Introduction
Prostate cancer (PCa) is the most common type of
cancer in males and the second most common cause of male
cancer-associated mortality in Western countries (1). It is necessary to stage PCa accurately
prior to surgery, to ensure appropriate treatment options, clinical
decisions and patient counseling are applied. PCa with adverse
pathology tends to be more aggressive and harbors poorer prognosis
(1). Adverse pathology is defined as
≥Grade Group 3 (GG 3)/Gleason score (GS) 4+3=7 radical
prostatectomy (RP), extraprostatic extension (EPE), seminal vesicle
invasion (SVI) or lymph node metastasis (LNM) (2). For patients with suspected EPE or SVI,
surgical techniques may be modified to include, for example, a
wider resection margin, enlarged lymph node dissection or nerve
resection, in order to reduce the risk of positive surgical margin.
In addition, for some patients with EPE or SVI, external beam
radiation therapy may be a more appropriate choice compared with
RP, which may have risks associated with incomplete resection
(3). Multi-parametric magnetic
resonance imaging (mpMRI) and MRI/ultrasound (US) fusion-targeted
biopsy (TBx) are relatively new techniques for PCa detection.
Studies have shown that TBx performs better in detecting clinically
significant PCa (csPCa) compared with systematic biopsy (SB) alone
(2,4). Ahmed et al (5) found that mpMRI had a higher sensitivity
compared with standard transrectal US (TRUS) in ruling out
clinically insignificant diseases. Under prostate imaging reporting
and data system (PI-RADS) version 2 (4), lesions receive an assessment PI-RADS
score of 1–5, with higher scores being associated with a higher
likelihood of clinically significant PCa (csPCa), which is when the
cancer volume is ≥0.5 ml and/or GS ≥3+4 and/or stage ≥pT3 (6). Certain studies have demonstrated that
the GS of PCa biopsy specimens is strongly associated with adverse
pathology at the time of RP, biochemical recurrence and poor
prognosis, with higher GS being associated with a worse treatment
response (5,7). Therefore, high-grade PCa is associated
with a less favorable prognosis and impacts clinical decisions,
such as determining the most appropriate treatment. Gleason pattern
(GP) 4 is a heterogeneous group that is classified into several
architectural patterns, including poorly formed, cribriform and
fused glands, by the International Society of Urological Pathology
(ISUP) (8). Compared with tumors
with a non-cribriform pattern 4 (poorly-formed and fused glands),
tumors with positive cribriform morphology are associated with a
higher likelihood of EPE and metastasis (9,10).
Previous studies have suggested that cribriform morphology may be
more aggressive and can optimize Gleason scoring (6,11). In
addition, studies have demonstrated that cribriform pattern in PCa
is associated with an increased risk of biochemical recurrence and
cancer-specific survival (12,13).
Currently, several clinical nomograms have been
developed, including GG and clinical factors, which can predict
adverse pathology in PCa. However, to the best of our knowledge, no
studies have incorporated pathological factors, such as cribriform
morphology, into a nomogram for adverse pathology prediction. The
aim of the present study was therefore to develop a novel nomogram
incorporating biopsy cribriform morphology, imaging parameters and
clinical factors to identify adverse pathology in PCa.
Materials and methods
Study population
A total of 657 patients who underwent RP at Nanjing
Drum Tower Hospital (Nanjing, China), due to
histologically-confirmed PCa, between August 2016 and March 2019
were retrospectively included in the present study. The following
patients were excluded: i) Patients with histologically-confirmed
PCa from other medical institutions (n=169); ii) GP 5 on biopsy
(n=27); iii) pure GP 3 on biopsy (n=142); iv) patients with a main
lesion of <PI-RADS 3 on MRI (n=6); and v) patients who had
undergone previous treatment, including hormone therapy (n=82) and
transurethral resection of the prostate (n=8; Fig. 1). Finally, 223 patients who had
undergone preoperative mpMRI, had GP 4 and absence of GP 5 on
biopsy specimens (GS 3+4, GS 4+3, GS 4+4) were included in the
present study. The median age was 69 years (age range, 50–84
years). The median age of adverse pathology-negative group and
adverse pathology-positive group were used to determine age
grouping. The study was approved by The Ethics Committee of the
Nanjing Drum Tower Hospital (approval no. 2017-147-01) and all
patients provided informed and signed consent.
mpMRI examination and image
evaluation
All patients underwent pelvic mpMRI using a 3.0-T MR
scanner (Achieva 3.0 TTX; Philips Healthcare) using a 16-channel
phased array coil, as previously described (14). Transverse/coronal/sagittal (18
slices; thickness, 3 mm/gap 0.5 mm; repetition time (TR), 3744 ms;
time of echo (TE), 120 ms; number of signals acquired, 2;
resolution, 1.49×1.51 mm) T2-weighted turbo spin-echo images were
acquired. Diffusion weighted spin-echo echo-planar images (18
slices; thickness; 3 mm; intersection gap, 1 mm; TR, 925/TE, 41 ms;
number of signals acquired, 1; resolution, 3×3 mm; b-factor,
0/800/1500 sec/mm2) were acquired. T1 high-resolution
isotropic volume with fat suppression, following a 30 ml gadolinium
injection was used for dynamic contrast-enhanced images (133
slices; thickness, 3 mm; no intersection gap; TR, 3.1/TE, 1.46 ms;
number of signals acquired, 1; resolution, 1.49×1.51 mm; dynamic
scan time, 00:06.9 min). All mpMRI imaging was reviewed by two
radiologists with >10 years of experience with prostate mpMRI,
who were aware of the PCa diagnosis for all cases but blinded to
the final pathology, including the GS. Regions of interest (ROI),
defined as regions with an abnormal signal on mpMRI, were contoured
and scored using PI-RADS (6). The
maximum length, width and height of the suspected lesion on
apparent-diffusion coefficient sequence were measured, and the
prostate volume was calculated using the following formula: 0.52 ×
width × lenth × height’.
Biopsy protocol and pathological
assessment
All biopsies were conducted using the mpMRI-US image
registration system (Esaote® and RVS®) that
provided real-time fusion of TRUS and mpMR images, in order to
guide the biopsy needles transperineally. The biopsy started with
TB, aiming at the center of suspicious lesions, using the free-hand
transperineal (7) method by a senior
urologist, and then standard systematic 12-core transperineal SB
(blinded to the MRI target lesions) was conducted in all patients
by another dedicated urologist. An 18-G automatic biopsy gun with a
specimen size of 18 mm (Gallini Medical; www.gallinimedical.com) was used to obtain biopsy
cores (14). Biopsy cores were
graded by two genitourinary pathologists at Nanjing Drum Tower
Hospital (Nanjing, China), in accordance with the ISUP guidelines
(15). GG is based on the highest GG
on TB and SB pathological outcomes. The GGs, used in parallel to
the modified Gleason grading system, translate GSs in five distinct
risk categories, where GG 1 is defined as GS 6, GG 2 as GS 3 + 4 =
7, GG 3 as GS 4 + 3 = 7, GG 4 as GS 8 and GG 5 as GS 9/10 (16). Cribriform morphology on biopsy was
identified by genitourinary pathologists at Nanjing Drum Tower
Hospital.
Whole-mount pathological
evaluation
Following robotic-assisted RP, whole-mount tissues
were fixed using 10% formalin, embedded in paraffin, microtome-cut
into 4–5 mm slices and hematoxylin & eosin-stained, according
to the standard protocol (17). All
whole mount histology slides were subsequently digitalized using a
scanning system (NanoZoomer Digital Pathology System; Hamamatsu
Photonics K.K.). All pathological images were interpreted by the
genitourinary pathologists. To identify pathological ROI, tumor
lesions were contoured, and corresponding GS were assigned.
Cribriform morphology was identified by the genitourinary
pathologists at Nanjing Drum Tower Hospital. For primary analyses,
adverse pathology was defined as ≥GG 3/GS 4+3=7 RP, EPE, SVI or LNM
(2).
Statistical analysis
All patient demographics, cribriform morphology on
biopsy, RP pathological results and MRI findings were analyzed
descriptively. Patients with or without adverse pathological
characteristics, cribriform morphology on biopsy, biopsy GG and
PI-RADS score were shown. Mann-Whitney U tests were used for
continuous variables and the χ2 test for categorical
variables. Univariate and multivariate logistic regression analyses
were performed to determine significant predictors of adverse
pathology for nomogram development. The performance of the novel
nomogram in predicting adverse pathology was assessed by receiver
operating characteristics (ROC) curves, area under the curve (AUC)
values and 95% confidence interval (CI), sensitivity and
specificity were calculated. The extent of over- or underestimation
of predicted probabilities relative to observed probabilities of
adverse pathology was explored graphically using calibration plots,
which were internally validated using bootstrapping with 1,000
iterations. Decision curve analysis (DCA) identified the optimal
approach for adverse pathology detection by comparing the net
benefit of each factor across different threshold probabilities.
Statistical analyses were performed using R version 3.5.2 (8) (packages rms, pROC and ggplot 2) and R
studio software [version 1.1.383; (9)]. DCA was performed using the DCA package
(18). All descriptive analyses were
performed using SPSS version 22.0 software (IBM Corp). For tests of
all variables, P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient characteristics
A total of 223 PCa patients, who underwent
preoperative mpMRI, had GP 4 and absence of GP 5 (GS 3+4, GS 4+3,
GS 4+4) in biopsy specimens, were retrospectively enrolled in the
present study. Table I includes the
characteristics of all enrolled patients with PCa. The median age
was 69 years, and the median PSA level was 13.16 ng/ml.
 | Table I.Characteristics of all 223 patients
with prostate cancer included in the present study. |
Table I.
Characteristics of all 223 patients
with prostate cancer included in the present study.
|
Characteristics | Value |
|---|
| Age, years
(range) | 69 (50–84) |
| PSA level, ng/ml
(range) | 13.16
(4.02–110.48) |
| Prostate volume, ml
(range) | 30.1
(8.20–119.00) |
| PSAD, ng/ml
(range) | 0.45
(0.05–3.44) |
| Maximum diameter
on | 1.70
(0.20–4.60) |
| MRI, cm
(range) |
| mpMRI findings, n
(%) |
|
| PI-RADS
score 3 | 23 (10.3) |
| PI-RADS
score 4 | 70 (31.4) |
| PI-RADS
score 5 | 130 (58.3) |
|
Suspected extraprostatic
extension | 113 (50.7) |
|
Suspected seminal vesicle
invasion | 20 (9.0) |
|
Suspected lymph node
invasion | 6
(2.7) |
| Grade Group on RP,
n (%) |
|
| 1 | 2 (0.9) |
| 2 | 97 (43.5) |
| 3 | 89 (39.9) |
| 4 | 17 (7.6) |
| 5 | 18 (8.1) |
| pT stage, n
(%) |
|
| 2 | 93 (41.7) |
| 3a | 97 (43.5) |
| 3b | 31 (13.9) |
| 4 | 2 (0.9) |
| pN stage, n
(%) |
|
| 0 | 208 (0.93) |
| 1 | 15
(0.07) |
| Cribriform
morphology on biopsy, n (%) |
|
|
Negative | 155 (69.5) |
|
Positive | 68
(30.5) |
Characteristics of patients with or
without adverse pathology
The demographics, biopsy GG, PI-RADS score and
cribriform architecture on biopsy with and without adverse
pathology are shown in Table II.
The patients harboring adverse pathology were older compared with
those who did not (71.00 vs. 68.00 years; P=0.04). The PSA level of
patients with adverse pathology was higher compared with patients
without (14.59 vs. 10.20 ng/ml; P<0.01). Compared with patients
without adverse pathology, the maximum lesion diameter was longer
in the MRI of patients with adverse pathology (1.90 vs. 1.20 cm,
P<0.01). The contribution of PI-RADS score and GG on biopsy was
also observed. For PI-RADS score 3 PCa, the detection rates of
adverse pathology were 7/23 (30.4%). In contrast, PI-RADS score 4
PCa was more likely to have adverse pathology [41/70 (58.6%) vs.
7/23 (30.4%) P=0.02]. However, the detection rates of adverse
pathology for PI-RADS score 5 PCa were the highest [110/130 (84.6%)
vs. 7/23 (30.4%); P<0.01]. The present study demonstrated that a
higher preoperative GG on biopsy was associated with a higher
likelihood of adverse pathology in PCa. The prediction rates of
biopsy GG 2, GG 3, GG 4 were 24/59 (40.7%), 57/79 (72.2%) and 77/85
(90.6%), respectively. For cribriform morphology-positive PCa, the
rates of adverse pathology detection were 60/68 (88.2%). By
contrast, cribriform morphology-negative PCa was associated with a
decreased likelihood of adverse pathology [98/155 (63.2%) vs. 60/68
(88.2%); P<0.01]. Representative radiopathological matching of
cribriform morphology-positive lesion on biopsy and RP are shown in
Fig. 2.
 | Table II.Characteristics of patients with or
without adverse pathology in the study (n=158 and 64,
respectively). |
Table II.
Characteristics of patients with or
without adverse pathology in the study (n=158 and 64,
respectively).
|
| Adverse
pathology |
|
|---|
|
|
|
|
|---|
|
Characteristics | Negative | Positive | P-value |
|---|
| Age, years
(range) | 68 (52–81) | 71 (50–84) |
0.04 |
| PSA level, ng/ml
(range) | 10.20
(4.02–100) | 14.59
(4.00–110.48) | <0.01 |
| Prostate volume, ml
(range) | 27.9
(15.8–85.90) | 33.2
(8.20–119.00) |
0.17 |
| PSAD, ng/ml
(range) | 0.34
(0.05–2.83) | 0.49
(0.05–3.44) | <0.01 |
| Maximum diameter on
MRI, cm (range) | 1.20
(0.20–3.80) | 1.90
(0.50–4.60) | <0.01 |
| PI-RADS score, n
(%) |
|
|
|
| 3 | 16 (69.6) | 7 (30.4) |
|
| 4 | 29 (41.4) | 41 (58.6) | 0.02a |
| 5 | 20 (15.4) | 110 (84.6) |
<0.01b |
| Grade Group on
biopsy, n (%) |
|
|
|
| 2 | 35 (59.3) | 24 (40.7) |
|
| 3 | 22 (27.8) | 57 (72.2) |
<0.01c |
| 4 | 8 (9.4) | 77 (90.6) |
<0.01d |
| Cribriform
morphology on biopsy, n (%) |
|
|
|
|
Negative | 57 (36.8) | 98 (63.2) |
<0.01e |
|
Positive | 8 (11.8) | 60 (88.2) |
|
Univariate and multivariate logistic
regression analyses for the detection of adverse pathology
In univariate logistic regression analysis for the
prediction of adverse pathology in PCa, prostate specific antigen
density (PSAD) (P<0.01), maximum lesion diameter on MRI
(P<0.01), cribriform morphology on biopsy (P<0.01), biopsy GG
(P<0.01) and PI-RADS score (P<0.01) were significant factors.
In multivariate logistic regression analysis for the prediction of
adverse pathology, PSAD (P=0.03), cribriform morphology (P=0.02),
biopsy GG (P<0.01) and PI-RADS score (P<0.01) were
significant factors and were included in the nomogram construction
(Table III; Fig. 3). The AUC of the novel nomogram was
0.88 (95% CI, 0.84–0.91), with a high specificity (0.91) and
moderate sensitivity (0.72; Fig. 4).
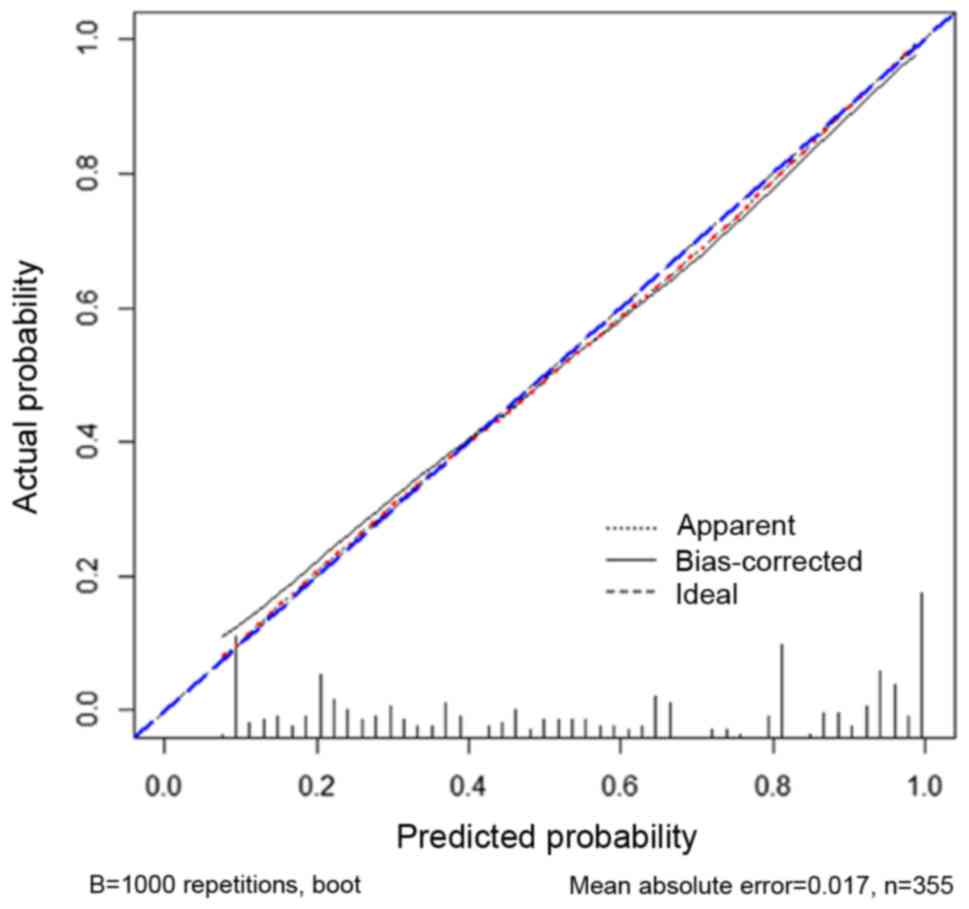
Bootstrapped calibration plots of the nomogram (Fig. 5) demonstrated that there were no
significant deviations of the predicted risk from the observed risk
of adverse pathology in PCa over the entire range (mean absolute
error=0.017). In DCA, compared with PSAD, cribriform morphology on
biopsy, biopsy GG, PI-RADS score, the nomogram had a higher net
benefit for the prediction of adverse pathology (Fig. 6).
 | Table III.Univariate and multivariate logistic
regression analyses for the detection of adverse pathology in
prostate cancer. |
Table III.
Univariate and multivariate logistic
regression analyses for the detection of adverse pathology in
prostate cancer.
|
| Univariate logistic
regression | Multivariate
logistic regression |
|---|
|
|
|
|
|---|
| Variable and
intercept | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Age, per 5
years | 1.09
(0.94–1.26) |
0.24 | NA | NA |
| PSAD, interquartile
OR | 2.26
(1.81–2.81) |
<0.01 | 1.40
(1.03–1.92) | 0.03 |
| Maximum diameter on
MRI, interquartile OR | 2.30
(1.84–2.87) |
<0.01 | 0.85
(0.54–1.32) | 0.49 |
| Cribriform
morphology on biopsy |
|
|
|
|
| No | 1 (Ref) |
| 1 (Ref) |
|
|
Yes | 20.70
(6.34–67.55) | <0.01 | 4.45
(1.24–15.90) | 0.02 |
| Grade Group on
biopsy |
|
|
|
|
| 2 | 1 (Ref) |
| 1 (Ref) |
|
| 3 | 3.14
(1.60–6.17) | <0.01 | 2.05
(0.97–4.36) | <0.01 |
| 4 | 9.00
(4.67–17.35) | <0.01 | 4.39
(2.11–9.17) | <0.01 |
| PI-RADS score |
|
|
|
|
| 3 | 1 (Ref) |
| 1 (Ref) |
|
| 4 | 4.53
(2.12–9.69) | <0.01 | 2.99
(1.27–7.07) | <0.01 |
| 5 | 21.77
(10.03–47.24) | <0.01 | 8.48
(2.47–29.14) | <0.01 |
Discussion
In the present study, a novel nomogram incorporating
PSAD, PI-RADS score, cribriform morphology on biopsy and biopsy GG
was developed, and showed a good prediction performance for PCa
harboring adverse pathology.
Prediction models that combine clinical stage, serum
PSA level and Gleason grade in biopsy specimens are commonly used
in clinical practice to predict the pathological stage of PCa. The
Partin (19) and Memorial Sloan
Kettering Cancer Center pre-RP (20)
nomograms are examples of predicted models widely used for
preoperative decision-making. However, imaging parameters and
pathological factors, such as mpMRI and cribriform pattern, are not
included in the nomogram, which are generally recognized as
valuable factors for improved detection of PCa harboring adverse
pathology. Rayn et al (21)
showed that MRI, alone or combined with standard clinical
nomograms, provides additional predictive value of adverse
pathology at the time of RP. Although MRI was incorporated, the
Rayn et al nomogram had certain limitations. First, PI-RADS
version 2 (v2) was not used to evaluate the effect of MRI
uniformly. PI-RADS v2 is widely accepted by peer experts for MRI
accessing and clinical practice as guidance (6). It was observed in the present study
that a higher PI-RADS score was associated with an increased
likelihood of adverse pathology. Junker et al (22), reported that PI-RADS 4 and PI-RADS 5
were associated with high-grade PCa. Lim et al (23), showed that PI-RADS 5 was associated
with a higher GSs and EPE compared with PI-RADS 4. Secondly,
pathological factors were not included in the nomogram of Rayn
et al. The present study incorporated pathological factors,
including cribriform morphology on biopsy, into the nomogram, and
observed a favorable prediction performance for adverse
pathology.
Sarbay et al (24), reported that cribriform morphology on
prostate biopsy was associated with an increased likelihood of
positive surgical margins and extraprostatic extension at the time
of RP. Another study showed that perineural invasion on biopsy was
associated with adverse pathology at the time of RP; however,
perineural invasion was inferior to cribriform pattern in
predicting non-organ-confined disease at the time of RP (25). Over the past few years, several
studies have shown that cribriform-positive PCa is associated with
an increased likelihood of lymph node invasion and distant
metastasis (10,26). The present study also demonstrated
that the presence of cribriform morphology on biopsy was associated
with an increased risk of adverse pathology in PCa compared with RP
pathological outcomes. Therefore, cribriform morphology on biopsy,
which was incorporated into the present nomogram, is a significant
factor for predicting PCa harboring adverse pathology. Although not
yet practiced clinically, and given that cribriform morphology has
not been extensively studied, reporting cribriform morphology on
prostate biopsies is strongly encouraged by some experts.
Suggestions for accommodating the presence of cribriform cancer
into the 2014 GG scheme have been made (6). The present team has recently been
trying to encourage pathologists to report cribriform morphology on
prostate biopsies at Nanjing Drum Tower Hospital. In addition, a
higher GG was found to be associated with an increased risk of
adverse pathology in PCa compared with RP pathological results.
Aminsharifi et al (27),
showed that a higher preoperative biopsy GG is linked to a higher
risk of adverse histopathological findings at the time of RP.
Hence, for a patient with a high biopsy GG and positive cribriform
morphology on biopsy, a careful evaluation should be conducted to
ensure suitable treatment strategies are followed.
A strength of the present study was that cribriform
morphology on prostate biopsy was incorporated into the nomogram.
To the best of our knowledge, the present study is the first to
incorporate pathological factors into the nomogram predicting
adverse pathology in PCa. In addition, the final pathological
results were based on whole-mount pathological analyses. However,
the present study had certain limitations. Firstly, it was a
retrospective study and therefore has the bias associated with this
type of study. Secondly, only a limited number of patients fitted
the inclusion criteria. Thirdly, the final pathology was used as a
reference standard, thus a selection bias is likely to have
occurred, in that patients with lower-risk disease are less likely
to have RP and negative- or low-risk patients were not included.
Nonetheless, the final RP specimen is the most accurate reference
standard to determine the presence or absence of PCa harboring
adverse pathology. Fourthly, according to the study aim, only
patients with a GP of 4 and without pure GP 3 and GP 5 were
enrolled in the study, which might restrict clinical application to
the general population. However, Stroup et al (28) showed that any GP 5 on biopsy, which
indicated a poor prognosis, is associated with a higher likelihood
of metastasis and PCa-specific mortality and adverse outcomes. In
addition, an increasing number of studies have demonstrated that
pure GP 3 diseases on biopsy have negligible PCa-specific mortality
(29) and metastasis (30). Therefore, the inclusion criteria for
the present study were considered suitable and in line with
clinical practice. Finally, because of short postoperative time,
follow-up of patients to evaluate the predictive value of the
nomogram in biochemical recurrence was not conducted. Therefore,
future studies should be conducted to verify the results of the
present imaging models.
The present study found that a novel nomogram
incorporating PSAD, PI-RADS score, cribriform morphology on biopsy
and biopsy GG provided a significant predictive ability for PCa
harboring adverse pathology at the time of RP. Urologists can use
this nomogram to counsel patients regarding surgery techniques and
future therapies and help them make significant management
decisions. In future, the present nomogram should be validated in
an independent cohort.
Acknowledgements
The authors of the present study would like to thank
Dr Guo at the Department of Urology, Nanjing Drum Tower Hospital,
The Affiliated Hospital of Nanjing University Medical School
(Nanjing, China) for his help in project development, analyzing the
data and editing the manuscript.
Funding
This research was supported by The National Natural
Science Foundation of China (grant nos. 81772710 and 81572519), The
Project of Invigorating Health Care through Science, Technology and
Education, Jiangsu Provincial Key Medical Discipline (grant no.
ZDXKB2016014), The National Natural Science Foundation of China
(grant no. 81802535), China Postdoctoral Fund (grant no. 223427)
and The Nanjing Medical Science and Technique Development
Foundation (grant no. YKK 18064).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BW, JG and QZ conceived the study and contributed
equally to data collection, data analysis, manuscript writing and
editing. DL performed radiological analysis. CZ and GL performed
clinical analyses associated with introducing the concept of csPCa,
the Gleason group, hormone therapy and prostate biopsy. WW and HH
performed MRI/ultrasound fusion-targeted biopsy and systematic
biopsy. YF and JS performed pathological analysis. HG was involved
in project development, data analysis and edited the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of the Nanjing Drum Tower Hospital (approval no.
2017-147-01) and all patients provided informed and signed
consent.
Patient consent for publication
Consent for publication was been obtained for each
participant according to federal and institutional guidelines
(14,31).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Center MM, Jemal A, Lortet-Tieulent J,
Ward E, Ferlay J, Brawley O and Bray F: International variation in
prostate cancer incidence and mortality rates. Eur Urol.
61:1079–1092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dean LW, Assel M, Sjoberg DD, Vickers AJ,
Al-Ahmadie HA, Chen YB, Gopalan A, Sirintrapun SJ, Tickoo SK,
Eastham JA, et al: Clinical usefulness of total length of Gleason
pattern 4 on biopsy in men with grade group 2 prostate cancer. J
Urol. 201:77–82. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boehmer D, Maingon P, Poortmans P, Baron
MH, Miralbell R, Remouchamps V, Scrase C, Bossi A and Bolla M;
EORTC radiation oncology group, : Guidelines for primary
radiotherapy of patients with prostate cancer. Radiother Oncol.
79:259–269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siddiqui MM, Rais-Bahrami S, Turkbey B,
George AK, Rothwax J, Shakir N, Okoro C, Raskolnikov D, Parnes HL,
Linehan WM, et al: Comparison of MR/ultrasound fusion-guided biopsy
with ultrasound-guided biopsy for the diagnosis of prostate cancer.
JAMA. 313:390–397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ahmed HU, El-Shater Bosaily A, Brown LC,
Gabe R, Kaplan R, Parmar MK, Collaco-Moraes Y, Ward K, Hindley RG,
Freeman A, et al: Diagnostic accuracy of multi-parametric MRI and
TRUS biopsy in prostate cancer (PROMIS): A paired validating
confirmatory study. Lancet. 389:815–822. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weinreb JC, Barentsz JO, Choyke PL, Cornud
F, Haider MA, Macura KJ, Margolis D, Schnall MD, Shtern F, Tempany
CM, et al: PI-RADS prostate imaging-reporting and data system:
2015, version 2. Eur Urol. 69:16–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou P, Chen MH, McLeod D, Carroll PR,
Moul JW and D'Amico AV: Predictors of prostate cancer-specific
mortality after radical prostatectomy or radiation therapy. J Clin
Oncol. 23:6992–6998. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Epstein JI, Allsbrook WC Jr, Amin MB,
Egevad LL and Committee IG; ISUP Grading Committee, : The 2005
international society of urological pathology (ISUP) consensus
conference on Gleason grading of prostatic carcinoma. Am J Surg
Pathol. 29:1228–1242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kweldam CF, Wildhagen MF, Steyerberg EW,
Bangma CH, van der Kwast TH and van Leenders GJ: Cribriform growth
is highly predictive for postoperative metastasis and
disease-specific death in Gleason score 7 prostate cancer. Mod
Pathol. 28:457–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Siadat F, Sykes J, Zlotta AR, Aldaoud N,
Egawa S, Pushkar D, Kuk C, Bristow RG, Montironi R and van der
Kwast T: Not all Gleason pattern 4 prostate cancers are created
equal: A study of latent prostatic carcinomas in a
cystoprostatectomy and autopsy series. Prostate. 75:1277–1284.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McKenney JK, Wei W, Hawley S, Auman H,
Newcomb LF, Boyer HD, Fazli L, Simko J, Hurtado-Coll A, Troyer DA,
et al: Histologic grading of prostatic adenocarcinoma can be
further optimized: Analysis of the relative prognostic strength of
individual architectural patterns in 1275 patients from the canary
retrospective Cohort. Am J Surg Pathol. 40:1439–1456. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kir G, Sarbay BC, Gümüş E and Topal CS:
The association of the cribriform pattern with outcome for
prostatic adenocarcinomas. Pathol Res Pract. 210:640–644. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harding-Jackson N, Kryvenko ON,
Whittington EE, Eastwood DC, Tjionas GA, Jorda M and Iczkowski KA:
Outcome of Gleason 3+5=8 prostate cancer diagnosed on needle
biopsy: Prognostic comparison with Gleason 4+4=8. J Urol.
196:1076–1081. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Q, Wang W, Zhang B, Shi J, Fu Y, Li
D, Guo S, Zhang S, Huang H, Jiang X, et al: Comparison of free-hand
transperineal mpMRI/TRUS fusion-guided biopsy with transperineal
12-core systematic biopsy for the diagnosis of prostate cancer: A
single-center prospective study in China. Int Urol Nephrol.
49:439–448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Epstein JI, Egevad L, Amin MB, Delahunt B,
Srigley JR and Humphrey PA; Grading Committee, : The 2014
international society of urological pathology (ISUP) consensus
conference on Gleason grading of prostatic carcinoma: Definition of
grading patterns and proposal for a new grading system. Am J Surg
Pathol. 40:244–252. 2016.PubMed/NCBI
|
|
16
|
Magi-Galluzzi C, Montironi R and Epstein
JI: Contemporary Gleason grading and novel grade groups in clinical
practice. Curr Opin Urol. 26:488–492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McNeal JE and Haillot O: Patterns of
spread of adenocarcinoma in the prostate as related to cancer
volume. Prostate. 49:48–57. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vickers AJ, Cronin AM, Elkin EB and Gonen
M: Extensions to decision curve analysis, a novel method for
evaluating diagnostic tests, prediction models and molecular
markers. BMC Med Inform Decis Mak. 8:532008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang Y, Isharwal S, Haese A, Chun FK,
Makarov DV, Feng Z, Han M, Humphreys E, Epstein JI, Partin AW and
Veltri RW: Prediction of patient-specific risk and percentile
cohort risk of pathological stage outcome using continuous
prostate-specific antigen measurement, clinical stage and biopsy
Gleason score. BJU Int. 107:1562–1569. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ohori M, Kattan MW, Koh H, Maru N, Slawin
KM, Shariat S, Muramoto M, Reuter VE, Wheeler TM and Scardino PT:
Predicting the presence and side of extracapsular extension: A
nomogram for staging prostate cancer. J Urol. 171:1844–1849. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rayn KN, Bloom JB, Gold SA, Hale GR,
Baiocco JA, Mehralivand S, Czarniecki M, Sabarwal VK, Valera V,
Wood BJ, et al: Added value of multiparametric magnetic resonance
imaging to clinical nomograms for predicting adverse pathology in
prostate cancer. J Urol. 200:1041–1047. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Junker D, Quentin M, Nagele U, Edlinger M,
Richenberg J, Schaefer G, Ladurner M, Jaschke W, Horninger W and
Aigner F: Evaluation of the PI-RADS scoring system for mpMRI of the
prostate: A whole-mount step-section analysis. World J Urol.
33:1023–1030. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lim CS, McInnes MDF, Lim RS, Breau RH,
Flood TA, Krishna S, Morash C, Shabana WM and Schieda N: Prognostic
value of prostate imaging and data reporting system (PI-RADS) v. 2
assessment categories 4 and 5 compared to histopathological
outcomes after radical prostatectomy. J Magn Reson Imaging.
46:257–266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sarbay BC, Kir G, Topal CS and Gumus E:
Significance of the cribriform pattern in prostatic
adenocarcinomas. Pathol Res Pract. 210:554–557. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Flood TA, Schieda N, Keefe DT, Morash C,
Bateman J, Mai KT, Belanger EC, Robertson SJ and Breau RH:
Perineural invasion on biopsy is associated with upstaging at
radical prostatectomy in Gleason score 3+4=7 prostate cancer.
Pathol Int. 66:629–632. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong F, Yang P, Wang C, Wu S, Xiao Y,
McDougal WS, Young RH and Wu CL: Architectural heterogeneity and
cribriform pattern predict adverse clinical outcome for Gleason
grade 4 prostatic adenocarcinoma. Am J Surg Pathol. 37:1855–1861.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Aminsharifi A, Schulman A, Howard LE, Tay
KJ, Amling CL, Aronson WJ, Cooperberg MR, Kane CJ, Terris MK,
Freedland SJ and Polascik TJ: Influence of African American race on
the association between preoperative biopsy grade group and adverse
histopathologic features of radical prostatectomy. Cancer.
125:3025–3032. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stroup SP, Moreira DM, Chen Z, Howard L,
Berger JH, Terris MK, Aronson WJ, Cooperberg MR, Amling CL, Kane CJ
and Freedland SJ: Biopsy detected Gleason pattern 5 is associated
with recurrence, metastasis and mortality in a cohort of men with
high risk prostate cancer. J Urol. 198:1309–1315. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eggener SE, Scardino PT, Walsh PC, Han M,
Partin AW, Trock BJ, Feng Z, Wood DP, Eastham JA, Yossepowitch O,
et al: Predicting 15-year prostate cancer specific mortality after
radical prostatectomy. J Urol. 185:869–875. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu JJ, Lichtensztajn DY, Gomez SL, Sieh
W, Chung BI, Cheng I and Brooks JD: Nationwide prevalence of lymph
node metastases in Gleason score 3+3=6 prostate cancer. Pathology.
46:306–310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao J, Zhang C, Zhang Q, Fu Y, Zhao X,
Chen M, Zhang B, Li D, Shi J, Wang F and Guo H: Diagnostic
performance of 68Ga-PSMA PET/CT for identification of
aggressive cribriform morphology in prostate cancer with
whole-mount sections. Eur J Nucl Med Mol Imaging. 46:1531–1541.
2019. View Article : Google Scholar : PubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBI
|