Introduction
Breast cancer is one of the most common malignant
tumors in the world and the leading cause of cancer-associated
death in women in recent years (1,2).
Numerous complex factors are involved in the proliferation and
invasion of tumor cells, including human epidermal growth factor
receptor 2 (HER-2), androgen receptor and estrogen receptor (ER)
(3–6). In previous years, significant advances
have been made in discerning the molecular drivers of breast cancer
and characterizing distinct subtypes based on gene expression
profiles (1).
The odd-skipped related transcription factor 1
(OSR1) gene belongs to the OSR family; it is located on
human chromosome 2 (2p24.1) and encodes a 266-amino acid protein
with three C2H2-type zinc fingers (7). OSR1 has multiple functions and is
essential for the development of the intermediate mesoderm. This
process is strictly regulated and is influenced by OSR1 in a number
of ways (8–17). Bone morphogenetic protein (BMP)
(8), retinoic acid (9), and 1,25-dihydroxyvitamin D3 (10) have been found to activate OSR1,
whereas IKAROS family zinc finger 1 (IKZF1) and RUNX family
transcription factor 2 (RUNX2) represses it (11). OSR1 suppresses the nodal signaling
pathway and SOX9 mRNA expression (12,13).
OSR1 also serves important roles in embryonic urogenital formation,
heart formation, and tongue development (13–17).
Zhang et al (18)
demonstrated that OSR1 was downregulated in renal cell carcinoma
(RCC) cells through promoter methylation. In addition, depletion of
OSR1 by small interfering (si)RNA repressed the expression level of
several tumor suppressor genes involved in the p53 pathway, such as
p53, p21, p27, p57 and RB, and suppressed the transcriptional
activity of p53 in RCC (18).
Furthermore, expression of OSR1 inhibited the invasion and
proliferation abilities of RCC cells (18). Otani et al (19) demonstrated that OSR1 was commonly
downregulated by siRNA by promoter methylation in gastric cancer.
In addition, expression of OSR1 was demonstrated to inhibit gastric
cancer cell growth, arrest the cell cycle and induce cell apoptosis
(19). The role and underlying
mechanism of OSR1 in other types of cancer, apart from renal and
gastric cancer has not been well characterized. It was reported
that OSR1-mediated tumor suppression in gastric cancer occurs by
repression of the Wnt/β-catenin signaling pathway and the
activation of p53 pathway (19). The
Wnt signaling pathway is regulated by multiple proteins, among
which, β-catenin serves a key role (20). Accumulation of β-catenin in the
cytoplasm and nucleus activates target genes of the Wnt pathway,
such as cyclin D1 and c-Myc (21).
Activation by β-catenin causes carcinogenesis and tumor progression
in numerous types of cancer, such as lung, gastric and intestine
cancers (21). Overall, the
expression and function of OSR1 in breast cancer remains
unclear.
In the present study, the expression of OSR1 in
breast cancer and corresponding normal adjacent tissues, and its
association with clinicopathological factors was examined. In
addition, the effects of OSR1 on the proliferative and invasive
abilities of breast cancer cells was investigated, as well as
identifying the regulating effects of OSR1 on the
epithelial-mesenchymal transition (EMT) process and activation of
the Wnt signaling pathway in breast cancer cells.
Materials and methods
Patient data and tissue specimens
Tissue samples from 70 female patients with breast
cancer who underwent complete surgical resection at the First
Affiliated Hospital of China Medical University between September
2013 and August 2016 were selected from the archival files in the
Department of Pathology. The 70 breast cancer samples were
accompanied by adjacent normal breast tissue specimens. and were
located >2 cm away from the tumor. The mean age of the patients
was 50-years-old (range, 31 to 70 years). The patients were graded
according to WHO (22) and TNM
staging systems (23), and divided
into ER, PR and HER2 positive and negative expression groups. The
histological grades of the specimens were evaluated as grade I
(n=18), II (n=44) and III (n=8). And patients were categorized into
stage I (n=32), II (n=21) or III (n=17). Lymph node metastases were
found in 30 cases. Estrogen receptor (ER)-, progesterone receptor
(PR)- and human epidermal growth factor receptor 2 (HER2)-positive
expression was found in 45, 43 and 24 cases, respectively. A total
of 20 pairs of fresh tumor and corresponding normal tissue
specimens were collected following resection between September 2013
and August 2016 and immediately stored at −80°C for subsequent use.
The study was conducted in accordance with the Declaration of
Helsinki and approved by the Institutional Review Board of the
First Hospital and College of Basic Medical Sciences of China
Medical University, China [approval no. LS(2018)016]. All patients
provided written informed consent.
The Cancer Genome Atlas (TCGA) data
collection and analysis
The mRNA expression data of OSR1 in breast cancer
and adjacent normal breast tissue was analyzed and downloaded
directly from the online database, UALCAN (http://ualcan.path.uab.edu) (24). The association between OSR1
expression and prognosis of breast cancer was analyzed and
downloaded directly from the online database, The Human Protein
Atlas (https://www.proteinatlas.org/)
(25), which is based on TCGA.
Immunohistochemistry
After fixation in 10% neutral formalin at room
temperature for 24 h, all resected specimens were embedded in
paraffin and cut into 4-µm sections. Immunostaining was performed
using a streptavidin-peroxidase method. All sections were
deparaffinized, rehydrated, and heated in 0.01 M citrate buffer for
2.5 min at 100°C in an autoclave. Then, the sections were incubated
with anti-OSR1 rabbit polyclonal antibody (cat. no. ab179612;
1:100; Abcam) and anti-ER mouse monoclonal antibody (cat. no.
MAB-0062; 1:200; Fuzhou Maixin Biotech Co.) overnight at 4°C.
Subsequently, the sections were incubated with the secondary
antibody and horseradish peroxidase-conjugated streptavidin-biotin
at 37°C for 2 h (cat. no. KIT 9002; Fuzhou Maixin Biotech Co.,
Ltd.). Expression was visualized using 3,3′-diaminobenzidine
chromogen (Fuzhou Maixin Biotech Co., Ltd.), as previously
described (26).
A total of 2 investigators, who were blinded to the
clinical data, evaluated the sections, using 5 fields of view
randomly per slide and 100 cells per view were observed at ×400
magnification using light microscope (Olympus Corporation). The
positive rate for each case was calculated from the percentage of
positively stained cells and scored as follows: 1, 1–25; 2, 26–50;
3, 51–75; and 4, 76–100%. The intensity of immunostaining was
scored as 0, 1, 2, or 3, for negative, weak, moderate or marked,
respectively. A final score ranging from 0 to 12 was obtained by
multiplying the scores from each sample. Based on their final
scores, the tumors were categorized as having low (≤6) or high (≥8)
expression of OSR1 and ER (26,27).
Cell culture and transfection
The human MCF-7 and MDA-MB-231 breast cancer cell
lines were purchased from American Type Culture Collection. The
MCF-7 cells were cultured in Dulbecco's modified Eagle's medium
(DMEM) and the MDA-MB-231 cells were cultured in Leibovitz 15
Medium (L15), both supplemented with 10% FBS (all from Thermo
Fisher Scientific, Inc.) in an atmosphere of 5% CO2 and
at 37°C. The cells were cultured in sterile culture dishes and
passaged every 1 or 2 days using 0.25% trypsin (Thermo Fisher
Scientific, Inc.).
For transfection, cells were seeded in a 6-well
plate 24 h at 37°C prior to the experiment. The empty control
vector, pCMV6, and pCMV6-OSR1 plasmids were purchased from
OriGene Technologies, Inc. The control siRNA and siRNA against
OSR1 (OSR1-siRNA) were synthesized by Guangzhou
RiboBio Co., Ltd). The plasmids (2.5 µg) were transfected into
cells when density of treated cells reached 80–90%, and the siRNAs
(5 nM) were transfected into cells when the density of treated
cells was 40–50% using Lipofectamine® 3000 (Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions.
Western blot analysis
Total protein from cells and tissue was extracted
from cells using a cell lysis buffer (Thermo Fisher Scientific,
Inc.) and quantified using the Bradford method. A total of 60 µg
total protein was separated using 10% SDS-PAGE, then transferred to
a PVDF membrane (EMD Millipore; Merck KGaA). Following blocking
with 5% skimmed milk at room temperature for 2 h, membranes were
incubated overnight at 4°C with antibodies against OSR1 (cat. no.
sc-376545; 1:150; Santa Cruz Biotechnology, Inc.), β-catenin
(sc-7963,1:100; Santa Cruz Biotechnology, Inc.), cyclin D1 (cat.
no. sc-8396; 1:100; Santa Cruz Biotechnology, Inc.), c-Myc (cat.
no. 554002; 1:200; BD Biosciences), axin (cat. no. sc-518090;
1:100; Santa Cruz Biotechnology, Inc.), Snail (cat. no. 3879,1:500;
Cell Signaling Technology, Inc.), transcription factor 4 (TCF4;
cat. no. sc-166699, 1:100; Santa Cruz Biotechnology, Inc.),
E-cadherin (cat. no. 14472; 1:500; Cell Signaling Technology,
Inc.), lymphoid enhancer-binding factor 1 (LEF1; cat. no.
sc-374522; 1:100; Santa Cruz Biotechnology, Inc.), N-cadherin (cat.
no. 13116; 1:500; Cell Signaling Technology, Inc.), β-actin (cat.
no. sc-8432; 1:1,000; Santa Cruz Biotechnology, Inc.) and GAPDH (s
cat. no. c-47724; 1:2,000; Santa Cruz Biotechnology, Inc.).
Following washing in TBST for 15 min, the membranes were incubated
with horseradish peroxidase-conjugated anti-mouse/rabbit IgG
secondary antibody (cat. no. SA00001-1/2; 1:2,000; ProteinTech
Group, Inc.) at 37°C for 2 h. Protein bands were visualized using
an ECL kit (Thermo Fisher Scientific, Inc.) and detected with a
bioimaging system (DNR Bio-Imaging Systems, Ltd.). The relative
protein levels were calculated using β-actin or GAPDH as the
loading control. The bands were quantified with Image J software
(X64; National Institutes of Health).
Cell proliferation assay
Cells (3-4×103 cells/well) were plated in
96-well plates at 37°C for 24 h following transfection and cultured
in medium containing 10% fetal bovine serum. Cell Counting Kit-8
(CCK-8; Dojindo Molecular Technologies, Inc.) reagent was added to
each well at 1:10 (v/v) per 100 µl and the cells were incubated for
2 h at 37°C according to the manufacturers' instructions. Cell
proliferation results were detected using spectrophotometric
quantitate on at 450 nm.
Matrigel invasion assay
To assess the invasive ability of the transfected
cells, Matrigel™ (BD Biosciences) and Transwell®
chambers (Costar; Corning, Inc.) with a pore size of 8 µm were
used, according to the manufacturers' instructions. Briefly, 100 µl
Matrigel™ (1:7 dilution) was added to each insert and the chambers
were placed at 37°C, for at least 2 h. Then, 8×104 cells
in 100 µl medium supplemented with 2% FBS were added to the upper
chamber. Medium supplemented with 20% FBS was added to the lower
chamber as the chemoattractant. After 20 h of incubation, the
filters were fixed at room temperature for 20 min and stained with
hematoxylin at room temperature for 10 min. The non-invading cells
on the upper surface were removed with a cotton swab. The number of
invasive cells in 10 high-power fields randomly was counted under
an inverted microscope (magnification, ×200). The experiments were
performed in triplicate.
Statistical analysis
Data are expressed as the mean ± standard deviation
of three independent experiments. Unpaired Student's t-test was
used to compare the mean values between two experimental groups.
Paired Student's t-test was used to compare the mean expression
levels of OSR1 in tumor vs. adjacent non-tumor samples of the same
individuals. The Kaplan-Meier curve was used to analyze the
prognosis value of OSR1 in breast cancer from the online database
The Human Protein Atlas. Associations between OSR1 expression level
and clinicopathological factors were examined using the
χ2 test. All statistical analyses were performed using
GraphPad Prism v6.0 software (GraphPad Software, Inc.) and SPSS
v17.0 software (SPSS, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
OSR1 expression is downregulated in
breast cancer tissue, and is negatively associated with lymph node
metastases, ER expression, and poor survival
The expression level of OSR1 was examined in 70
pairs of breast cancer and adjacent normal breast tissue using
immunohistochemistry. In normal breast tissue, 55 cases (78.6%)
demonstrated high expression levels of OSR1 (Fig. 1A) and 15 cases (21.4%) demonstrated
low expression levels. However, in breast cancer tissue, 30 cases
(42.9%) had high expression levels of OSR1 (Fig. 1B), whereas 40 cases (57.1%) had low
levels (Fig. 1C). The expression
level of OSR1 was lower in breast cancer tissue compared with that
in normal breast tissue (P<0.001; Table I). The low expression level of OSR1
in breast cancer was also significantly associated with lymph node
metastases (P=0.004) and ER expression (P=0.031; Fig. 1D). However, the expression level of
OSR1 was not significantly associated with patient age (P=0.266),
maximum diameter of the tumor (P=0.785), histological
classification (P=0.554), TNM stage (P=0.165), PR
expression (P=0.436) or HER2 expression (P=0.095;
Table I).
 | Table I.Association between OSR1 expression
level and clinicopathological factors in breast cancer. |
Table I.
Association between OSR1 expression
level and clinicopathological factors in breast cancer.
|
|
| OSR1
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
factor | Number of
patients | High, n (%) | Low, n (%) | P-value |
|---|
| Tissue |
|
|
| <0.001 |
|
Normal | 70 | 55 (78.6) | 15 (21.4) |
|
| Breast
cancer | 70 | 30 (42.9) | 40 (57.1) |
|
| Age, years |
|
|
| 0.266 |
|
<50 | 31 | 11 (35.5) | 20 (64.5) |
|
|
≥50 | 39 | 19 (48.7) | 20 (51.3) |
|
| Maximum diameter,
cm |
|
|
| 0.785 |
| ≤2 | 34 | 16 (47.1) | 18 (52.9) |
|
|
2-5 | 26 | 10 (38.5) | 16 (61.5) |
|
|
>5 | 10 | 4 (40.0) | 6 (60.0) |
|
| Histological
classification |
|
|
| 0.554 |
| I | 18 | 8 (44.4) | 10 (55.6) |
|
| II | 44 | 20 (45.5) | 24 (54.5) |
|
|
III | 8 | 2 (25.0) | 6 (75.0) |
|
| TNM stages |
|
|
| 0.165 |
|
I–II | 53 | 21 (39.6) | 32 (60.4) |
|
|
III–IV | 17 | 10 (58.8) | 7 (41.2) |
|
| Lymphatic
metastasis |
|
|
| 0.004a |
|
Yes | 30 | 7 (23.3) | 23 (76.7) |
|
| No | 40 | 23 (57.5) | 17 (42.5) |
|
| ER expression |
|
|
| 0.031b |
|
Positive | 45 | 15 (33.3) | 30 (66.7) |
|
|
Negative | 25 | 15 (60.0) | 10 (40.0) |
|
| PR expression |
|
|
| 0.436 |
|
Positive | 43 | 20 (46.5) | 23 (53.5) |
|
|
Negative | 27 | 10 (37.0) | 17 (63.0) |
|
| HER-2
expression |
|
|
| 0.095 |
|
Positive | 24 | 7 (29.2) | 17 (70.8) |
|
|
Negative | 46 | 23 (50.0) | 23 (50.0) |
|
It was also confirmed that the expression level of
OSR1 was significantly higher in normal breast tissue compared with
that in breast cancer tissue, using western blot analysis
(2.12±0.18 vs. 0.97±0.15; n=20; P<0.01; Fig. 1E and F). The significant reduction of
OSR1 mRNA expression levels in breast cancer tissue was confirmed
using the UALCAN web resource, based on the TCGA database
(P<0.001; Fig. S1A).
In addition, a search of the online database The
Human Protein Atlas revealed that patients with breast cancer and
low expression levels of OSR1 had significantly shorter overall
survival rates compared with patients with high expression levels
(P<0.01; Fig. SIB).
OSR1 regulates the expression level of
proteins in the Wnt signaling pathway and inhibits the
proliferation of breast cancer cells
The expression level of OSR1 was low in MCF-7 cells
and high in MDA-MB-231 cells; therefore, for a more appropriate
comparison and visualization of the effects of OSR1, the
OSR1 gene was overexpressed in MCF-7 cells
(MCF7-OSR1), while OSR1 expression was knocked down in
MDA-MB-231 cells (MDA-MB-231-siOSR1).
Compared with that in the control cells, the protein
expression levels of β-catenin and the Wnt target genes, cyclin D1
and c-Myc were significantly decreased in MCF7-OSR1 cells
(P<0.05), whereas there was no significant difference in the
protein expression levels of axin, TCF4 and LEF1 (P>0.05;
Fig. 2A). By contrast, compared with
that in the control cells, the protein expression level of
β-catenin, cyclin D1 and c-Myc was significantly increased in
MDA-MB-231-siOSR1 cells (P<0.05), whereas there was no
significant difference in the expression levels of axin, TCF4 and
LEF1 (P>0.05; Fig. 2B). In
addition, overexpression of OSR1 significantly inhibited
proliferation of MCF-7 cells from day two (P<0.05; Fig. 2C) and downregulation of OSR1
significantly increased proliferation of MDA-MB-231 cells from day
two (P<0.05; Fig. 2D).
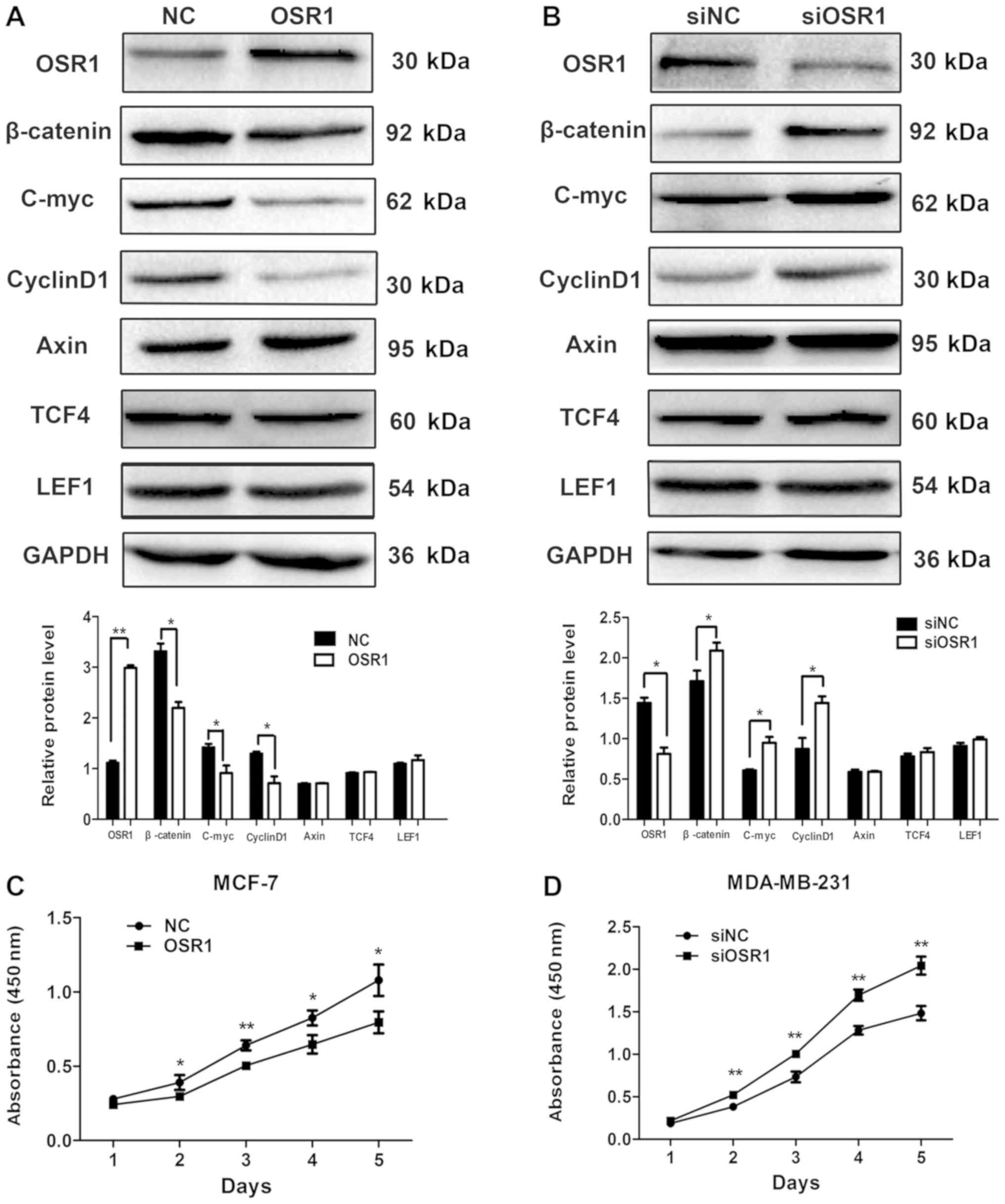 | Figure 2.OSR1 inhibits the expression of Wnt
target proteins and the proliferative abilities of breast cancer
cells. Western blot analysis and relative protein levels for OSR1,
β-catenin, c-Myc, cyclin D1, Axin, TCF4, and LEF1 in (A)
MCF7-OSR1 and NC cells and (B) in MDA-MB-231 cells
transfected with an OSR1 overexpression plasmid,
siOSR1 or siNC. GAPDH served as an internal control. The
cell growth curve of (C) MCF7 cells transfected with an OSR1
overexpression plasmid or its NC and (D) MDA-MB-231 cells
transfected with siOSR1 or siNC. *P<0.05, **P<0.01.
OSR1, odd-skipped related transcription factor 1; TCF4,
transcription factor 4; LEF1, lymphoid enhancer binding factor 1;
NC, negative control; si, small interfering; siNC, scramble control
siRNA. |
OSR1 regulates the expression of
EMT-related proteins and inhibits the invasive ability of breast
cancer cells
Overexpression of OSR1 significantly inhibited the
invasive ability of MCF-7 cells (P<0.01), while downregulation
of OSR1 promoted the invasive ability of MDA-MB-231 cells
(P<0.01; Fig. 3A and B).
Compared with that in the control cells, the
expression level of E-cadherin in MCF7-OSR1 cells was
significantly increased (P<0.01), whereas the expression levels
of N-cadherin and Snail were significantly decreased (P<0.05;
Fig. 3C). By contrast, compared with
that in the control cells, the expression level of E-cadherin in
MDA-MB-231-siOSR1 cells was significantly decreased
(P<0.01), whereas the expression levels of N-cadherin and Snail
were significantly increased (P<0.05; Fig. 3D).
Discussion
Previous studies on OSR1 were focused on the field
of embryonic development (12–17),
while the studies on OSR1 in tumors have been limited (18,19).
Therefore, the association between OSR1 and tumorigenesis, and the
possible mechanisms, have not been discussed. The results of the
present study demonstrated that the expression level of OSR1 was
significantly reduced in breast cancer tissue compared with that in
normal breast tissue and negatively associated with lymph node
metastases and ER expression level. Therefore, reduced expression
of OSR1 may be involved in the progression of breast cancer.
Notably, the present study demonstrated negative association
between ER expression and OSR1 in breast cancer. Thus, high level
of ER may be involved in the downregulation of OSR1 expression and
may be one of the potential reasons for low expression of OSR1;
however, further investigation is required. To the best of our
knowledge, this was the first time that the expression pattern and
clinical significance of OSR1 in breast cancer was examined. Data
from TCGA database confirmed that OSR1 expression is significantly
reduced in breast cancer and associated with poor prognosis.
Furthermore, the in vitro experiments in the
present study confirmed that overexpression of OSR1 inhibited the
proliferative and invasive abilities of breast cancer cells. Otani
et al (19) demonstrated that
OSR1 suppresses the protein expression of cytoplasmic β-catenin,
TCF-1 and LEF1, which are part of the Wnt signaling pathway. OSR1
acts as a functional tumor suppressor through the transcriptional
repression of TCF/LEF in gastric cancer (19). The present study demonstrated that
overexpression of OSR1 inhibited the expression of β-catenin and
Wnt target genes, cyclin D1 and c-Myc, in breast cancer cells. As a
cell cycle regulator, cyclin D1 is essential for progression
through the G1 phase and is a candidate proto-oncogene
(28). Mutation, amplification and
overexpression of cyclin D1 has been found to alter cell cycle
progression and may contribute to the proliferation of tumor cells
(29,30). As such, OSR1 inhibits the
proliferative abilities of breast cancer cells by inhibiting the
activity of the Wnt signaling pathway.
In addition, the results of the present study
demonstrated that overexpression of OSR1 inhibited the protein
expression level of Snail. An integrated and complex signaling
network of pathways, including Wnt, TGF-β, Notch and BMP, are known
to activate Snail (31). As an
EMT-inducing transcription factor, Snail has been found to regulate
the protein expression of the cell adhesion molecules, E-cadherin
and N-cadherin in the EMT process (31). The present study examined whether the
protein expression levels of E-cadherin, N-cadherin and Snail were
altered through regulation of OSR1 and the results demonstrated
that overexpression of OSR1 increased expression level of
E-cadherin and decreased expression of N-cadherin, thereby
suppressing the EMT process in breast cancer cells. A previous
study has suggested that the EMT process contributes to early stage
dissemination of cancer cells and is important for invasion and
metastasis (32). Thus, OSR1 may
inhibit the invasive abilities of breast cancer cells by
restricting the EMT process. Apart from the Wnt signaling pathway
and EMT, there may be other pathways involved in the regulating
mechanism of OSR1, which will be investigated in future studies. In
addition, the lack of a normal breast cell line as a normal control
is also a limitation to the present study, while, in vivo
tumorigenicity assay is also required to further confirm the
function of OSR1, which will be performed in the future.
In conclusion, OSR1 is a novel tumor suppressor
gene, which is downregulated in breast cancer tissue, which
suggests it could be a potential marker for tumor malignancy and
prognosis, as well as a possible target for drug treatment. OSR1
downregulates the invasive and proliferative abilities of breast
cancer by suppressing the EMT process and activity of the Wnt
signaling pathway.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Program for Liaoning
Excellent Talents in University (grant no. LR2015067), the Program
for The Doctoral Scientific Research Foundation of Liaoning
Province (grant no. 2019-BS-094) and the Natural Science Foundation
Funding Scheme of Liaoning Province (grant no. 2019-MS-145).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HX and YW conceived and designed the experiments. YW
performed some experiments, analyzed the data and wrote the
manuscript. LL and FX performed some experiments and analyzed the
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Institutional Review
Board of the First Hospital and College of Basic Medical Sciences
of China Medical University [approval no. iLS(2018)016]. Written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cadoo KA, Fornier MN and Morris PG:
Biological subtypes of breast cancer: Current concepts and
implications for recurrence patterns. Q J Nucl Med Mol Imaging.
57:312–321. 2013.PubMed/NCBI
|
|
2
|
Yip CH and Rhodes A: Estrogen and
progesterone receptors in breast cancer. Future Oncol.
10:2293–2301. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fidler IJ and Kripke ML: Genomic analysis
of primary tumors does not address the prevalence of metastatic
cells in the population. Nat Genet. 34:232003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cedolini C, Bertozzi S, Londero AP,
Bernardi S, Seriau L, Concina S, Cattin F and Risaliti A: Type of
breast cancer diagnosis, screening, and survival. Clin Breast
Cancer. 14:235–240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carey LA, Perou CM, Livasy CA, Dressler
LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S,
et al: Race, breast cancer subtypes, and survival in the carolina
breast cancer study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McNamara KM, Moore NL, Hickey TE, Sasano H
and Tilley WD: Complexities of androgen receptor signalling in
breast cancer. Endocr Relat Cancer. 21:T161–T181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katoh M: Molecular cloning and
characterization of OSR1 on human chromosome 2p24. Int J Mol Med.
10:221–225. 2002.PubMed/NCBI
|
|
8
|
James RG and Schultheiss TM: Bmp signaling
promotes intermediate mesoderm gene expression in a dose-dependent,
cell-autonomous and translation-dependent manner. Dev Biol.
288:113–125. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mae S, Shirasawa S, Yoshie S, Sato F,
Kanoh Y, Ichikawa H, Yokoyama T, Yue F, Tomotsune D and Sasaki K:
Combination of small molecules enhances differentiation of mouse
embryonic stem cells into intermediate mesoderm through
BMP7-positive cells. Biochem Biophys Res Commun. 393:877–882. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Verlinden L, Kriebitzsch C, Eelen G, Camp
MV, Leyssens C, Tan BK, Beullens I and Verstuyf A: The odd-skipped
related genes Osr1 and Osr2 are induced by 1,25-dihydroxyvitamin
D3. J Steroid Biochem Mol Biol. 136:94–97. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamauchi M, Kawai S, Kato T, Ooshima T and
Amano A: Odd-skipped related 1 gene expression is regulated by
Runx2 and Ikzf1 transcription factors. Gene. 426:81–90. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Terashima AV, Mudumana SP and Drummond IA:
Odd skipped related 1 is a negative feedback regulator of
nodal-induced endoderm development. Dev Dyn. 243:1571–1580. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu H, Lan Y, Xu J, Chang CF, Brugmann SA
and Jiang R: Odd-skipped related-1 controls neural crest
chondrogenesis during tongue development. Proc Natl Acad Sci USA.
110:18555–18560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Q, Lan Y, Cho ES, Maltby KM and Jiang
R: Odd-skipped related 1 (Odd 1) is an essential regulator of heart
and urogenital development. Dev Biol. 288:582–594. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tena JJ, Neto A, de la Calle-Mustienes E,
Bras-Pereira C, Casares F and Gómez-Skarmeta JL: Odd-skipped genes
encode repressors that control kidney development. Dev Biol.
301:518–531. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stricker S, Mathia S, Haupt J, Seemann P,
Meier J and Mundlos S: Odd-skipped related genes regulate
differentiation of embryonic limb mesenchyme and bone marrow
mesenchymal stromal cells. Stem Cells Dev. 21:623–633. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
James RG, Kamei CN, Wang Q, Jiang R and
Schultheiss TM: Odd-skipped related 1 is required for development
of the metanephric kidney and regulates formation and
differentiation of kidney precursor cells. Development.
133:2995–3004. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Yuan Y, Liang P, Guo X, Ying Y,
Shu XS, Gao M Jr and Cheng Y: OSR1 is a novel epigenetic silenced
tumor suppressor regulating invasion and proliferation in renal
cell carcinoma. Oncotarget. 8:30008–30018. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Otani K, Dong Y, Li X, Lu J, Zhang N, Xu
L, Go MYY, Ng EKW, Arakawa T, Chan FKL, et al: Odd-skipped related
1 is a novel tumour suppressor gene and a potential prognostic
biomarker in gastric cancer. J Pathol. 234:302–315. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie XM, Zhang ZY, Yang LH, Yang DL, Tang
N, Zhao HY, Xu HT, Li QC and Wang EH: Aberrant hypermethylation and
reduced expression of disabled-2 promote the development of lung
cancers. Int J Oncol. 43:1636–1642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu HT, Yang LH, Li QC, Liu SL, Liu D, Xie
XM and Wang EH: Disabled-2 and Axin are concurrently colocalized
and underexpressed in lung cancers. Hum Pathol. 42:1491–1498. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Allison KH, Brogi E, Ellis LO, et al: WHO
Classification of Tumours of Breast Tumors. (5th). IARC. (Lyon,
France). 2019.
|
|
23
|
Gabriel NH, Stephen BE and Armando G: New
and important changes in the TNM staging system for breast cancer.
Am Soc Clin Oncol Educ Book. 38:457–467. 2018.PubMed/NCBI
|
|
24
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:eaan25072017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Lei L, Zheng YW, Zhang L, Li ZH,
Shen HY, Jiang GY, Zhang XP, Wang EH and Xu HT: Odd-skipped related
1 inhibits lung cancer proliferation and invasion by reducing Wnt
signaling through the suppression of SOX9 and β-catenin. Cancer
Sci. 109:1799–1810. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng YW, Zhang L, Wang Y, Chen SY, Lei L,
Tang N, Yang DL, Bai LL, Zhang XP, Jiang GY, et al: Thyroid cancer
1 (C8orf4) shows high expression, no mutation and reduced
methylation level in lung cancers, and its expression correlates
with β-catenin and DNMT1 expression and poor prognosis. Oncotarget.
8:62880–62890. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Zhu JF, Liu YY and Han GP: An
analysis of cyclin D1, cytokeratin 5/6 and cytokeratin 8/18
expression in breast papillomas and papillary carcinomas. Diagn
Pathol. 8:82013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Horvai AE, Kramer MJ and O'Donnell R:
Beta-catenin nuclear expression correlates with cyclin D1
expression in primary and metastatic synovial sarcoma: A tissue
microarray study. Arch Pathol Lab Med. 130:792–798. 2006.PubMed/NCBI
|
|
30
|
Lin L, Hicks D, Xu B, Sigel JE, Bergfeld
WF, Montgomery E, Fisher C, Hartke M, Tubbs R and Goldblum JR:
Expression profile and molecular genetic regulation of cyclin D1
expression in epithelioid sarcoma. Mod Pathol. 18:705–709. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Shi J, Chai K, Ying X and Zhou BP:
The Role of snail in EMT and tumorigenesis. Curr Cancer Drug
Targets. 13:963–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng X, Carstens JL, Kim J, Scheible M,
Kaye J, Sugimoto H, Wu CC, LeBleu VS and Kalluri R:
Epithelial-to-mesenchymal transition is dispensable for metastasis
but induces chemoresistance in pancreatic cancer. Nature.
527:525–530. 2015. View Article : Google Scholar : PubMed/NCBI
|

















