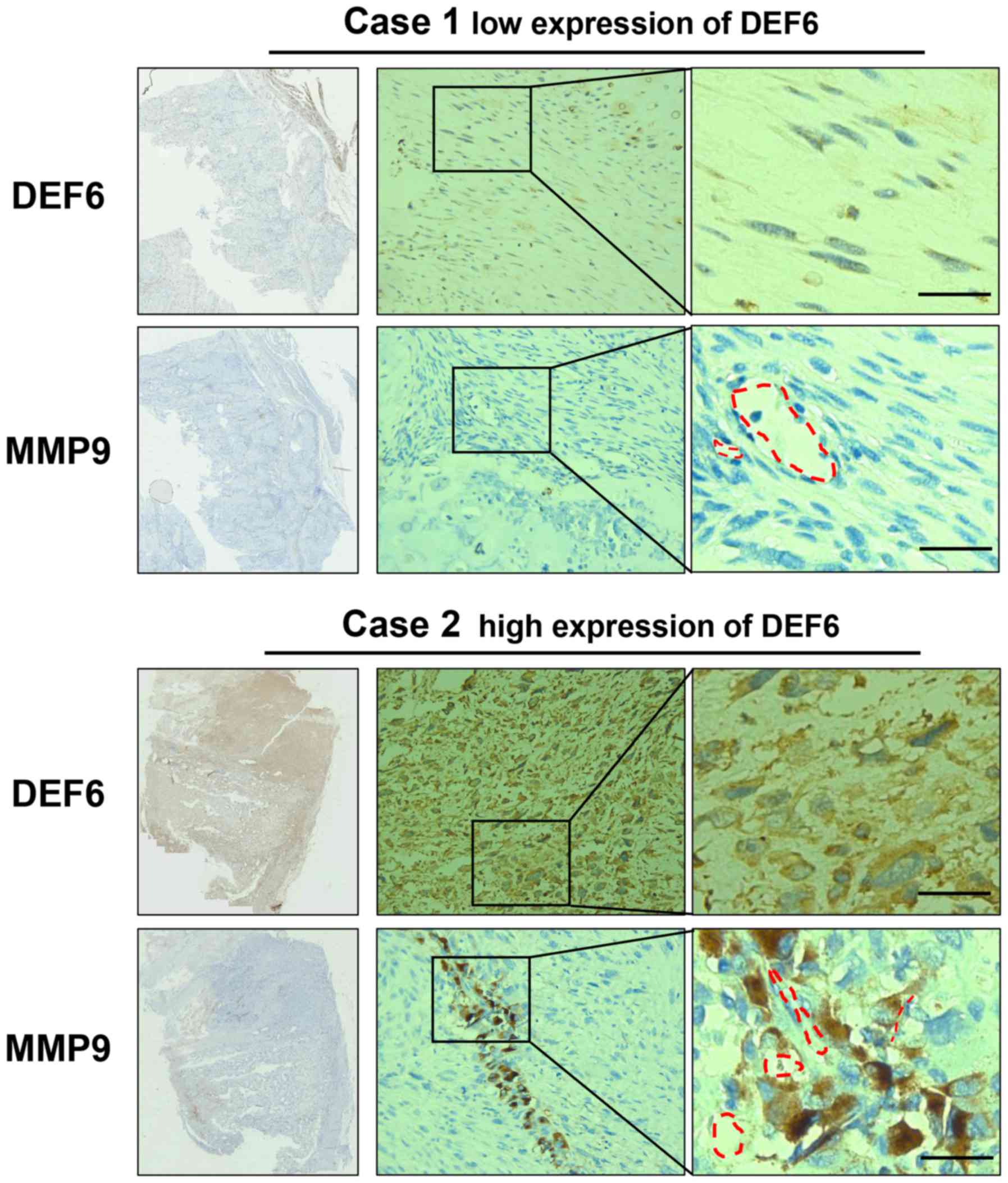
A total of 58 human OS tissue samples were analyzed
to further substantiate the association between MMP9 and DEF6
expression in vivo. Fig. 6
demonstrated the results of positive or negative IHC staining of
DEF6 and corresponding MMP9 levels. A higher positive staining rate
(61.76%) was detected for MMP9 in patients with OS exhibiting high
DEF6 levels compared with patients with OS exhibiting low DEF6
levels (25.00%; P<0.05). Notably, MMP9-positive OS cells were
demonstrated to populate the perivascular space instead of
throughout the tissue in OS cases with high DEF6 expression, which
may indicate a pre-invasive condition of OS cells (Fig. 6). Further analysis demonstrated that
DEF6 expression was positively correlated with MMP9 expression
(Table II). The IHC and in
vitro results of the current study indicated that MMP9 serves a
crucial role in DEF6-promoted metastasis in OS.
 | Table II.Correlation between DEF6 and MMP9
expression in human osteosarcoma. |
Table II.
Correlation between DEF6 and MMP9
expression in human osteosarcoma.
|
| MMP9
expression |
|---|
|
|
|
|---|
| DEF6
expression | Positive | Negative |
|---|
| High | 21 | 13 |
| Low | 6 | 18 |
| P-value | 0.006a |
|
Discussion
DEF6, also known as IBP, is a pivotal driver
responsible for regulating immune cell biology and autoimmunity,
and has been frequently been reported to be amplified in several
types of cancer (8,11–23). The
results of the current study determined whether DEF6 is a novel
oncogene in OS. Analysis of 58 patients with OS revealed that DEF6
expression was associated with metastasis and worse clinical stage
in OS. Furthermore, high DEF6 levels were positively associated
with poor prognosis. DEF6 knockdown demonstrated that metastasis
was significantly suppressed in vitro in MTF and MMNG/HOS
cells; however, knockdown did not demonstrate a significant
difference in cell proliferation. Furthermore, DEF6 expression was
positively correlated with MMP9 expression in OS cells and human OS
tissues. These data provided preliminary evidence that DEF6 is a
potential anti-metastatic target for OS.
Due to the clinical importance of metastasis, a
major cause of therapeutic failure in patients with OS (7), early prediction and inhibition of OS
metastasis is an appealing curative approach. However, a robust
biomarker for predicting metastasis and prognosis in patients with
OS does not exist (37).
Accumulating evidence suggests that in various types of cancer,
such as ovarian carcinoma and colorectal cancer, DEF6 is associated
with disease progression and poor prognosis (12,17–20). In
the present study, the results demonstrated that DEF6 was widely
expressed in human OS tissues and that its accumulation was
positively correlated with advanced Enneking stage and distant
metastases. These results were consistent with those of previous
studies (20,22). Moreover, DEF6 was validated to be a
potential negative prognostic marker in patients with OS and this
result was similar to those of previous studies in ovarian cancer
and colorectal cancer (18,20). To the best of our knowledge, the
current study is the first to report that increased DEF6 expression
may be an indicator of tumor progression and poor prognosis in
patients with OS.
DEF6 has been previously reported to serve a role in
cancer cell proliferation (19,22).
However, the results of current study did not report a significant
difference in OS cell proliferation following the knockdown of DEF6
expression via siRNA transfection. Bioinformatics analyses further
supported the observation that DEF6 has a negligible impact on OS
cell death or growth. Differences in observations between the
current and previous studies may be attributed to heterogeneity in
different types of cancers.
DEF6 is reportedly an upstream activator of
Rho-family GTPases (14,38). In breast cancer cells, DEF6 activates
Ras-related C3 botulinum toxin substrate 1 (Rac1), Ras homolog
family member A and cell division control protein 42 homolog to
promote filipodium and lamellipodia formation, cell migration and
MMP secretion (12). Moreover, DEF6
selectively binds to Rac1 to regulate cell morphology (14). These data indicate that DEF6 serves a
key role in cancer metastases (22,39).
Consistent with these results, the current study demonstrated that
DEF6 significantly promoted motility and MMP9 expression in OS
cells. Furthermore, a positive correlation was observed between
DEF6 and MMP9 expression in human OS tissues. The results also
demonstrated that tumor tissues with a high expression of DEF6,
which may indicate a high risk of OS metastasis, did not contain
MMP9-positive OS cells throughout. Instead, MMP9 cells populated
the perivascular space. Tumor angiogenesis, one of the most common
routes for OS dissemination, has also been demonstrated to promote
OS metastasis within the cancer microenvironment (25,40).
Thus, the presence of MMP9-positive OS cells around tumor vessels
may represent invasion and dissemination through the vessels, which
may eventually develop into distant metastasis.
As Rho-family GTPases also reportedly participates
in regulating cell migration, cell invasion and actin stress fiber
formation in OS cells (3,9). There is a strong possibility that their
activation is the potential underlying mechanism through which DEF6
controls OS metastasis. Furthermore, in addition to being a
regulator of Rho-family GTPases, DEF6 is reportedly involved in
mTOR and Wnt signaling pathway regulation, and DEF6 expression has
been directly associated with p53 activation (22,41–43).
However, in the current study, whether DEF6 significantly promotes
motility in OS cells in vivo and the mechanism via which
DEF6 serves a role in OS cell migration and invasion could not be
determined; thus, further studies are warranted.
In summary, the current study revealed a new
function of DEF6 as an oncogene in OS progression. Upregulated
expression of DEF6 may contribute to distant metastasis and poor
prognosis in patients with OS, making DEF6 a potential oncogene of
OS. Furthermore, the results demonstrated that MMP9 is a potential
downstream target of DEF6 and facilitates OS cell invasion.
Furthermore, MMP9 expression was positively correlated with DEF6 in
OS tissues. Further studies should aim to explore the mechanism by
which DEF6 regulates OS metastasis.
Acknowledgements
The authors of the current study would like to thank
Professor Chuanmin Hu and his research group from the Army Medical
University for supplying mouse anti-human DEF6 antibodies. The
authors would also like to thank Professor Qing Yin from the
Xinqiao Hospital of Army Medical University (Chongqing, China) for
her technical and theoretical support, and Mr Jia-Shen Ye and Ms
Ya-Li Wang from the Xinqiao Hospital of Army Medical University
(Chongqing, China) for their assistance with immunohistochemistry
techniques.
Funding
The current study was supported by the National
Natural Science Foundation of China (grant nos. 81972519 and
81672653).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ and QG designed the experiments. QZ and GZ
conducted the experiments, collected and analyzed the data, and
drafted the initial manuscript. YC performed cell culture and some
experiments. XT and LL contributed to data collection of human
specimens. GZ and QT performed IHC staining analyses. QG provided
financial support, administrative support and final approval of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All human tissues experiments were approved by the
Institutional Ethics Committee of Xinqiao Hospital and Army Medical
University, Chongqing, China (approval nos. 27-2011 and
2018-069-01). Written informed consent for the experimental studies
was obtained from the patients or their guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment - where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Landis SH, Murray T, Bolden S and Wingo
PA: Cancer statistics, 1998. CA Cancer J Clin. 48:6–29. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Zhang L, Qu R and Huang W: Rho A
regulates epidermal growth factor-induced human osteosarcoma MG63
cell migration. Int J Mol Sci. 19:14372018. View Article : Google Scholar
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao GS, Zhang Q, Cao Y, Wang Y, Lv YF,
Zhang ZS, Zhang Y, Tan QL, Chang Y, Quan XZ, et al: High expression
of ID1 facilitates metastasis in human osteosarcoma by regulating
the sensitivity of anoikis via PI3K/AKT depended suppression of the
intrinsic apoptotic signaling pathway. Am J Transl Res.
11:2117–2139. 2019.PubMed/NCBI
|
|
6
|
Briccoli A, Rocca M, Salone M, Guzzardella
GA, Balladelli A and Bacci G: High grade osteosarcoma of the
extremities metastatic to the lung: Long-term results in 323
patients treated combining surgery and chemotherapy, 1985–2005.
Surg Oncol. 19:193–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao GS, Gao ZR, Zhang Q, Tang XF, Lv YF,
Zhang ZS, Zhang Y, Tan QL, Peng DB, Jiang DM and Guo QN: TSSC3
promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR
pathway to suppress tumorigenesis and metastasis in osteosarcoma,
and predicts a favorable prognosis. J Exp Clin Cancer Res.
37:1882018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu JL, Li J, Xu JJ, Xiao F, Cui PL, Qiao
ZG, Chen XD, Tao WD and Zhang XL: MiR-144 Inhibits Tumor Growth and
Metastasis in Osteosarcoma via Dual-suppressing RhoA/ROCK1
Signaling Pathway. Mol Pharmacol. 95:451–461. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin H, Hao Y, Zhao Z and Tong Y: Sirtuin 6
contributes to migration and invasion of osteosarcoma cells via the
ERK1/2/MMP9 pathway. FEBS Open Bio. 7:1291–1301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hotfilder M, Baxendale S, Cross MA and
Sablitzky F: Def-2, −3, −6 and −8, novel mouse genes differentially
expressed in the haemopoietic system. Br J Haematol. 106:335–344.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Z, Yang M, Chen R, Su W, Li P, Chen
S, Chen Z, Chen A, Li S and Hu C: IBP regulates
epithelial-to-mesenchymal transition and the motility of breast
cancer cells via Rac1, RhoA and Cdc42 signaling pathways. Oncogene.
33:3374–3382. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Manni M, Ricker E and Pernis AB:
Regulation of systemic autoimmunity and CD11c(+) Tbet(+) B cells by
SWEF proteins. Cell Immunol. 321:46–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oka T, Ihara S and Fukui Y: Cooperation of
DEF6 with activated Rac in regulating cell morphology. J Biol Chem.
282:2011–2018. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Joshi RN, Binai NA, Marabita F, Sui Z,
Altman A, Heck AJ, Tegnér J and Schmidt A: Phosphoproteomics
reveals regulatory T cell-mediated DEF6 dephosphorylation that
affects cytokine expression in human conventional T cells. Front
Immunol. 8:11632017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Binder N, Miller C, Yoshida M, Inoue K,
Nakano S, Hu X, Ivashkiv LB, Schett G, Pernis A, Goldring SR, et
al: Def6 restrains osteoclastogenesis and inflammatory bone
resorption. J Immunol. 198:3436–3447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Subramanian S, West RB, Marinelli RJ,
Nielsen TO, Rubin BP, Goldblum JR, Patel RM, Zhu S, Montgomery K,
Ng TL, et al: The gene expression profile of extraskeletal myxoid
chondrosarcoma. J Pathol. 206:433–444. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liew PL, Fang CY, Lee YC, Chen CL and Chu
JS: DEF6 expression in ovarian carcinoma correlates with poor
patient survival. Diagn Pathol. 11:682016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jian CX, Yang MZ, Li P, Xiong J, Zhang ZJ,
Li CJ, Chen A, Hu CM, Zhou JX and Li SH: Ectopically expressed IBP
promotes cell proliferation in oral squamous cell carcinoma. Cancer
Invest. 30:748–756. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Z, Wang Q, Li P, Zhou Y, Li S, Yi W,
Chen A, Kong P and Hu C: Overexpression of the Interferon
regulatory factor 4-binding protein in human colorectal cancer and
its clinical significance. Cancer Epidemiol. 33:130–136. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li P, Zhang Z, Wang Q, Li S, Zhang Y, Bian
X, Chen A and Hu C: The ectopic expression of IFN regulatory factor
4-binding protein is correlated with the malignant behavior of
human breast cancer cells. Int Immunopharmacol. 9:1002–1009. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen S, Han Q, Wang X, Yang M, Zhang Z, Li
P, Chen A, Hu C and Li S: IBP-mediated suppression of autophagy
promotes growth and metastasis of breast cancer cells via
activating mTORC2/Akt/FOXO3a signaling pathway. Cell Death Dis.
4:e8422013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Enneking WF, Spanier SS and Goodman MA: A
system for the surgical staging of musculoskeletal sarcoma. 1980.
Clin Orthop Relat Res. 4–18. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Meng G and Guo QN: Changes in
genomic imprinting and gene expression associated with
transformation in a model of human osteosarcoma. Exp Mol Pathol.
84:234–239. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Z, Zhao GS, Lv Y, Peng D, Tang X, Song
H and Guo QN: Anoikisresistant human osteosarcoma cells display
significant angiogenesis by activating the Src kinasemediated MAPK
pathway. Oncol Rep. 41:235–245. 2019.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dai H, Huang Y, Li Y, Meng G, Wang Y and
Guo QN: TSSC3 overexpression associates with growth inhibition,
apoptosis induction and enhances chemotherapeutic effects in human
osteosarcoma. Carcinogenesis. 33:30–40. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng G, Lv Y, Dai H, Zhang X and Guo QN:
Epigenetic silencing of methyl-CpG-binding protein 2 gene affects
proliferation, invasion, migration, and apoptosis of human
osteosarcoma cells. Tumour Biol. 35:11819–11827. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lv YF, Yan GN, Meng G, Zhang X and Guo QN:
Enhancer of zeste homolog 2 silencing inhibits tumor growth and
lung metastasis in osteosarcoma. Sci Rep. 5:129992015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo J, Liu Q, Li Z, Guo H, Bai C and Wang
F: miR-222-3p promotes osteosarcoma cell migration and invasion
through targeting TIMP3. Onco Targets Ther. 11:8643–8653. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kunz P, Sähr H, Lehner B, Fischer C,
Seebach E and Fellenberg J: Elevated ratio of MMP2/MMP9 activity is
associated with poor response to chemotherapy in osteosarcoma. BMC
Cancer. 16:2232016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li H, Cui J, Xu B, He S, Yang H and Liu L:
Long non-coding RNA XIST serves an oncogenic role in osteosarcoma
by sponging miR-137. Exp Ther Med. 17:730–738. 2019.PubMed/NCBI
|
|
33
|
Waresijiang N, Sun J, Abuduaini R, Jiang
T, Zhou W and Yuan H: The downregulation of miR125a5p functions as
a tumor suppressor by directly targeting MMP11 in osteosarcoma. Mol
Med Rep. 13:4859–4864. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hirahata M, Osaki M, Kanda Y, Sugimoto Y,
Yoshioka Y, Kosaka N, Takeshita F, Fujiwara F, Kawai A, Ito H, et
al: PAI-1, a target gene of miR-143, regulates invasion and
metastasis by upregulating MMP-13 expression of human osteosarcoma.
Cancer Med. 5:892–902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fan H, Lu S, Wang S and Zhang S:
Identification of critical genes associated with human osteosarcoma
metastasis based on integrated gene expression profiling. Mol Med
Rep. 20:915–930. 2019.PubMed/NCBI
|
|
36
|
Su Y, Wan D and Song W: Dryofragin
inhibits the migration and invasion of human osteosarcoma U2OS
cells by suppressing MMP-2/9 and elevating TIMP-1/2 through
PI3K/AKT and p38 MAPK signaling pathways. Anticancer Drugs.
27:660–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roberts RD, Lizardo MM, Reed DR, Hingorani
P, Glover J, Allen-Rhoades W, Fan T, Khanna C, Sweet-Cordero EA,
Cash T, et al: Provocative questions in osteosarcoma basic and
translational biology: A report from the children's oncology group.
Cancer. 125:3514–3525. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Q, Gupta S and Pernis AB: Regulation
of TLR4-mediated signaling by IBP/Def6, a novel activator of Rho
GTPases. J Leukoc Biol. 85:539–543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu Y, Hou Y, Liu T and Lou G:
Overexpression and clinical significance of IBP in epithelial
ovarian carcinoma. Oncol Lett. 15:6604–6610. 2018.PubMed/NCBI
|
|
40
|
Wang SW, Liu SC, Sun HL, Huang TY, Chan
CH, Yang CY, Yeh HI, Huang YL, Chou WY, Lin YM and Tang CH:
CCL5/CCR5 axis induces vascular endothelial growth factor-mediated
tumor angiogenesis in human osteosarcoma microenvironment.
Carcinogenesis. 36:104–114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu X, Shuen WH, Chen C, Goudevenou K,
Jones P and Sablitzky F: Swap70b is required for convergent and
extension cell movement during zebrafish gastrulation linking Wnt11
signalling and RhoA effector function. Dev Biol. 386:191–203. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Goudevenou K, Martin P, Yeh YJ, Jones P
and Sablitzky F: Def6 is required for convergent extension
movements during zebrafish gastrulation downstream of Wnt5b
signaling. PLoS One. 6:e265482011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang M, Yuan F, Li P, Chen A, Li S and Hu
C: Interferon regulatory factor 4 binding protein is a novel p53
target gene and suppresses cisplatin-induced apoptosis of breast
cancer cells. Mol Cancer. 11:542012. View Article : Google Scholar : PubMed/NCBI
|