Introduction
Gastric cancer is a major cause of cancer-associated
mortality. It ranks as the 5th most common neoplasm and the 3rd
most deadly cancer, and was responsible for >1,000,000 new cases
with an estimated 783,000 deaths in 2018 (1). However, analysis of the clinical and
genetic characteristics of gastric cancer has been complicated by
its etiological and histological heterogeneity (2). Etiologically, gastric cancer is often
accompanied by infectious agents such as Helicobacter pylori
or Epstein-Barr virus, susceptible genetic variants and
environmental factors, along with the accumulation of epigenetic
and genetic changes (3).
Histopathological classification systems such as the Lauren's
classification and World Health Organization classification system
(4) have limited clinical usefulness
as to which classifications unify a clinical correlation with a
high validity and practicability in diagnosis and prognostic
outcome, making the development of relevant classifiers that can
help patient care an urgent priority (5). Thus, molecular classification of
gastric cancer has been developed, and candidate drivers and
dysregulated pathways of notable subtypes of gastric cancer have
been identified (6). Several
molecular targeted therapies associated with survival outcomes in
other cancer types are currently in clinical research for the
treatment of gastric cancer, including the inhibitors of epidermal
growth factor receptor (EGFR), fibroblast growth factor receptor
(FGFR), Met proto-oncogene receptor tyrosine kinase (MET),
phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) and vascular
endothelial growth factor (VEGF) (7).
The application of next-generation sequencing (NGS)
technologies exploiting whole genome sequencing to targeted
sequencing has served an important role in the identification of
genetic variations and anomalies in patients with gastric cancer,
which has improved our understanding of the molecular profiles and
heterogeneity of gastric cancers (1,2).
Targeted NGS represents a resource- and cost-efficient approach,
enabling the detection of somatic alterations of potential
interest. The Oncomine Focus Assay (OFA) is a targeted NGS assay
for the simultaneous and rapid identification of single nucleotide
variants (SNVs), short insertions and deletions (indels; 35 genes),
copy number variations (CNVs; 19 genes) and gene rearrangements (23
genes) in 52 cancer genes with therapeutic relevance, and can
detect potential targets and current actionable genetic variants
for personalized medicine (8). The
OFA is designed for use with the Ion Torrent Personal Genome
Machine (PGM™) that generates 10–100 Mb pairs (Mbp) of sequence
data on various chips within several hours of instrument run time
and leverages the uniquely minimal total of DNA or RNA input (10
ng), which is useful for frequent analysis of small amounts of
clinical samples (9). Combined with
Ion AmpliSeq technology, this approach enables highly accurate and
reproducible sequence analysis of various types of tumor specimens
(10).
The present study aimed to compare the
clinicopathological characteristics with the mutation profiles of
50 Korean patients with advanced gastric cancer (AGC) by targeted
NGS assay along with the OFA panel.
Materials and methods
DNA isolation and quantification
The study protocol was approved by the Institutional
Review Board of The Catholic University of Korea (approval no.
DC18SESI0113). All subjects provided written informed consent for
clinical and molecular analyses and publication before the study. A
total of 50 patients with AGC who received surgical resection
between January 2015 and February 2019 at the Department of
Surgery, Chungnam National University Hospital (Daejeon, Republic
of Korea) were enrolled in the present study. The cohort comprised
of 72% (36/50) male and 28% (14/50) female Korean patients with a
mean age of 66 years (age range, 39–91 years). Genomic DNA was
isolated from 50 frozen human AGC tissues using the RecoverAll
Total Nucleic Acid Isolation kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Amplifiable genomic
DNA was determined by fluorometric quantitation using Qubit 2.0
Fluorometer with Qubit dsDNA HS Assay kits and the TaqMan RNase P
Detection Reagents kit (Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocols and was considered appropriate when
the nucleic acid concentration was >30 ng/µl.
Library preparation
DNA libraries were constructed using the Ion
AmpliSeq™ Library kit 2.0 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The Oncomine Focus
DNA Assay (Thermo Fisher Scientific, Inc.) was used to generate
sequencing libraries from 10 ng input genomic DNA per specimen. The
OFA panel can identify hotspot mutations, including SNVs, indels
(35 genes) and CNVs (19 genes) that are commonly implicated in
human cancers and relevant to targeted treatment of solid tumors
(9). Unique Ion Xpress Barcode 1–16
and Ion P1 Adapter (Thermo Fisher Scientific, Inc.) were ligated to
the amplicons and subsequently purified to ensure that each
individual sample had a unique ID. The final amplicon libraries
were amplified, purified and equalized to ~100 pM using an AMPure
XP Reagent (Beckman Coulter, Inc.).
Semiconductor sequencing
A total of six uniquely barcoded library samples
were pooled for sequencing per run on an Ion 318™ v2 chip (Thermo
Fisher Scientific, Inc.). The Ion Chef™ System (Thermo Fisher
Scientific, Inc.) was applied using the Ion PGM™ Hi-Q™ Chef Kit for
fully automated template preparation and Ion 318™ v2 chip loading.
Single-end sequence analysis was performed using the Ion PGM™ Hi-Q™
Sequencing Kit on the Ion Torrent PGM™ (Thermo Fisher Scientific,
Inc.) for 200 base-read sequencing.
Variant calling and data analysis
Raw data from the DNA panel was generated for
sequence reads, collected, processed and trimmed using the Ion
Torrent platform-specific pipeline software as follows. Removal of
polyclonal and poor signal profile reads as well as 3′ quality
trimming of reads was performed using Torrent Suite Assay
Development Mode v5.0 (Thermo Fisher Scientific, Inc.). Reads were
aligned to the human genome hg19 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/)
and Ion Reporter v5.1 software package (Thermo Fisher Scientific,
Inc.) was used for data analysis of the DNA panel. A cut off of
500× coverage was applied to all analyses in the present study; the
target regions with >500× demonstrated sufficient and uniform
amplification and sequencing coverage, with mutant alleles detected
at >5% allele frequency. Briefly, the ‘Oncomine
Focus-520-w2.4-DNA-Single Sample’ automatic workflow in Ion
Reporter was used to identify and annotate the SNVs, indels and
CNVs from the OFA with the following Torrent Variant Caller
parameter settings: Frequency cutoff for supporting a variant, SNV
0.04, indel 0.07, Hotspot 0.03; total coverage required of reads or
no-call, SNV 15, indel 15, Hotspot 15; proportion of variant
alleles coming overwhelmingly from one strand, SNV 0.96, indel 0.9,
Hotspot 0.96 for SNV and indel calls; and median of the absolute
values of all pairwise differences <0.4; 5% confidence interval
CNV ploidy ≥ gain of 2 over normal for CNV calling.
Candidate variant prioritization
Pathogenic impact of missense mutations other than
hotspot mutations on gene function was estimated using in
silico prediction tools such as ‘Damaging’ (score 0) by SIFT
(11) and ‘Probably damaging’ (score
>0.8) by Polyphen-2 (12).
Conservation change of an affected amino acid was compared by
aligning protein sequences of various vertebrate species obtained
from the Evolutionary Annotation Database (http://www.h-invitational.jp/evola/). In addition, the
candidate mutation was investigated whether it has been reported as
pathogenic for gastric cancer in the sequence databases including
COSMIC (https://cancer.sanger.ac.uk/cosmic) (13) or ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
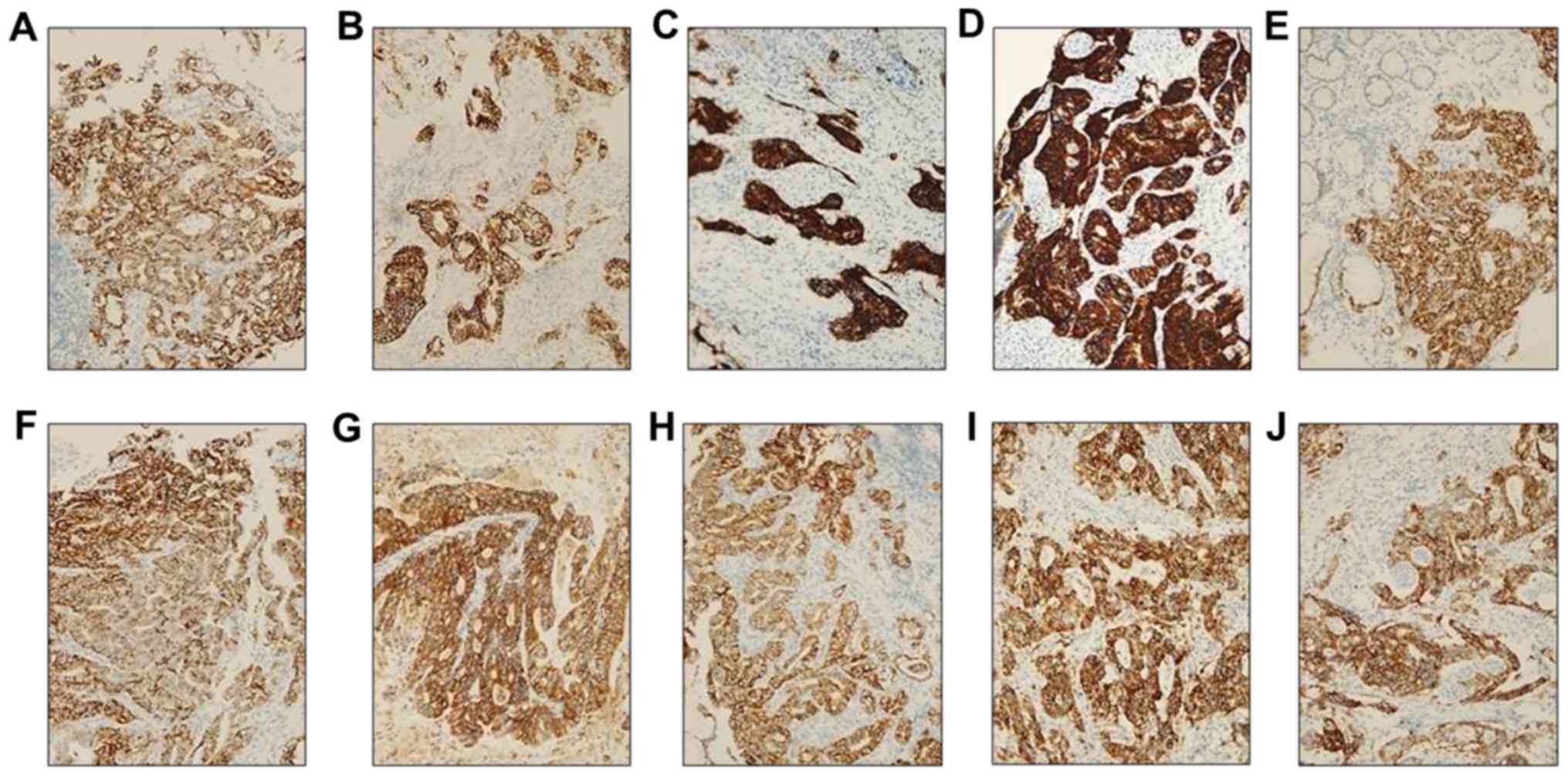
Immunohistochemistry (IHC) for CDK4,
EGFR, ERBB2, FGFR2, KRAS, MET, MYC and PIK3CA
A total of 10 tissue samples with the gene
amplification identified by the OFA assay were fixed in buffered
10% formalin at 65°C for 10 min and embedded in paraffin.
Formalin-fixed, paraffin-embedded (FFPE) samples were sectioned at
a thickness of 4 µm. The BenchMark XT automated slide processing
system (Ventana Medical Systems, Inc.) was used for
deparaffinization pretreatment with Cell Conditioning 1 solution
(Ventana) and ultraviolet irradiation to abrogate endogenous
hydroperoxidase activity according to the manufacturer's
instructions. These sections were incubated at 37°C for 24 min with
primary antibodies (1:100; Abcam) against CDK4 (cat. no. ab108357),
EGFR (cat. no. ab52894), ERBB2 (cat. no. ab16662), FGFR2 (cat. no.
ab58201), KRAS (cat. no. ab180772), MET (cat. no. ab216574), MYC
(cat. no. ab32072), PIK3CA (cat. no. ab40776). Sections, were then
incubated with horseradish peroxidase-conjugated goat anti-rabbit
IgG heavy & light chain secondary antibody (Abcam) at 37°C for
10 min. Sections were counterstained with hematoxylin II (Ventana)
for 5 min and bluing reagent (Ventana) for 5 min at 37°C. Slides
were imaged under a light microscope (BX51; Olympus Corporation).
The intensity of immunostaining for protein expression was scored
as follows: 0, negative; 1+, weak; 2+, moderate; and 3+, strong in
>10% of tumor cells; only 2+ or 3+ were interpreted as being
positive as previously described (14).
Statistical analysis
Data were presented as the means ± standard
deviation. Statistical analysis was performed using MedCalc
Statistical Software Version 17.6 (MedCalc Software, Ltd.).
Normality was assessed using Kolmogorov-Smirnov and Shapiro-Wilk
tests, and one-way analysis of variance followed by Tukey's post
hoc test was used to compare the means of age between three groups
categorized by Lauren's classification or mutation profiles by
targeted NGS. The Fisher's exact test was used to compare the
clinicopathological characteristics and mutation profiles between
two or three groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Mutation analysis
A median sequencing coverage depth of 1,845× (range,
129–2,000×) was achieved for the 50 gastric cancer tissues.
Integrative analysis using the Ion Reporter identified somatic
mutations with allele frequencies between 6 and 47% in tumor DNA
samples without matched normal controls, in which a variant was
classified as germline origin if its mutation frequency was near
50% (heterozygous) or 100% (homozygous), or otherwise classified as
somatic. Null mutations (nonsense, frameshift or canonical ±1 or 2
splice sites) and missense variants with allele frequency
<0.00001 predicted to be deleterious or damaging that were not
registered in COSMIC database were also included. After applying
stringent parameters for reliable variant calling (coverage depth
>500×; allele frequency >5%) by filtering out unlikely
pathogenic variants or potential raw base calling errors, at least
one somatic alteration including SNVs, indels and amplification was
detected in 22/50 (44%) patients. Details of the somatic alteration
profiles identified by targeted NGS in these 22 cases of AGC are
summarized in Tables I and II. Of the 35 hotspot genes and 19 genes
for copy number variations (CNVs), 18 single nucleotide variants
(SNVs) or small indels (14 missense and four frameshift mutations)
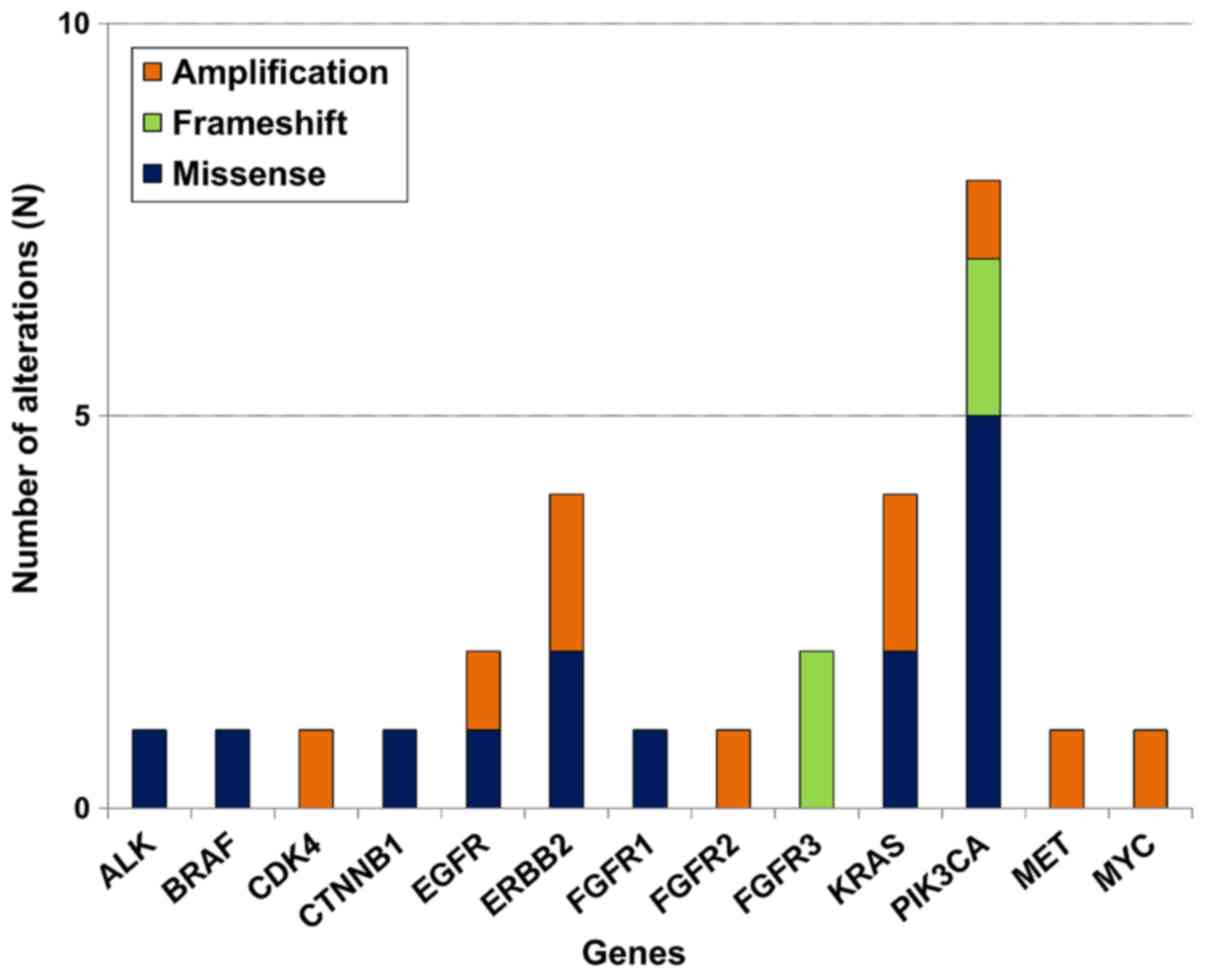
and 10 amplifications were identified (Fig. 1). Amplification of the CDK4, EGFR,
ERBB2, FGFR2, KRAS, MET, MYC and PIK3CA genes was
confirmed to be graded as ≥2 by additional IHC (Fig. 2). Somatic alterations in the
PIK3CA gene were the most frequently identified genetic
alteration, occurring in 8/22 (36%) samples, followed by four
ERBB2, four KRAS, two EGFR, two FGFR3
and one each for the other tested genes (Fig. 3).
 | Table I.Results of somatic alteration
profiles identified by the Oncomine focus assay in 22 patients with
advanced gastric cancer. |
Table I.
Results of somatic alteration
profiles identified by the Oncomine focus assay in 22 patients with
advanced gastric cancer.
| Sample | Gene | Transcript | Base change | Codon change | Effect | Freq % | COSMIC ID | dbSNP |
|---|
| CN03 | ERBB2 | NM_004448.3 | c.2524G>A | p.Val842Ile | Missense | 10 | COSM14065 | rs1057519738 |
| CN11 | EGFR | NM_005228.4 | c.2227G>A | p.Ala743Thr | Missense | 22 | – | rs759256622 |
| CN11 | KRAS | NM_033360.3 | c.38G>A | p.Gly13Asp | Missense | 47 | COSM532 | rs112445441 |
| CN13 | FGFR3 | NM_000142.4 | c.274delC | p.Gln92Serfs*6 | Frameshift | 28 | – | – |
| CN14 | ALK | NM_004304.4 | c.4061G>T | p.Cys1354Phe | Missense | 16 | – | rs963770969 |
| CN15 | PIK3CA | NM_006218.3 |
c.328_330delGAA | p.Glu109del | Frameshift | 41 | COSM24710 | rs1060500031 |
| CN16 | PIK3CA | NM_006218.3 | c.323G>A | p.Arg108His | Missense | 27 | COSM27497 | rs886042002 |
| CN23 | PIK3CA | NM_006218.3 |
c.328_330delGAA | p.Glu109del | Frameshift | 20 | COSM24710 | – |
| CN27 | ERBB2 | NM_004448.3 | c.2033G>A | p.Arg678Gln | Missense | 9 | COSM436498 | rs1057519862 |
| CN28 | PIK3CA | NM_006218.3 | c.1633G>A | p.Glu545Lys | Missense | 8 | COSM763 | rs104886003 |
| CN34 | CTNNB1 | NM_001904.3 | c.98C>T | p.Ser33Phe | Missense | 6 | COSM5669 | rs121913400 |
| CN38 | FGFR1 | NM_001174067.1 | c.2359C>T | p.Arg787Cys | Missense | 20 | – | – |
| CN40 | KRAS | NM_033360.3 | c.34G>A | p.Gly12Ser | Missense | 24 | COSM517 | rs121913530 |
|
| PIK3CA | NM_006218.3 | c.1633G>A | p.Glu545Lys | Missense | 7 | COSM763 | rs104886003 |
| CN44 | FGFR3 | NM_000142.4 | c.274delC | p.Gln92Serfs*6 | Frameshift | 18 | – | – |
|
| PIK3CA | NM_006218.3 | c.1390T>G | p.Ser464Ala | Missense | 39 | – | – |
| CN46 | PIK3CA | NM_006218.3 | c.1633G>A | p.Glu545Lys | Missense | 23 | COSM763 | rs104886003 |
| CN49 | BRAF | NM_004333.4 | c.1780G>A | p.Asp594Asn | Missense | 6 | COSM27639 | rs397516896 |
 | Table II.Results of copy number variations
identified by the Oncomine Focus DNA Assay in 22 patients with
advanced gastric cancer. |
Table II.
Results of copy number variations
identified by the Oncomine Focus DNA Assay in 22 patients with
advanced gastric cancer.
| Sample | Gene | Length, kb | Variant class | CytoBand |
|---|
| CN02 | MYC | 4.4 | Amplification |
8q24.21(128,748,885-128,753,261)×13.51 |
| CN14 | ERBB2 | 15.1 | Amplification |
17q12(37,868,126-37,883,249)×24.58 |
| CN16 | ERBB2 | 15.1 | Amplification |
17q12(37,868,126-37,883,249)×14.25 |
| CN17 | FGFR2 | 107.0 | Amplification |
10q26.13(123,247,505-123,354,466)×13.78 |
| CN18 | PIK3CA | 35.4 | Amplification |
3q26.32(178,916,683-178,952,097)×13.23 |
| CN20 | KRAS | 35.5 | Amplification |
12p12.1(25,364,761-25,400,274)×19.43 |
| CN30 | CDK4 | 4.0 | Amplification |
12q14.1(58,142,052-58,146,026)×14.39 |
| CN38 | MET | 121.0 | Amplification |
7q31.2(116,313,480-116,434,565)×9.82 |
| CN41 | EGFR | 60.6 | Amplification |
7p11.2(55,198,956-55,259,538)×15.35 |
| CN42 | KRAS | 35.5 | Amplification |
12p12.1(25,364,761-25,400,274)×14.69 |
Comparison of clinicopathological
characteristics according to Lauren's classification
The 50 AGC cases were categorized into three
subtypes according to Lauren's classification: Diffuse, intestinal
and mixed type, and the clinicopathological characteristics and
mutation profiles of patients in these three groups were compared.
The mixed type was more frequently associated with a younger age
compared with the diffuse or intestinal type (P=0.003; Table III). By contrast, the intestinal
type more frequently exhibited moderate differentiation compared
with the diffuse or mixed type (P<0.001). The frequency of
mutations identified by targeted NGS with the OFA was not
statistically different among the three groups (P=0.240).
 | Table III.Comparison of clinicopathologic
findings according to Lauren's classification subtypes in 50
patients with advanced gastric cancer. |
Table III.
Comparison of clinicopathologic
findings according to Lauren's classification subtypes in 50
patients with advanced gastric cancer.
| Characteristic | Diffuse (n=15) | Intestinal
(n=30) | Mixed (n=5) | P-value |
|---|
| Male | 8 (53%) | 24 (80%) | 4 (80%) | 0.138 |
| Age, years
(range) | 62 (39–91) | 71 (52–89) | 56 (53–59) | 0.003 |
|
Differentiation |
|
|
| <0.001 |
|
Moderate | 0 (0%) | 24 (80%) | 1 (20%) |
|
|
Poor | 15 (100%) | 6 (20%) | 4 (80%) |
|
| TNM T stage |
|
|
| 0.830 |
| 2 | 1 (7%) | 3 (10%) | 0 (0%) |
|
| 3 | 3 (20%) | 9 (30%) | 1 (20%) |
|
| 4a | 11 (73%) | 18 (60%) | 4 (80%) |
|
| TNM N stage |
|
|
| 0.378 |
| 0 | 3 (20%) | 7 (23%) | 0 (0%) |
|
| 1 | 1 (7%) | 9 (30%) | 1 (20%) |
|
| 2 | 4 (27%) | 7 (23%) | 1 (20%) |
|
| 3a | 5 (33%) | 7 (23%) | 2 (40%) |
|
| 3b | 2 (13%) | 0 (0%) | 1 (20%) |
|
| TNM M stage |
|
|
| 0.088 |
| 0 | 14 (93%) | 30 (100%) | 4 (80%) |
|
| 1 | 1 (7%) | 0 (0%) | 1 (20%) |
|
| Any mutations (SNV
+ indel) |
|
|
| 0.240 |
| No | 13 (87%) | 19 (63%) | 3 (60%) |
|
|
Yes | 2 (13%) | 11 (37%) | 2 (40%) |
|
| Any
amplifications |
|
|
| 1.000 |
| No | 12 (80%) | 24 (80%) | 4 (80%) |
|
|
Yes | 3 (20%) | 6 (20%) | 1 (20%) |
|
| Any mutations or
amplifications |
|
|
| 0.522 |
| No | 10 (67%) | 16 (53%) | 2 (40%) |
|
|
Yes | 5 (33%) | 14 (47%) | 3 (60%) |
|
Comparison of clinicopathological
characteristics according to mutation profiles
To examine the association between mutation profiles
and clinicopathological characteristics, each element in the
clinicopathological characteristics was categorized into three
groups: No alteration, PIK3CA alterations and alterations
other than PIK3CA. By Fisher's exact test, there were no
statistical differences between clinicopathological findings except
TNM staging (T) between the three groups (Table IV). AGCs with somatic alterations
but no PIK3CA showed statistical difference in TNM staging
(T), compared to AGCs without or with PIK3CA alterations
(P=0.044). In addition, AGC with the PIK3CA alterations was
categorized by Lauren's classification to the intestinal type only.
The distribution of Lauren's classification in AGC with
PIK3CA alterations was statistically different compared with
AGC with alterations other than PIK3CA (P=0.028), but not
with AGC with no alterations (P=0.076).
 | Table IV.Comparison of clinicopathologic
characteristics according to mutation profiles identified by
Oncomine focus assay. |
Table IV.
Comparison of clinicopathologic
characteristics according to mutation profiles identified by
Oncomine focus assay.
|
Characteristics | No alterations
(n=28) | Other
alterationsa
(n=14) | PIK3CA
alterations (n=8) | P-value |
|---|
| Male | 19 (68%) | 9 (64%) | 8 (100%) | 0.152 |
| Age, years
(range) | 62 (39–84) | 64 (53–91) | 77 (52–89) | 0.156 |
|
Differentiation |
|
|
| 0.675 |
|
Moderate | 14 (50%) | 6 (43%) | 5 (63%) |
|
|
Poor | 14 (50%) | 8 (57%) | 3 (37%) |
|
| TNM T stage |
|
|
| 0.044 |
| 2 | 1 (4%) | 3 (21%) | 0 (0%) |
|
| 3 | 11 (39%) | 1 (7%) | 1 (12%) |
|
| 4a | 16 (57%) | 10 (72%) | 7 (88%) |
|
| TNM N stage |
|
|
| 0.892 |
| 0 | 5 (18%) | 3 (21%) | 2 (25%) |
|
| 1 | 6 (21%) | 3 (21%) | 2 (25%) |
|
| 2 | 5 (18%) | 5 (37%) | 2 (25%) |
|
|
3a/ | 10 (36%) | 2 (14%) | 2 (25%) |
|
| 3b | 2 (7%) | 1 (7%) | 0 (0%) |
|
| TNM M stage |
|
|
| 0.702 |
| 0 | 27 (96%) | 13 (93%) | 8 (100%) |
|
| 1 | 1 (4%) | 1 (7%) | 0 (0%) |
|
| Lauren's
classification |
|
|
| 0.028b |
|
|
|
|
| 0.076c |
|
Diffuse | 10 (36%) | 5 (36%) | 0 (0%) |
|
|
Intestinal | 16 (57%) | 6 (43%) | 8 (100%) |
|
|
Mixed | 2 (7%) | 3 (21%) | 0 (0%) |
|
Discussion
Molecular characterization of gastric cancer may
offer new tools for effective therapeutic strategies for
well-defined sets of patients, as well as new clinical trial
designs leading to an improvement of medical management of this
disease (15). These novel
classifications allow the identification of relevant gastric cancer
genomic subsets by using techniques such as genomic screening,
functional studies and molecular or epigenetic characterization
(16). The large scale study of
molecular profiling on gastric cancer in The Cancer Genome Atlas
(TCGA), including a report from TCGA (5) and an independent study from the Asian
Cancer Research Group (17), provide
an outstanding opportunity to establish advanced molecular
classifiers and predictors for the diagnosis and treatment of
gastric cancer. In addition to these studies, several smaller
studies have performed NGS to thoroughly establish the genomics of
gastric cancer (18–21). Similar to these small-scale studies,
the results of the present study demonstrated that AGC with
PIK3CA alterations was associated with the intestinal type
in Lauren's classification. The PIK3CA mutations activate
the PI3K/Akt signaling pathway, have been reported in several types
of carcinoma and are associated with negative outcome (22). PIK3CA amplification is
associated with increased Akt phosphorylation levels, suggesting
that this genetic alteration may serve a significant role in
activating the PI3K/Akt signaling pathway that contributes to
gastric carcinogenesis (23). Kim
et al (24) have suggested
that PIK3CA-mutated gastric cancer is a distinct disease
entity that may require a different therapeutic approach.
PIK3CA mutations were associated with Akt activation and
high tumor aggressiveness in gastric cancer (24). In addition, high PIK3CA
expression was significantly associated with tumor invasiveness,
phenotype and poor patient survival (25). Unlike previous studies using
quantitative PCR (24) or IHC
(25) for the PIK3CA
alterations only, the present study confirmed that PIK3CA
mutation and amplification in gastric cancer were associated with
adverse clinical manifestation using multi-gene analysis. The
results of the present gene panel study demonstrated that AGC with
mutated PIK3CA tended to be of an advanced TNM T stage (T4a,
88%), compared with AGC with wild-type PIK3CA (57%) or with
mutations other than PIK3CA (72%), although Epstein-Barr
virus (EBV) in situ hybridization was not investigated; a
previous study demonstrated that PIK3CA mutations were more
dispersed in EBV-positive cancer, but localized in the kinase
domain (exon 20) in EBV-negative cancer (5).
The plethora of data obtained from recent NGS
studies has resulted in the discovery of other candidate genes with
similar functions to those of CDH1 and TP53 as
classic driver genes of gastric cancer that may have valuable
influence on therapeutic decisions and clinical outcomes (6). The novel main categories of driver
mutations that have been ascertained by NGS include cell
motility/cytoskeleton (26),
chromatin remodeling (27), receptor
tyrosine kinase pathway genes (28)
and Wnt signaling (29). A recent
study using NGS demonstrated notable mutation distributions in
seven candidate genes (A-kinase anchoring protein 6, cyclic
nucleotide binding domain containing 1, collagen type XIV alpha 1
chain, -box and WD repeat domain containing 7, integrin subunit
alpha V, neurobeachin and xin actin binding repeat containing 2)
that had not been previously report to be prominently mutated in
gastric cancer (30). For medical
genetic testing, which is crucial for precision medicine in cancer
treatment, target NGS with a gene panel is advantageous due to cost
savings, enhanced depth of coverage and precise target enhancement
(31). Therefore, clinically helpful
molecular classification based on targeted sequencing with a gene
panel may enable the use of precise medicine in gastric cancer
(21,30,32).
The identification of specific cancer subgroups is
also enabling precise selection of patients who are likely to
respond to immunotherapy (33).
Through conventional methods for EBV and microsatellite
instability, as well as the use of emerging genetics testing that
focuses on a gene panel for mutations and amplifications, the
proposed genetic group may be applied to new cases of AGC (5). Tumor heterogeneity and the incomplete
understanding of the complex tumor biology represent an obstacle to
the overcoming of the ‘one size fits all’ era of gastric cancer
treatment (33). The most disturbed
pathways in gastric cancer include adherens junction and focal
adhesion (18). The clustered
mutations in recurrent hotspots influence the functional domain and
produce defective RHOA signaling, facilitating escape from
anoikis in organoid cultures (18).
In addition, gastric cancers with different Lauren's
classifications exhibit diverse characteristics, and EGF containing
fibulin extracellular matrix protein 1 (EFEMP1), frizzled
related protein (FRZB) and keratin 23 (KRT23)
have been identified as prognostic factors for gastric cancer
subtypes (34). EFEMP1 and
FRZB may be involved in diffuse gastric cancer-specific
pathways, such as cell adhesion; KRT23 may serve a critical
role in intestinal gastric cancer, considering that it has been
demonstrated to be an oncogene that can influence DNA damage and
proliferation response of colon cancer cells (35).
There were several limitations to the present study.
The most notable limitation was the small sample size, as it was
difficult to investigate significant relationships for the genetic
landscape of gastric cancers from the present data. Thus, it is
essential to further improve the molecular characterization of
gastric cancer subtypes in order to provide researchers and medical
oncologists with new tools for patient selection and stratification
in future clinical development programs and subsequent trials
(36). Comprehensive large-scale
studies on the molecular classification in gastric cancer covering
recent genomic, transcriptomic, proteomic and epigenomic features
are required (37). Another
limitation of the present study was that due to the inherent
problems with the OFA, particularly important variants may have not
been called; the OFA does not identify the mutations of previously
known such as AT-rich interaction domain 1A (ARID1A),
cadherin 1 (CDH1) and tumor protein p53 (TP53) as
well as new, such as catenin alpha 2, GLI family zinc finger 3,
mucin 6, and ring finger protein 43) significantly mutated driver
genes. The OFA applied in the present study was a relatively small
gene panel to be used for identification of complicated genetic
alterations in AGC. Although PIK3CA gene alterations were
the most frequently identified in the present study, frequencies of
genetic alteration in ARID1A, LDL receptor related protein
1B and TP53 are higher compared with PIK3CA in public
cancer genome databases such as cBioPortal for Cancer Genomics
(www.cbioportal.org). Since PIK3CA
alterations are significantly enriched in EBV-positive gastric
cancer samples (28), EBV in
situ hybridization should be required in a future study to
confirm whether the previously published data may be extrapolated
to the cohort of the present study. Similarly, although 15 diffuse
type gastric cancer samples were included in the present study, no
CDH1 mutations were reported, as the OFA did not cover
coding region of the CDH1 gene. The identification of
CDH1 gene mutations in diffuse type gastric cancer is
important since most diffuse type gastric cancers are known to
harbor pathogenic CDH1 mutations.
In conclusion, the present study demonstrated a
molecular profiling approach that identified the potential
molecular classifications for gastric cancer and suggested a
framework for precision medicine in AGC. The improvements in this
field may influence the discovery of novel driver mutations as well
as sophisticated classification systems for gastric cancer that may
be crucial for its pathogenesis if they can be effectively applied
to improve the clinical outcome and therapeutic paradigm of
AGC.
Acknowledgements
The authors would like to thank Mr. Taekyu Lee
(Thermo Fisher Scientific, Inc.) for their contribution in
providing technical support for Ion Torrent next generation
sequencing.
Funding
This work was supported by The Catholic University
of Korea Daejeon St. Mary's Hospital, Clinical Research Institute
Grant (grant nos. CMCDJ-P-2018-010 and CMCDJ-P-2019-012) and the
Chungnam National University Hospital Research Fund 2009. The
bio-specimens and data used for this study were provided by the
Biobank of Chungnam National University Hospital (Daejeon, Republic
of Korea), which is a member of the Korea Biobank Network.
Availability of data and materials
The data generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
JP and SL drafted the manuscript and revised it
critically for important intellectual content. JP and SS performed
the majority of the experiments and analyzed data. JH and HY
performed the molecular experiments and interpreted data. SL and JK
contributed to the conception and design of the work. JK gave the
final approval of the version to be published. All authors agreed
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Institutional
Review Board of The Catholic University of Korea (DC18SESI0113).
All subjects provided written informed consent for clinical and
molecular analyses and publication before the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang H and Kim YH: Identifying molecular
drivers of gastric cancer through next-generation sequencing.
Cancer Lett. 340:241–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan P and Yeoh KG: Genetics and molecular
pathogenesis of gastric adenocarcinoma. Gastroenterology.
149:1153–1162.e1153. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Berlth F, Bollschweiler E, Drebber U,
Hoelscher AH and Moenig S: Pathohistological classification systems
in gastric cancer: Diagnostic relevance and prognostic value. World
J Gastroenterol. 20:5679–5684. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Katona BW and Rustgi AK: Gastric Cancer
Genomics: Advances and future directions. Cell Mol Gastroenterol
Hepatol. 3:211–217. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arkenau HT: Gastric cancer in the era of
molecularly targeted agents: Current drug development strategies. J
Cancer Res Clin Oncol. 135:855–866. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paasinen-Sohns A, Koelzer VH, Frank A,
Schafroth J, Gisler A, Sachs M, Graber A, Rothschild SI, Wicki A,
Cathomas G and Mertz KD: Single-center experience with a targeted
next generation sequencing assay for assessment of relevant somatic
alterations in solid tumors. Neoplasia. 19:196–206. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Williams HL, Walsh K, Diamond A, Oniscu A
and Deans ZC: Validation of the Oncomine™ focus panel for
next-generation sequencing of clinical tumour samples. Virchows
Arch. 473:489–503. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee A, Lee SH, Jung CK, Park G, Lee KY,
Choi HJ, Min KO, Kim TJ, Lee EJ and Lee YS: Use of the Ion AmpliSeq
cancer hotspot panel in clinical molecular pathology laboratories
for analysis of solid tumours: With emphasis on validation with
relevant single molecular pathology tests and the Oncomine Focus
Assay. Pathol Res Pract. 214:713–719. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sim NL, Kumar P, Hu J, Henikoff S,
Schneider G and Ng PC: SIFT web server: Predicting effects of amino
acid substitutions on proteins. Nucleic Acids Res. 40:W452–W457.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Adzhubei I, Jordan DM and Sunyaev SR:
Predicting functional effect of human missense mutations using
PolyPhen-2. Curr Protoc Hum Genet. 7(Unit7.20)2013.PubMed/NCBI
|
|
13
|
Forbes SA, Beare D, Bindal N, Bamford S,
Ward S, Cole CG, Jia M, Kok C, Boutselakis H, De T, et al: COSMIC:
High-resolution cancer genetics using the catalogue of somatic
mutations in cancer. Curr Protoc Hum Genet. 91:10.11.1–10.11.37.
2016. View
Article : Google Scholar
|
|
14
|
Matsusaka S, Kobunai T, Yamamoto N, Chin
K, Ogura M, Tanaka G, Matsuoka K, Ishikawa Y, Mizunuma N and
Yamaguchi T: Prognostic impact of KRAS mutant type and MET
amplification in metastatic and recurrent gastric cancer patients
treated with first-line S-1 plus cisplatin chemotherapy. Genes
Cancer. 7:27–35. 2016.PubMed/NCBI
|
|
15
|
Serra O, Galán M, Ginesta MM, Calvo M,
Sala N and Salazar R: Comparison and applicability of molecular
classifications for gastric cancer. Cancer Treat Rev. 77:29–34.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alessandrini L, Manchi M, De Re V,
Dolcetti R and Canzonieri V: Proposed molecular and miRNA
classification of gastric cancer. Int J Mol Sci. 19:16832018.
View Article : Google Scholar
|
|
17
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi
ST, Siu HC, Deng S, Chu KM, Law S, et al: Whole-genome sequencing
and comprehensive molecular profiling identify new driver mutations
in gastric cancer. Nat Genet. 46:573–582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salomao M, Luna AM, Sepulveda JL and
Sepulveda AR: Mutational analysis by next generation sequencing of
gastric type dysplasia occurring in hyperplastic polyps of the
stomach: Mutations in gastric hyperplastic polyps. Exp Mol Pathol.
99:468–473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ge S, Li B, Li Y, Li Z, Liu Z, Chen Z, Wu
J, Gao J and Shen L: Genomic alterations in advanced gastric cancer
endoscopic biopsy samples using targeted next-generation
sequencing. Am J Cancer Res. 7:1540–1553. 2017.PubMed/NCBI
|
|
21
|
Ichikawa H, Nagahashi M, Shimada Y, Hanyu
T, Ishikawa T, Kameyama H, Kobayashi T, Sakata J, Yabusaki H,
Nakagawa S, et al: Actionable gene-based classification toward
precision medicine in gastric cancer. Genome Med. 9:932017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi J, Yao D, Liu W, Wang N, Lv H, Zhang
G, Ji M, Xu L, He N, Shi B and Hou P: Highly frequent PIK3CA
amplification is associated with poor prognosis in gastric cancer.
BMC Cancer. 12:502012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim JW, Lee HS, Nam KH, Ahn S, Kim JW, Ahn
SH, Park DJ, Kim HH and Lee KW: PIK3CA mutations are associated
with increased tumor aggressiveness and Akt activation in gastric
cancer. Oncotarget. 8:90948–90958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jang SH, Kim KJ, Oh MH, Lee JH, Lee HJ,
Cho HD, Han SW, Son MW and Lee MS: Clinicopathological significance
of elevated PIK3CA expression in gastric cancer. J Gastric Cancer.
16:85–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thumkeo D, Watanabe S and Narumiya S:
Physiological roles of Rho and Rho effectors in mammals. Eur J Cell
Biol. 92:303–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang K, Kan J, Yuen ST, Shi ST, Chu KM,
Law S, Chan TL, Kan Z, Chan AS, Tsui WY, et al: Exome sequencing
identifies frequent mutation of ARID1A in molecular subtypes of
gastric cancer. Nat Genet. 43:1219–1223. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deng N, Goh LK, Wang H, Das K, Tao J, Tan
IB, Zhang S, Lee M, Wu J, Lim KH, et al: A comprehensive survey of
genomic alterations in gastric cancer reveals systematic patterns
of molecular exclusivity and co-occurrence among distinct
therapeutic targets. Gut. 61:673–684. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pai P, Rachagani S, Dhawan P and Batra SK:
Mucins and Wnt/β-catenin signaling in gastrointestinal cancers: An
unholy nexus. Carcinogenesis. 37:223–232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, Wu WK, Xing R, Wong SH, Liu Y, Fang
X, Zhang Y, Wang M, Wang J, Li L, et al: Distinct subtypes of
gastric cancer defined by molecular characterization include novel
mutational signatures with prognostic capability. Cancer Res.
76:1724–1732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Horak P, Frohling S and Glimm H:
Integrating next-generation sequencing into clinical oncology:
Strategies, promises and pitfalls. ESMO Open. 1:e0000942016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuboki Y, Yamashita S, Niwa T, Ushijima T,
Nagatsuma A, Kuwata T, Yoshino T, Doi T, Ochiai A and Ohtsu A:
Comprehensive analyses using next-generation sequencing and
immunohistochemistry enable precise treatment in advanced gastric
cancer. Ann Oncol. 27:127–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tirino G, Pompella L, Petrillo A, Laterza
MM, Pappalardo A, Caterino M, Orditura M, Ciardiello F, Galizia G
and De Vita F: What's new in gastric cancer: The therapeutic
implications of molecular classifications and future perspectives.
Int J Mol Sci. 19:26592018. View Article : Google Scholar
|
|
34
|
Min L, Zhao Y, Zhu S, Qiu X, Cheng R, Xing
J, Shao L, Guo S and Zhang S: Integrated analysis identifies
molecular signatures and specific prognostic factors for different
gastric cancer subtypes. Transl Oncol. 10:99–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Birkenkamp-Demtroder K, Hahn SA, Mansilla
F, Thorsen K, Maghnouj A, Christensen R, Oster B and Orntoft TF:
Keratin23 (KRT23) knockdown decreases proliferation and affects the
DNA damage response of colon cancer cells. PLoS One. 8:e735932013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pellino A, Riello E, Nappo F, Brignola S,
Murgioni S, Djaballah SA, Lonardi S, Zagonel V, Rugge M, Loupakis F
and Fassan M: Targeted therapies in metastatic gastric cancer:
Current knowledge and future perspectives. World J Gastroenterol.
25:5773–5788. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ho SWT and Tan P: Dissection of gastric
cancer heterogeneity for precision oncology. Cancer Sci.
110:3405–3414. 2019. View Article : Google Scholar : PubMed/NCBI
|