Introduction
Esophageal cancer is considered to be one of the
most malignant tumors, with a high mortality rate that ranks sixth
worldwide. Esophageal cancer is categorized into two subtypes: i)
Esophageal squamous cell carcinoma (ESCC), which accounts for ~90%
of all cases; and ii) adenocarcinoma (EADC), which accounts for the
remaining ~10% of cases (1,2). ESCC is a common malignant
gastrointestinal tumor with a high incidence rate in certain rural
areas of China, for example in Linxian, Cixian, Shexian and Yanting
the incidence rates of esophageal cancer (per 100,000 population)
for 2011 were 83.8, 91.5, 64.2 and 99.0, respectively, and is
characterized by a unique geographical distribution along the
Taihang Mountains (3,4). The risk factors of ESCC include
nutritional deficiency, and the consumption of betel quid, tobacco,
pickled vegetables and hot food and drinks (2,5). With
changes in socioeconomic status, living conditions and eating
habits, the incidence of esophageal cancer has declined, but the
situation remains severe (3,6).
Surgical resection is the first choice for the
treatment of early ESCC. However, the majority of patients have
already developed advanced ESCC before symptoms become apparent,
which is usually indicated by difficulties in swallowing. At this
point, radiotherapy is considered to be the primary treatment
option, although the outcome is often unsatisfactory; following
radiotherapy alone, the 5-year survival rate is ~10% due to poor
local control and distant metastasis (7), and as such, the development of novel
anticancer strategies is highly warranted.
Telomerase maintains telomere length by adding
TTAGGG hexamers and inhibiting cellular senescence (8,9).
Telomerase has been reported to be closely associated with cancer,
serving crucial roles in tumor growth and progression, in part
through the maintenance of telomere structure (10). Telomerase reactivation is an
essential step in malignant tumor progression, and almost 90% of
cancer in humans exhibit telomerase activity (11). Therefore, the expression and activity
of telomerase are indispensable for tumor formation (12).
Telomerase consists of telomerase RNA (hTR) and
telomerase reverse transcriptase (hTERT). hTERT is the core
catalytic subunit of telomerase, and has historically been
considered to play an important role in telomere length maintenance
(13,14). hTERT is distinguishable from other
reverse transcriptases by its unique mode of action, which promotes
template realignment to enable continued synthesis of multiple DNA
sequence repeats (15). hTERT
transcription induces telomerase activity, which is critical for
cellular proliferation, differentiation and senescence (16,17).
Moreover, hTERT is expressed in proliferative cells such as germ
and stem cells, as well as in tumors, but is not found in the
majority of somatic cells (16,17). The
hTERT promoter is located within a dense CG-rich CpG-island,
indicating a role for methylation in the regulation of hTERT
expression (18). hTERT promoter
hypermethylation has been observed in numerous cancer cell types
(compared with normal, noncancerous cells), which correlates with
hTERT expression, especially in epithelial tumors (19). Furthermore, hypermethylation of the
human telomerase catalytic subunit (hTERT) gene correlates with
telomerase activity (20). hTERT has
also been noted as enzymatically active in different cancer types,
and serves a crucial role in tumorigenesis and progression. As well
as maintaining telomere length, hTERT was found to be associated
with migration and invasion in various cancer cell types (21). hTERT expression is also the
rate-limiting factor for telomerase activity, and increased
telomerase activity can be blocked by inhibiting hTERT (22). Genome-wide studies of clinical
samples have also highlighted the importance of hTERT in numerous
malignancies (23). However,
previous studies have largely focused on the enzymatic activity of
hTERT, rather than its other biological functions (24). Studies in which telomerase was
inhibited by genetic, antisense and pharmacological strategies
indicate that telomerase, especially hTERT, is an ideal target for
cancer therapy (25).
Metastasis is the leading cause of death in patients
with esophageal cancer, and the liver, lung and bones are the most
common sites of metastasis (26).
Tumor metastasis involves a series of complex processes, including
cellular migration. Abnormal regulation of cellular migration
results in tumor cell invasion and metastasis, which enables tumor
cell escape from the primary site, invasion into lymphatic and
blood vessels, and ultimately, colonization at distant sites
(27). Cellular migration is a
dynamic, multi-step process (28).
Key molecules in cancer cell migration are of great importance for
tumor metastasis, and may therefore serve as potential targets for
cancer treatment. To date, a large number of studies have reported
the role of hTERT in malignancies such as urological tumors
(29–32), melanomas (30,33,34),
gastric cancer (35–37), gliomas (38–41) and
hepatocellular tumors (42).
However, few studies have reported the association between hTERT
expression and the migration and invasion of ESCC cells (43,44).
In the present study, hTERT expression was analyzed
in ESCC tissues, and its association with specific
clinicopathological characteristics was determined. Moreover, hTERT
knockdown using RNA interference (RNAi) methods resulted in
decreased cellular migration and invasiveness. It was therefore
concluded that targeting hTERT may highlight novel approaches for
the treatment of ESCC.
Materials and methods
Patients and samples
The present study included 100 patients with ESCC,
of which 75 also donated paired paracancerous tissues (located
<3 cm away from the cancer tissue). The mean age of the patients
was 65.29 years (range, 48–82 years), including 74 males and 26
females. All the samples in the tissue microarrays (TMAs) were
purchased (Shanghai Outdo Biotech Co., Ltd.) and the use of the
TMAs for research purposes was approved by the Institutional
Research Ethics Committee, The Second Hospital of Nanjing (approval
no. 2018-LY-KY068). Specimens from patients with incomplete
clinical data were not included in the statistical analysis.
Immunohistochemistry analysis
The expression of Ki67 (proliferation
cell-associated nuclear antigen in tumor tissues), p53 (tumor
suppressor) and hTERT was assessed by immunohistochemistry. TMAs
were embedded with paraffin at 63°C for 1 h, deparaffinized in
xylene at room temperature for 30 min and then rehydrated in
absolute ethanol for 14 min (solvent refreshed at 7 min), then
rehydrated in 90, 80 and 70 ethanol (7 min each), and finally
placed in distilled water for 9 min (solvent refreshed every 3
min). The TMAs were then immersed in boiling sodium citrate buffer
for 5 min and left to cool at room temperature. Following
incubation in 10% BSA (Sangon Biotech Co., Ltd.) for 1 h at room
temperature, the TMAs were incubated with primary antibodies
specific to hTERT (rabbit anti-human TERT, polyclonal; 1:100; cat.
no. ab183105; Abcam), p53 (rabbit anti-human, polyclonal; 1:100;
cat. no. 9282; Cell Signaling Technology, Inc.) and Ki-67 (mouse
anti-human, monoclonal; 1:100; cat. no. sc-23900; Santa Cruz
Biotechnology, Inc.) at 4°C overnight; this was followed by a
further incubation with goat anti-mouse IgG (H+L) (1:1,000; cat.
no. SA00001-1; ProteinTech Group, Inc.) or goat anti-rabbit IgG
(H+L) (1:1,000; cat. no. SA00001-2; ProteinTech Group, Inc.)
secondary antibodies at 37°C for 20 min. The sections were stained
at room temperature with DAB for 5 min and counterstained with
hematoxylin and eosin (H&E) for 2 min at room temperature. The
TMAs were then dehydrated and dried, and subsequently mounted using
neutral gum. The TMAs were scanned using an Aperio ScanScope system
(Leica Microsystems, Inc.), and staining was analyzed using Aperio
Imagescope version 12.4.0.5043 (Aperio Technologies, Inc.). The
results were verified by a senior pathologist who was blinded to
the clinicopathological data of the patients. Three fields with
different staining intensities were analyzed at ×20 magnification.
Approximately 100 cells in each field of view were analyzed and the
percentage of positive cells in nucleus and cytoplasm were
calculated manually. The final staining positive rate of the tissue
point was the average number of three fields.
Cell culture and transfection
Kyse410 and Kyse520 ESCC cell lines were purchased
from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH,
and cultured in RPMI-1640 with 10% fetal bovine serum, 100 µg/ml
streptomycin and 100 U/ml penicillin (all Gibco; Thermo Fisher
Scientific, Inc.) at 37°C (5% CO2). The sequences of the
negative control (NC) and hTERT siRNAs (Guangzhou RiboBio Co.,
Ltd.) were as follows: NC forward, 5′-UUCUCCGAACGUGUCACGUdTdT-3′;
NC reverse, 5′-ACGUGACACGUUCGGAGAAdTdT-3′; anti-hTERT forward,
5′-GCGACGACGUGCUGGUUCAdTdT-3′; and anti-hTERT reverse,
5′-dTdTCGCUGCUGCACGACCAAGU-3′. On the day before transfection,
5×105 cells were seeded into 6-well plates without
antibiotics and cultured until 70% confluence was achieved. The NC
or hTERT siRNA (25 nmol/l) and Lipofectamine® RNAiMAX
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) were diluted
in Opti-MEM® Reduced Serum Medium (Gibco; Thermo Fisher
Scientific, Inc.) and gently mixed; both reagents were then
combined, mixed gently and incubated for 10–20 min at room
temperature. The complexes were added to each well of the 6-well
plates, and incubated at 37°C for the times indicated in each
subsequent experimental section.
Western blot analysis
Following transfection for 48 h, Kyse410 and Kyse520
cells were harvested and lysed with RIPA buffer (KeyGEN BioTECH);
protein concentration was quantified using a bicinchoninic acid
protein assay kit (Nanjing KeyGen Biotech Co., Ltd.). The samples
were boiled with 5X loading buffer, and 20-µg aliquots of total
protein were separated using 8% SDS-PAGE gels, prior to transfer
onto PVDF membranes (EMD Millipore). The membranes were blocked at
room temperature in 5% nonfat milk for 2 h and incubated with GAPDH
(1:500; cat.no. GB11002; Wuhan Servicebio Technology Co., Ltd.) or
hTERT (rabbit anti-human TERT, polyclonal; 1:1,000; cat. no.
ab183105; Abcam) primary antibodies overnight at 4°C. The membranes
were than washed, and subsequently incubated with goat anti-rabbit
IgG (H+L) secondary antibody (1:10,000; cat. no. SA00001-2;
ProteinTech Group, Inc.) for 1 h at room temperature. Proteins were
visualized using the FluorChem M System (ProteinSimple).
MTT assays
The effects of hTERT knockdown on cell proliferation
were detected using an MTT Cell Proliferation and Cytotoxicity
Assay Kit (Beijing Leagene Biotechnology Co., Ltd.). The cells were
separated into three groups: i) Untreated; ii) NC; and iii) hTERT
siRNA. Kyse410 and Kyse520 cells were seeded into 96-well plates
(2×103 cells/well) 12 h post-transfection, and left to
adhere for a further 6 h. Following 0-, 24-, 48- and 72-h
incubation periods at 37°C, 10 µl MTT solution was added to each
well and the cells were incubated for an additional 4 h. The
culture supernatants were carefully removed and 110 µl formazan
solvent was added per well to dissolve the blue–purple formazan
crystals. Absorbance was detected at 570 nm using a
spectrophotometer (SN.1510-05687; Thermo Fisher scientific,
Inc.).
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed to analyze the effect
of hTERT knockdown on cellular proliferation, according the
manufacturer's protocol (Dojindo Molecular Technologies, Inc.).
Cells were seeded into 96-well plates at a density of
2×103 cells/well (n=5). After incubation for 24, 48 and
72 h, 10 µl CCK-8 reagent was added to each well and incubated at
37°C (5% CO2) for 3 h. Absorbance was detected at 450 nm
using the aforementioned spectrophotometer.
Clonogenic assay
To assess clonogenic cell survival, Kyse410 and
Kyse520 cells were harvested with 0.25% trypsin (Gibco; Thermo
Fisher Scientific, Inc.) and seeded into 3.5-cm tissue culture
dishes at various cell densities. The cells were incubated at 37°C
for 7–12 days, fixed with 95% ethanol and stained using
hematoxylin. Clonogenic cells were defined as those able to form a
colony of ≥50 cells. Colony images were captured using a commercial
digital camera (PowerShot S110; Canon, Inc), and the colonies from
four independent replicates were counted using Adobe Photoshop CS5
software version 12.01 (Adobe Systems, Inc.).
Invasion assays
For the invasion assays (12 h post-transfection),
1×105 cells were resuspended in 100 µl RPMI-1640 medium
(without serum) and seeded onto the upper surfaces of 24-well
Transwell filter inserts (pore diameter, 8 µm; Corning Inc.); the
inserts were pre-coated with Matrigel (BD Biosciences). The lower
chamber was loaded with 600 µl RPMI-1640 medium supplemented with
10% FBS as a chemoattractant; each sample was assessed in
triplicate. After 48 h, cells in the upper chamber were removed
with wet cotton swab. The cells in the lower chamber were fixed
with 99.99% methanol at 4°C for 30 min, stained with 0.1% crystal
violet at 37°C for 15 min (Invitrogen; Thermo Fisher Scientific,
Inc.), and observed and images were captured under an inverted
phase contrast microscope (magnification, ×100) (Olympus
Corporation).
Wound healing assay
Following 24 h of transfection, a straight-line
wound was made in each cell monolayer using a 100-µl pipette tip.
Detached cells were gently removed with PBS. Each well was
replenished with RPMI-1640, and then observed using a phase
contrast microscope (magnification, ×40) and images were captured
at 0 and 24-h time intervals.
Statistical analysis
Data analysis was performed using SPSS version 25
(IBM Corp.). The χ2 test was used to determine
significant differences between the frequency of different
categories. Fisher's exact test was used to assess if high
hTERT-expression rate in ESCC tissues and paracancerous tissues
different. ANOVA followed by Fisher's LSD post hoc test was used to
assess whether the mean difference between groups was significant.
The Kaplan-Meier method and log-rank test were used to calculate
survival rate, and Spearman's rank correlation was used to assess
the degree of association between two variables. P<0.05 was
considered to indicate a statistically significantly
difference.
Results
HTERT is highly expressed in tumors
and is associated with patient survival
The present study included 100 patients with ESCC
(74 males and 26 females). The assessed patient clinicopathological
characteristics included sex, age, tumor size, pathological grade
and tumor stage (Table I). The
incidence of ESCC among males (74%) was higher compared with that
of females (26%), which corresponds with the findings of a previous
study (69 and 31%, respectively) (2). Patient follow-up revealed that
following surgery, the survival time of females was significantly
higher compared with that of males (P=0.024; Fig. 1B), and that the combined 5-year
survival rate was 20%, consistent with data the National Cancer
Institute (NIH) (5-30%; http://www.cancer.gov/types/esophageal/hp/esophageal-treatment-pdq).
There was no significant difference in the survival time of
patients with ESCC among different age groups and pathological
grades (Fig. 1A and D). However,
significant differences were observed in survival times according
to different tumor sizes, T stages, N stages and TNM stages of
patients with ESCC (Fig. 1C and
E-G).
 | Figure 1.Survival times of patients with
esophageal squamous cell carcinoma according to different
clinicopathological characteristics. Survival curves according to
(A) age, (B) sex, (C) tumor size, (D) pathological grade, (E)
T-staging, (F) N-staging and (G) TNM-staging. TNM,
Tumor-Node-Metastasis; T, primary tumor; N, regional lymph nodes;
M, distant metastasis; Cum, cumulative. |
 | Table I.Clinicopathological characteristics
of patients with esophageal squamous carcinoma. |
Table I.
Clinicopathological characteristics
of patients with esophageal squamous carcinoma.
| Clinicopathological
features | Patients, n | Censored patients,
n | P-value |
|---|
| Sex |
|
| 0.024 |
|
Male | 74 | 9 |
|
|
Female | 26 | 10 |
|
|
Total | 100 | 19 |
|
| Age, years |
|
| 0.723 |
|
≤65 | 51 | 8 |
|
|
>65 | 49 | 11 |
|
|
Total | 100 | 19 |
|
| Tumor size, cm |
|
| 0.021 |
| ≤5 | 56 | 14 |
|
|
>5 | 29 | 4 |
|
|
Total | 85 | 18 |
|
| Pathological
grade |
|
| 0.631 |
| I | 6 | 2 |
|
| II | 66 | 12 |
|
|
III | 28 | 5 |
|
|
Total | 94 | 19 |
|
| T-stage |
|
| 0.026 |
| T0 | 4 | 3 |
|
| T1 | 11 | 4 |
|
| T2 | 79 | 11 |
|
| T3 | 3 | 0 |
|
|
Total | 97 | 18 |
|
| N-stage |
|
| 0.001 |
| N0 | 45 | 13 |
|
| N1 | 31 | 5 |
|
| N2 | 17 | 0 |
|
| N3 | 5 | 0 |
|
|
Total | 98 | 18 |
|
| TNM-stage |
|
| <0.001 |
|
TNM1 | 4 | 3 |
|
|
TNM2 | 42 | 12 |
|
|
TNM3 | 50 | 2 |
|
|
Total | 96 | 17 |
|
Pathological changes in esophageal carcinoma were
evaluated by H&E staining. hTERT staining is exhibited as a
brown color (Fig. 2A-C). Ki67 and
p53 were also assessed using immunohistochemistry. hTERT expression
level of ≤90% was defined as low expression, and >90% was
defined as high expression. No significant differences were
observed in the proportions of high nuclear hTERT expression
between esophageal carcinoma and paracancerous tissues (85 and 84%
respectively; Fig. 2A). However, a
significant difference was observed in the proportions of high
hTERT expression in the cytoplasm between these tissue types (66
and 6.7% respectively; P<0.05; Table
II, Fig. 2A). The follow-up
revealed no significant differences between the survival times of
patients in different hTERT expression groups (Fig. 3B-D). However, the P-value between the
survival time of patients with different cytoplasmic hTERT
expression levels was 0.061, which is close to 0.05, indicting a
potential association between hTERT expression and poor prognosis
in patients with ESCC (Fig. 3A).
 | Table II.hTERT expression in 100 esophageal
squamous carcinoma tissues and 75 paracancerous tissues. |
Table II.
hTERT expression in 100 esophageal
squamous carcinoma tissues and 75 paracancerous tissues.
|
| Expression, n
(%) |
|
|---|
|
|
|
|
|---|
| Tissue type | High | Low | P-value |
|---|
| Cancer tissues | 67 (67) | 33 (33) | <0.0001 |
| Paracancerous
tissues | 5 (6.7) | 70 (93.3) |
|
Considering the increased hTERT expression in the
cytoplasm of cancer tissue cells, the association between positive
cytoplasmic hTERT expression and clinical data are presented in
Table III. The hTERT-positive rate
in the cytoplasm of esophageal carcinoma cells was significantly
correlated with pathological grade (r=0.243, P=0.015), N stage
(r=0.290, P=0.004) and TNM stage (r=0.298, P=0.003), but not with
age, sex or tumor size.
 | Table III.Correlation of the hTERT-positive
rate in the cytoplasm of esophageal squamous carcinoma cells with
clinical parameters. |
Table III.
Correlation of the hTERT-positive
rate in the cytoplasm of esophageal squamous carcinoma cells with
clinical parameters.
| Clinical
parameter | N | ρ | P-value |
|---|
| Age | 100 | 0.051 | 0.612 |
| Sex | 100 | −0.056 | 0.577 |
| Tumor size | 85 | 0.014 | 0.896 |
| Pathological
grade | 100 | 0.243a | 0.015 |
| N stage | 98 | 0.290b | 0.004 |
| TNM stage | 96 | 0.298b | 0.003 |
hTERT knockdown does not significantly
inhibit Kyse410 or Kyse520 cell proliferation
To investigate the mechanism by which high hTERT
expression correlates with the poor prognosis of patients with
esophageal carcinoma, hTERT knockdown was performed in Kyse410 and
Kyse520 esophageal carcinoma cell lines using RNAi methods. Western
blotting was used to confirm successful knockdown in both cell
lines (Fig. 4A and B). The effect of
hTERT knockdown on proliferation was analyzed using MTT assays
(Fig. 4C and D). At 48 and 72 h, the
proliferation of hTERT-knockdown Kyse410 and Kyse520 cells was
slightly lower compared with that of the control cells, however no
significant differences were observed. The results of the CCK-8
assays (Fig. 4E and F) were
consistent with those of the MTT assays.
hTERT knockdown inhibits Kyse410 cell
colony formation
To investigate whether hTERT expression influences
the colony-formation ability of esophageal carcinoma cells, hTERT
expression was knocked down in Kyse410 and Kyse520 cells using
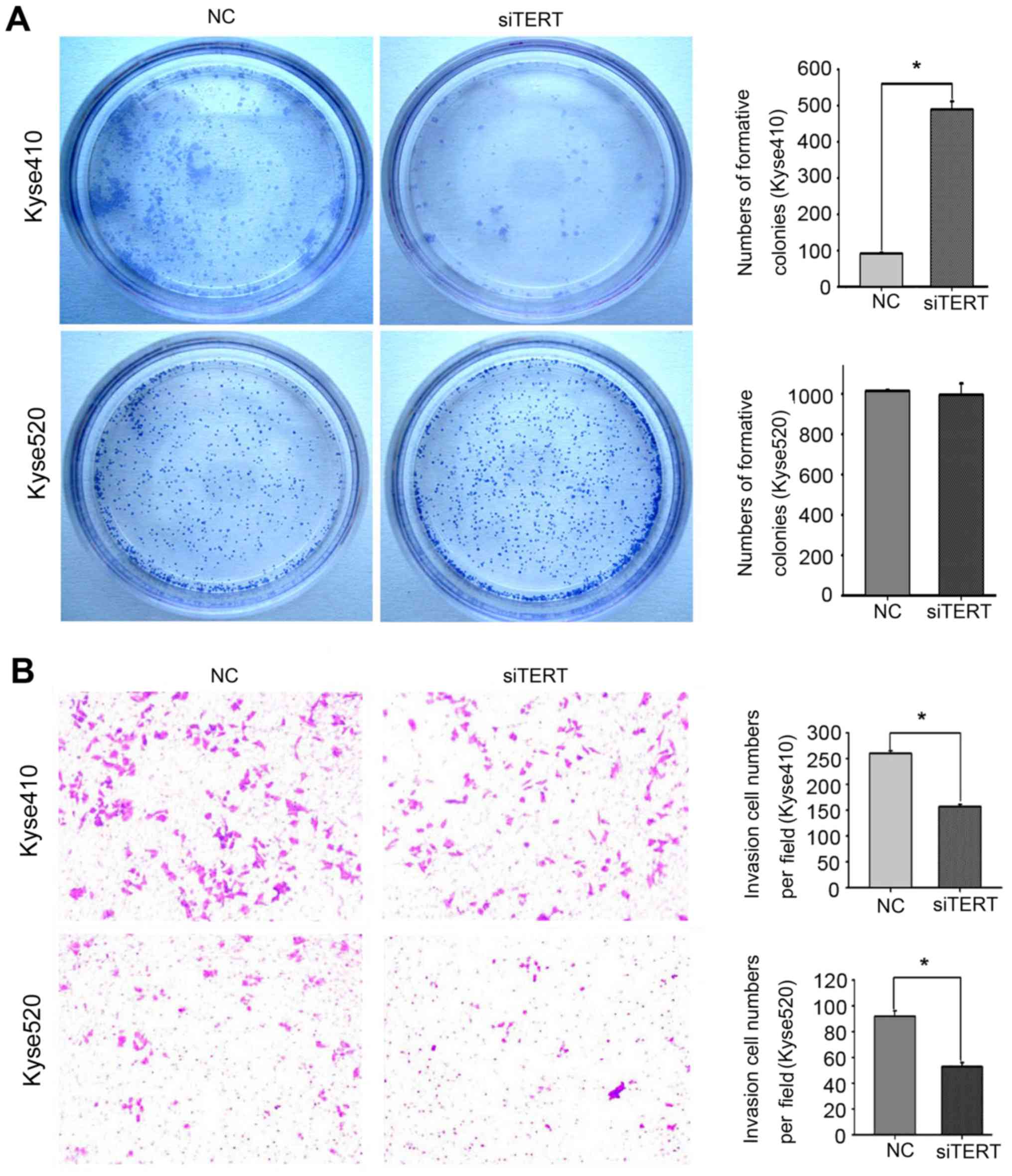
RNAi. Colony-formation ability was decreased by ~80% following
hTERT knockdown in Kyse410 cells (Fig.
5A, upper panel), but no significant decrease was observed in
Kyse520 cells (Fig. 5A, lower
panel).
hTERT knockdown inhibits Kyse410 and
Kyse520 cell invasion and migration
To confirm the effect of hTERT knockdown on the
migration and invasion abilities of Kyse410 and Kyse520 cells,
Transwell assays were used to assess invasion, and wound-healing
assays were used to assess migration. Cellular invasion was
measured after transfection for 48 h. The number of invading cells
from three random fields was determined using light microscopy. As
shown in Fig. 5B, the invasion
ability of Kyse410 cells (upper panel) was higher compared with
that of Kyse520 cells (lower panel), and hTERT knockdown
significantly decreased the invasiveness of both cell types
(*P<0.05). Similar results were observed for the wound healing
assay. The data revealed that the migration ability of Kyse410
cells was higher than that of Kyse520 cells (Fig. 6A and C, left panel), and that hTERT
knockdown significantly inhibited the migration of both cell types
(Fig. 6B and D; P<0.05).
Discussion
Abnormalities in hTERT are considered to be
associated with the tumorigenesis of 85% of all cancer types tested
(45), and hTERT has been found to
be overexpressed in a number of different cancer types, such as
cervical cancer and gastric cancer (35,46,47).
hTERT plays critical roles in tumorigenesis by preventing
apoptosis, and enhancing motility and invasiveness (48,49).
RNAi is a gene-silencing technology that was developed by Fire
et al (50). Previous studies
have reported that hTERT knockdown inhibits cellular proliferation
and induces apoptosis in numerous types of cancer cells, for
example anaplastic thyroid cancer and osteosarcoma cells (51,52).
RNAi-induced silencing of hTERT is considered to be a promising
strategy for cancer gene therapy by inhibiting tumorigenesis and
progression, and the results of the present study provide insights
into the development of novel therapeutic approaches for esophageal
cancer.
In the present study, H&E staining was used to
evaluate alterations in ESCC tissues compared with adjacent normal
tissues. p53 is expressed at low levels in most normal fetal and
adult tissues, and also has a short half-life. Since p53 and ki67
are reportedly highly expressed in esophageal cancer tissues
(53,54), their expression was evaluated in
pathological sections in the present study to further confirm this
observation. High hTERT expression was observed in the cytoplasm of
ESCC tissues, indicating that esophageal cell carcinogenesis is
accompanied by an increase in hTERT synthesis in the cytoplasm.
Following synthesis, hTERT is trafficked to the nucleus, where it
is then assembled and activated. The biogenesis, trafficking,
recruitment and activation of hTERT affects the development of ESCC
in a complex manner (55).
Therefore, further studies are required to assess the progress of
nuclear-cytoplasmic hTERT trafficking for telomerase maturation and
activity in esophageal cell carcinogenesis (55).
Next, the correlation between hTERT expression and
the clinicopathological data of patients with ESCC was analyzed;
hTERT expression was found to be significantly correlated with
pathological grade, T stage, N stage and TNM stage. Survival curve
analysis also revealed significant differences in the survival
rates of postoperative patients at different T, N and TNM stages.
It was therefore speculated that a high expression level of hTERT
in the cytoplasm of ESCC cells is associated with poor patient
prognosis.
A preliminary study on the effects of hTERT on the
occurrence and development of ESCC was subsequently conducted.
hTERT was knocked down in Kyse410 and Kyse520 cells using RNAi
methods, which was found to inhibit the proliferation of both cell
types at the 72-h timepoint, although no significant difference was
observed; this was consistent with a previous study in which
imetelstat was found to block hTERT activity and consequently
inhibit Kyse410 and Kyse520 cell proliferation after 3–4 weeks
(1). A possible reason for this
increased onset period is that cellular proliferation within a few
days does not cause a great degree of telomere shortening. It was
speculated that cell proliferation would not be affected until the
telomeres were shortened below a specific threshold, thus hTERT
knockdown did not immediately cause a significant inhibition in
proliferation. The effects of hTERT knockdown on migration and
invasion were then evaluated using cell lines with the longest
telomere length (Kyse410,6.32Kb) and the shortest telomere length
(Kyse520, 3.50Kb) among several esophageal cancer cell lines
(56). Notably, the results
demonstrated that the migration and invasion abilities of Kyse410
cells were stronger compared with those of Kyse520 cells, thus
Kyse410 appears to be the more aggressive ESCC cell line.
Matrix metalloproteinases (MMPs) represent a major
group of extracellular matrix regulatory proteins, which serve an
important role in tumor invasion and metastasis (57). A previous study demonstrated that
hTERT regulates MMP expression in U2OS cells independently of
telomerase activity (24). hTERT
knockdown also significantly inhibits the colony-formation ability
of Kyse410 cells, confirming its effects on the tumor-formation
ability of these cells. In addition, hTERT promotor
hypermethylation was observed in esophageal adenocarcinoma and its
precancerous lesions in a number of different studies (58,59).
According to the aforementioned observations, hTERT may regulate
esophageal cancer by targeting numerous different molecular
pathways. In the present study, hTERT was found to be associated
with ESCC cell migration, invasion and colony formation, but the
associated mechanisms remain to be elucidated.
In conclusion, the present study determined the
association between high hTERT expression in the cytoplasm and the
poor prognosis of patients with ESCC. In vitro experiments
confirmed that hTERT knockdown suppresses the migration and
invasion abilities of ESCC cells, and suppresses the
colony-formation ability of Kyse410 cells. The previous study
reported that Wnt5a promotes ESCC cell invasion via retinoic
acid-related orphan receptors (ROR)1 and ROR2 and
disheveled-associated activator of morphogenesis 1/RhoA signaling
pathway (60), thus the potential
association between hTERT, Wnt5a and ROR needs to be further
studied.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from National
Nature Science Foundation of China (Grant No. 81301938), and
Medical Science and Technology Development Foundation, Nanjing
Department of Health (Grant No. JQX14007) to Xuping Wu.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW designed the study. JL, JS and GT performed the
experiments. JL and GD analyzed and interpreted the data. JL, XW
and GD drafted and edited the manuscript. JL, GD and XW revised the
manuscript critically and approved the final of version of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The use of the clinical specimens for research
purposes was approved by the Institutional Research Ethics
Committee, The Second Hospital of Nanjing (approval no.
2018-LY-KY068).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ESCC
|
esophageal squamous cell carcinoma
|
|
hTERT
|
telomerase reverse transcriptase
|
|
EADC
|
esophageal adenocarcinoma
|
|
hTR
|
telomerase RNA
|
|
TMAs
|
tissue microarrays
|
|
FBS
|
fetal bovine serum
|
|
NC
|
negative control
|
References
|
1
|
Wu X, Zhang J, Yang S, Kuang Z, Tan G,
Yang G, Wei Q and Guo Z: Telomerase antagonist imetelstat increases
radiation sensitivity in esophageal squamous cell carcinoma.
Oncotarget. 8:13600–13619. 2017.
|
|
2
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018.
|
|
3
|
Lin Y, Totsuka Y, Shan B, Wang C, Wei W,
Qiao Y, Kikuchi S, Inoue M, Tanaka H and He Y: Esophageal cancer in
high-risk areas of China: Research progress and challenges. Ann
Epidemiol. 27:215–221. 2017.
|
|
4
|
Zuo J, Wang DH, Zhang YJ, Liu L, Liu FL
and Liu W: Expression and mechanism of PinX1 and telomerase
activity in the carcinogenesis of esophageal epithelial cells.
Oncol Rep. 30:1823–1831. 2013.
|
|
5
|
Domper Arnal MJ, Ferrández Arenas Á and
Lanas Arbeloa Á: Esophageal cancer: Risk factors, screening and
endoscopic treatment in Western and Eastern countries. World J
Gastroenterol. 21:7933–7943. 2015.
|
|
6
|
Zeng H, Zheng R, Zhang S, Zuo T, Xia C,
Zou X and Chen W: Esophageal cancer statistics in China, 2011:
Estimates based on 177 cancer registries. Thorac Cancer. 7:232–237.
2016.
|
|
7
|
Zhu LL, Yuan L, Wang H, Ye L, Yao GY, Liu
C, Sun NN, Li XJ, Zhai SC, Niu LJ, et al: A Meta-analysis of
concurrent chemoradiotherapy for advanced esophageal cancer. PLoS
One. 10:e01286162015.
|
|
8
|
Baker DJ and Petersen RC: Cellular
senescence in brain aging and neurodegenerative diseases: Evidence
and perspectives. J Clin Invest. 128:1208–1216. 2018.
|
|
9
|
Maciejowski J and de Lange T: Telomeres in
cancer: Tumour suppression and genome instability. Nat Rev Mol Cell
Biol. 18:175–186. 2017.
|
|
10
|
Nomikos NN, Nikolaidis PT, Sousa CV,
Papalois AE, Rosemann T and Knechtle B: Exercise, telomeres, and
cancer: ‘The exercise-telomere hypothesis’. Front Physiol.
9:17982018.
|
|
11
|
Jafri MA, Ansari SA, Alqahtani MH and Shay
JW: Roles of telomeres and telomerase in cancer, and advances in
telomerase-targeted therapies. Genome Med. 8:692016.
|
|
12
|
Gunes C, Avila AI and Rudolph KL:
Telomeres in cancer. Differentiation. 99:41–50. 2018.
|
|
13
|
Jaiswal RK, Kumar P, Kumar M and Yadava
PK: hTERT promotes tumor progression by enhancing TSPAN13
expression in osteosarcoma cells. Mol Carcinog. 57:1038–1054.
2018.
|
|
14
|
Aviv A, Anderson JJ and Shay JW:
Mutations, cancer and the telomere length paradox. Trends Cancer.
3:253–258. 2017.
|
|
15
|
Farooqi AA, Mansoor Q, Alaaeddine N and Xu
B: MicroRNA regulation of telomerase reverse transcriptase (TERT):
Micro machines pull strings of papier-mâché puppets. Int J Mol Sci.
19:10512018.
|
|
16
|
Kim NW, Piatyszek MA, Prowse KR, Harley
CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL and Shay
JW: Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994.
|
|
17
|
Alberti L, Losi L, Leyvraz S and Benhattar
J: Different Effects of BORIS/CTCFL on stemness gene expression,
sphere formation and cell survival in epithelial cancer stem cells.
PLoS One. 10:e01329772015.
|
|
18
|
Horikawa I, Cable PL, Afshari C and
Barrett JC: Cloning and characterization of the promoter region of
human telomerase reverse transcriptase gene. Cancer Res.
59:826–830. 1999.
|
|
19
|
Devereux TR, Horikawa I, Anna CH, Annab
LA, Afshari CA and Barrett JC: DNA methylation analysis of the
promoter region of the human telomerase reverse transcriptase
(hTERT) gene. Cancer Res. 59:6087–6090. 1999.
|
|
20
|
Renaud S, Loukinov D, Abdullaev Z,
Guilleret I, Bosman FT, Lobanenkov V and Benhattar J: Dual role of
DNA methylation inside and outside of CTCF-binding regions in the
transcriptional regulation of the telomerase hTERT gene. Nucleic
Acids Res. 35:1245–1256. 2007.
|
|
21
|
Park YJ, Kim EK, Bae JY, Moon S and Kim J:
Human telomerase reverse transcriptase (hTERT) promotes cancer
invasion by modulating cathepsin D via early growth response
(EGR)-1. Cancer Lett. 370:222–231. 2016.
|
|
22
|
Lavanya C, Venkataswamy MM, Sibin MK,
Srinivas Bharath MM and Chetan GK: Down regulation of human
telomerase reverse transcriptase (hTERT) expression by BIBR1532 in
human glioblastoma LN18 cells. Cytotechnology. 70:1143–1154.
2018.
|
|
23
|
Maida Y and Masutomi K: Telomerase reverse
transcriptase moonlights: Therapeutic targets beyond telomerase.
Cancer Sci. 106:1486–1492. 2015.
|
|
24
|
Ding D, Xi P, Zhou J, Wang M and Cong YS:
Human telomerase reverse transcriptase regulates MMP expression
independently of telomerase activity via NF-κB-dependent
transcription. FASEB J. 27:4375–4383. 2013.
|
|
25
|
Jäger K and Walter M: Therapeutic
targeting of telomerase. Genes (Basel). 7:392016.
|
|
26
|
Shi H, Shi D, Wu Y, Shen Q and Li J:
Qigesan inhibits migration and invasion of esophageal cancer cells
via inducing connexin expression and enhancing gap junction
function. Cancer Lett. 380:184–190. 2016.
|
|
27
|
Duff D and Long A: Roles for RACK1 in
cancer cell migration and invasion. Cell Signal. 35:250–255.
2017.
|
|
28
|
Su P, Tian Y, Yang C, Ma X, Wang X, Pei J
and Qian A: Mesenchymal stem cell migration during bone formation
and bone diseases therapy. Int J Mol Sci. 19:23432018.
|
|
29
|
Huang DS, Wang Z, He XJ, Diplas BH, Yang
R, Killela PJ, Meng Q, Ye ZY, Wang W, Jiang XT, et al: Recurrent
TERT promoter mutations identified in a large-scale study of
multiple tumour types are associated with increased TERT expression
and telomerase activation. Eur J Cancer. 51:969–976. 2015.
|
|
30
|
Vinagre J, Almeida A, Pópulo H, Batista R,
Lyra J, Pinto V, Coelho R, Celestino R, Prazeres H, Lima L, et al:
Frequency of TERT promoter mutations in human cancers. Nat Commun.
4:21852013.
|
|
31
|
Kinde I, Munari E, Faraj SF, Hruban RH,
Schoenberg M, Bivalacqua T, Allaf M, Springer S, Wang Y, Diaz LA
Jr, et al: TERT promoter mutations occur early in urothelial
neoplasia and are biomarkers of early disease and disease
recurrence in urine. Cancer Res. 73:7162–7167. 2013.
|
|
32
|
Vail E, Zheng X, Zhou M, Yang X, Fallon
JT, Epstein JI and Zhong M: Telomerase reverse transcriptase
promoter mutations in glandular lesions of the urinary bladder. Ann
Diagn Pathol. 19:301–305. 2015.
|
|
33
|
Pópulo H, Lopes JM, Sobrinho-Simões M and
Soares P: RE: TERT promoter mutation status as an independent
prognostic factor in cutaneous melanoma. J Natl Cancer Inst.
107:djv042015.
|
|
34
|
Horn S, Figl A, Rachakonda PS, Fischer C,
Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al:
TERT promoter mutations in familial and sporadic melanoma. Science.
339:959–961. 2013.
|
|
35
|
Qu Y, Shi L, Wang D, Zhang B, Yang Q, Ji
M, Shi B and Hou P: Low frequency of TERT promoter mutations in a
large cohort of gallbladder and gastric cancers. Int J Cancer.
134:2993–2994. 2014.
|
|
36
|
Liu Z, Li Q, Li K, Chen L, Li W, Hou M,
Liu T, Yang J, Lindvall C, Björkholm M, et al: Telomerase reverse
transcriptase promotes epithelial-mesenchymal transition and stem
cell-like traits in cancer cells. Oncogene. 32:4203–4213. 2013.
|
|
37
|
Tang B, Xie R, Qin Y, Xiao YF, Yong X,
Zheng L, Dong H and Yang SM: Human telomerase reverse transcriptase
(hTERT) promotes gastric cancer invasion through cooperating with
c-Myc to upregulate heparanase expression. Oncotarget.
7:11364–11379. 2016.
|
|
38
|
Arita H, Narita Y, Fukushima S, Tateishi
K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP,
Kawahara N, et al: Upregulating mutations in the TERT promoter
commonly occur in adult malignant gliomas and are strongly
associated with total 1p19q loss. Acta Neuropathol. 126:267–276.
2013.
|
|
39
|
Nonoguchi N, Ohta T, Oh JE, Kim YH,
Kleihues P and Ohgaki H: TERT promoter mutations in primary and
secondary glioblastomas. Acta Neuropathol. 126:931–937. 2013.
|
|
40
|
Koelsche C, Sahm F, Capper D, Reuss D,
Sturm D, Jones DT, Kool M, Northcott PA, Wiestler B, Böhmer K, et
al: Distribution of TERT promoter mutations in pediatric and adult
tumors of the nervous system. Acta Neuropathol. 126:907–915.
2013.
|
|
41
|
Killela PJ, Reitman ZJ, Jiao Y, Bettegowda
C, Agrawal N, Diaz LA Jr, Friedman AH, Friedman H, Gallia GL,
Giovanella BC, et al: TERT promoter mutations occur frequently in
gliomas and a subset of tumors derived from cells with low rates of
self-renewal. Proc Natl Acad Sci USA. 110:6021–6026. 2013.
|
|
42
|
Liu T, Li W, Lu W, Chen M, Luo M, Zhang C,
Li Y, Qin G, Shi D, Xiao B, et al: RBFOX3 promotes tumor growth and
progression via hTERT signaling and predicts a poor prognosis in
hepatocellular carcinoma. Theranostics. 7:3138–3154. 2017.
|
|
43
|
Lu C, Yang L, Chen H and Shan Z:
Upregulated long non-coding RNA BC032469 enhances carcinogenesis
and metastasis of esophageal squamous cell carcinoma through
regulating hTERT expression. Tumour Biol. 37:16065–16075. Oct
10–2016.(Epub ahead of print).
|
|
44
|
Okawa T, Michaylira CZ, Kalabis J, Stairs
DB, Nakagawa H, Andl CD, Johnstone CN, Klein-Szanto AJ, El-Deiry
WS, Cukierman E, et al: The functional interplay between EGFR
overexpression, hTERT activation, and p53 mutation in esophageal
epithelial cells with activation of stromal fibroblasts induces
tumor development, invasion, and differentiation. Genes Dev.
21:2788–2803. 2007.
|
|
45
|
Akincilar SC, Unal B and Tergaonkar V:
Reactivation of telomerase in cancer. Cell Mol Life Sci.
73:1659–1670. 2016.
|
|
46
|
Jaiswal RK, Kumar P, Sharma A, Mishra DK
and Yadava PK: Proteomic identification of proteins differentially
expressed following overexpression of hTERT (human telomerase
reverse transcriptase) in cancer cells. PLoS One.
12:e01810272017.
|
|
47
|
Yang H, Zhang H, Zhong Y, Wang Q, Yang L,
Kang H, Gao X, Yu H, Xie C, Zhou F and Zhou Y: Concomitant
underexpression of TGFBR2 and overexpression of hTERT are
associated with poor prognosis in cervical cancer. Sci Rep.
7:416702017.
|
|
48
|
Kim SH, Cho KH, Kim YN, Jeong BY, Park CG,
Hur GM and Lee HY: Resveratrol attenuates norepinephrine-induced
ovarian cancer invasiveness through downregulating hTERT
expression. Arch Pharm Res. 39:240–248. 2016.
|
|
49
|
Li Z, Liu YH, Diao HY, Ma J and Yao YL:
MiR-661 inhibits glioma cell proliferation, migration and invasion
by targeting hTERT. Biochem Biophys Res Commun. 468:870–876.
2015.
|
|
50
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998.
|
|
51
|
Maggisano V, Celano M, Lombardo GE, Lepore
SM, Sponziello M, Rosignolo F, Verrienti A, Baldan F, Puxeddu E,
Durante C, et al: Silencing of hTERT blocks growth and migration of
anaplastic thyroid cancer cells. Mol Cell Endocrinol. 448:34–40.
2017.
|
|
52
|
Chen P, Gu WL, Gong MZ, Wang J and Li DQ:
shRNA-mediated silencing of hTERT suppresses proliferation and
promotes apoptosis in osteosarcoma cells. Cancer Gene Ther.
24:325–332. 2017.
|
|
53
|
da Costa AM, Fregnani JHTG, Pastrez PRA,
Mariano VS, Silva EM, Neto CS, Guimarães DP, Villa LL, Sichero L,
Syrjanen KJ and Longatto-Filho A: HPV infection and p53 and p16
expression in esophageal cancer: Are they prognostic factors?
Infect Agent Cancer. 12:542017.
|
|
54
|
Wang L, Yu X, Li J, Zhang Z, Hou J and Li
F: Prognostic significance of p53 expression in patients with
esophageal cancer: A meta-analysis. BMC Cancer. 16:3732016.
|
|
55
|
Schmidt JC and Cech TR: Human telomerase:
Biogenesis, trafficking, recruitment, and activation. Genes Dev.
29:1095–1105. 2015.
|
|
56
|
Wu X, Smavadati S, Nordfjall K, Karlsson
K, Qvarnström F, Simonsson M, Bergqvist M, Gryaznov S, Ekman S and
Paulsson-Karlsson Y: Telomerase antagonist imetelstat inhibits
esophageal cancer cell growth and increases radiation-induced DNA
breaks. Biochim Biophys Acta. 1823:2130–2135. 2012.
|
|
57
|
Kapoor C, Vaidya S, Wadhwan V, Hitesh;
Kaur G and Pathak A: Seesaw of matrix metalloproteinases (MMPs). J
Cancer Res Ther. 12:28–35. 2016.
|
|
58
|
Guilleret I, Losi L, Chelbi ST, Fonda S,
Bougel S, Saponaro S, Gozzi G, Alberti L, Braunschweig R and
Benhattar J: DNA methylation profiling of esophageal adenocarcinoma
using Methylation Ligation-dependent Macroarray (MLM). Biochem
Biophys Res Commun. 479:231–237. 2016.
|
|
59
|
Clement G, Braunschweig R, Pasquier N,
Bosman FT and Benhattar J: Methylation of APC, TIMP3, and TERT: A
new predictive marker to distinguish Barrett's oesophagus patients
at risk for malignant transformation. J Pathol. 208:100–107.
2006.
|
|
60
|
Wu X, Yan T, Hao L and Zhu Y: Wnt5a
induces ROR1 and ROR2 to activate RhoA in esophageal squamous cell
carcinoma cells. Cancer Manag Res. 11:2803–2815. 2019.
|