Introduction
Ovarian clear cell carcinoma (OCCC) was identified
as a histopathological type of ovarian cancer by the World Health
Organization in 1973 (1). OCCC
accounts for 5–10% of epithelial carcinomas in North America and
for ~25% of epithelial ovarian cancer cases in Japan (2). Furthermore, OCCC is chemoresistant, and
thus patient prognosis is poorer, compared with serous or
endometrioid carcinoma (2–4).
Epigenetic mechanisms, such as histone modification,
regulate gene expression by altering chromatin structure (5,6).
Mutations in the chromatin remodeling gene AT rich interactive
domain 1A (ARID1A) are present in >50% of patients with OCCC
(4). Histone methylation can include
mono-, di- or trimethylation and is regulated by histone
methyltransferases. Dysregulation of histone methylation has been
implicated in cancer development and progression (6), and several types of histone
methyltransferases are overexpressed in various types of cancer
(6). For example, our previous study
reported that histone methyltransferase suppressor of variegation
3–9 homolog 2 methylates histone H2AX and regulates H2AX
phosphorylation during DNA repair (7). Other studies have indicated that
histone methyltransferases could also represent potential
therapeutic targets for OCCC (8,9). For
instance, inhibition of enhancer of zeste 2 polycomb repressive
complex 2 subunit (EZH2) has been shown to induce synthetic
lethality in ARID1A-mutant OCCC cells, and sensitivity to EZH2
inhibitors is associated with ARID1A mutation status (8). Moreover, our previous study suggested
that Wolf-Hirschhorn syndrome candidate 1 (WHSC1) was overexpressed
and promoted cancer cell proliferation in OCCC. The expression of
WHSC1 was attenuated via the knockdown or inhibition of EZH2
(9). EZH2 suppression attenuates
cell proliferation and induces apoptosis, suggesting the
possibility of a novel molecular target drug in endometrial cancer
(10).
SET and MYND domain containing 2 (SMYD2) is a
methyltransferase that methylates histones H3K4 and H3K36 (11). The SMYD2 protein can also methylate
proteins other than histones, including p53, estrogen receptor α
and RB transcriptional corepressor 1 (RB1), poly ADP ribose
polymerase (PARP)-1, echinoderm microtubule associated protein like
4-ALK receptor tyrosine kinase fusion gene, heat shock protein 90
and β-catenin (12–17).
Our previous study suggested that SMYD2 was
upregulated in clinical samples of high-grade serous ovarian cancer
(HGSOC); additionally, SMYD2 suppression induced apoptosis in HGSOC
cells (18). However, the role of
SMYD2 in OCCC remains poorly understood. Therefore, the aim of the
present study was to determine the role of SMYD2 in OCCC and
evaluate its suitability as a potential therapeutic target. The
expression of SMYD2 was measured in clinical OCCC specimens and
normal ovarian tissues. Moreover, cell proliferation was examined
in OCCC cell lines following SMYD2 gene silencing. The findings of
the present study may provide insight into novel and effective
therapeutic strategies for OCCC with.
Materials and methods
Clinical specimens and OCCC cell
lines
Surgical specimens were collected from 23 patients
with OCCC and 3 pathologically normal ovarian tissues removed for
other diseases (Table SI). Samples
were collected during surgery at The University of Tokyo Hospital
between January 2006 and December 2016 after obtaining written
patient consent and approval from the Human Genome, Gene Analysis
Research Ethics Committee at the University of Tokyo (approval no.
G0683-18). Tumor specimens were immediately snap-frozen in liquid
nitrogen and stored at −80°C prior to RNA extraction.
The OCCC cell lines OVISE (clone no. JCRB1043),
OVTOKO (clone no. JCRB1048) and OVMANA (clone no. JCRB1045) were
purchased from the Japanese Cancer Research Resources Bank. The
TOV-21G OCCC cell line was purchased from American Type Culture
Collection (clone no. ATCC® CRL-11730™). OVISE, OVTOKO
and TOV-21G cells were cultured in RPMI-1640 medium (FUJIFILM Wako
Pure Chemical Corporation) with 10% FBS (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2. OVMANA cells were
cultured under the same conditions using RPMI-1640 medium
supplemented with 20% FBS. All cultures were confirmed
Mycoplasma-free before and after the experiments using a
MycoAlert™ Mycoplasma Detection kit (cat. no. LT07-218; Lonza
Group, Ltd.).
Total RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Frozen specimens were embedded in
Tissue-Tek® OCT™ compound (Sakura Finetek Japan Co.,
Ltd.), and 500-µm sections were cut using a CM1520 Cryostat (Leica
Microsystems, Inc.). Then, 50–100 slices per specimen were placed
in MagNA Lyser Green Beads and homogenized using a MagNA Lyser
(both from Roche Diagnostics). Total RNA extraction from the
supernatant (centrifugation at 8,000 × g for 1 min at 24°C) was
carried out using an RNeasy Mini kit (Qiagen, Inc.). Total RNA was
reverse transcribed using ReverTra Ace-α-(Toyobo Life Science) at
42°C for 20 min and 99°C for 5 min. The mRNA expression levels of
eight histone methyltransferases were measured using One-Step SYBR
PrimeScript RT-PCR kit (Takara Bio, Inc.) on a Light Cycler
instrument (Roche Diagnostics) using the following thermocycling
conditions: Initial denaturation step at 95°C for 30 sec, followed
by 45 cycles at 95°C for 10 sec, 55°C for 30 sec and 72°C 30 sec,
and a final extension step at 72°C for 10 min. The primer sequences
are given in Table SII. Gene
expression was normalized to GAPDH mRNA levels, using the second
derivative method (19).
Gene knockdown
SMYD2 knockdown was performed using small
interfering (si)RNA (Sigma-Aldrich; Merck KGaA; Table SIII). OCCC cells
(1×105/well in 6-well plates; 2×104/well in
24-well plates) were transfected with 100 nM siRNA using
Lipofectamine® RNAi MAX transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The Mission siRNA
Universal Negative Control (cat. no. SIC001; Sigma-Aldrich; Merck
KGaA) was used as a negative control (siNC). After 24 h from the
start of cell culture, the cells were transfected with a specific
siRNA and incubated for 48–96 h at 37°C before subsequent
experiments.
Cell proliferation assay
Cell proliferation was assessed using the Cell
Counting Kit-8 (CCK-8) method. OCCC cell lines were plated at a
density of 2×104 cells/well in 24-well plates and
incubated for 24 h. The cells were then transfected with siSMYD2 or
siNC. CCK-8 (Dojindo Molecular Technologies, Inc.) was used 48–96 h
following transfection according to the manufacturer's protocol,
and the absorbance at 450 nM was measured using an Epoch™
Microplate Spectrophotometer BioTek Instruments, Inc.). The
experiment was repeated three times.
Western blotting
OCCC cells were plated at a density of
1×105 cells/wells in 6-well plates and transfected with
siSMYD2 or siNC. After transfection, cells were resuspended in
lysis buffer (50 mmol/l Tris-HCl, pH 7.5; 150 mmol/l sodium
chloride; 5 mmol/l EDTA; 2 mmol/l sodium orthovanadate; 10 mmol/l
sodium fluoride). The lysate was centrifuged at 12,000 × g for 20
min at 4°C, and the protein concentration was then measured in the
supernatant using a Bradford assay (Bio-Rad Laboratories, Inc.).
Protein samples (30 µg/lane) were separated via SDS-PAGE(Any kD™
Mini-PROTEAN TGX Precast Gels; cat. no. 4569033; Bio-Rad
Laboratories, Inc.) and transferred to polyvinylidene difluoride
membranes (Bio-Rad Laboratories, Inc.) using the trans blot Turbo
blotting system (Bio-Rad Laboratories, Inc.). The membranes were
incubated with 5% skimmed milk in TBS-Tween (0.1% Tween 20) for 1 h
with shaking at room temperature. After being blocked, the
membranes were incubated overnight at 4°C with the following
primary antibodies: Rabbit anti-SMYD2 (1:1,000; cat. no. 9734; Cell
Signaling Technology, Inc.), rabbit anti-cleaved PARP (1:1,000;
cat. no. 5625; Cell Signaling Technology, Inc.) and mouse
anti-β-actin (1:3,000; cat. no. A2228; Sigma-Aldrich; Merck KGaA).
Anti-rabbit IgG HRP-linked antibody (1:3,000; cat. no. 7074S; Cell
Signaling Technology, Inc.) and anti-mouse IgG HRP-linked antibody
(1:3,000; cat. no. 7076S; Cell Signaling Technology, Inc.) were
used as secondary antibodies and were incubated at room temperature
for 1 h with shaking. Proteins were detected using ImageQuant
LAS4000 (Cytiva) exposed under X-ray using an ECL select Western
Blotting detection kit (Cytiva). Protein bands were quantified
using ImageJ software (v1.52; National Institutes of Health).
Cell cycle analysis
OCCC cell lines were plated at a density
1×105cells/well in 6-well plates and cultured for 24 h.
The cells were then transfected with siSMYD2 or siNC. Following
48–72 h incubation, the cells were digested with trypsin, washed
with PBS and then added to ice-cold 70% ethanol for 2 h at 4°C. The
samples were then incubated overnight at 4°C, washed with PBS, then
treated with 0.25 mg/ml RNase A (Sigma-Aldrich; Merck KGaA) at 37°C
for 30 min. Propidium iodide (final concentration, 50 µg/ml) was
then added to each sample and incubated for 30 min in the dark at
4°C. Cell cycle progression was then evaluated using a FACSCalibur
HG flow cytometer (BD Biosciences). The data were analyzed using
the CellQuest Pro software (version 3.1; BD Biosciences).
IC50 determination
The SMYD2 inhibitor LLY-507 was purchased from
Selleck Chemicals (cat. no. S7575) and dissolved in <0.1% DMSO
for all experiments. OCCC cells were treated with LLY-507 at
concentrations ranging from 1 nmol/l to 10 µmol/l or DMSO and
cultured for 8 days. After adding CCK-8 solution, the number of
viable cells was determined by measuring the absorbance at 450 nm.
Cell viability data in each experimental group were normalized the
untreated cells. The IC50 was calculated using
non-linear regression, as per the logistic curve equation. The cell
viability, for both the control and the different LLY-507 treatment
conditions, was plotted, and a logistic curve was drawn. The
equation is as follows: IC50=10^[LOG(A/B)*(50-C)/(D-C) +
LOG(B)], where A is the higher concentration considering the two
values that sandwich 50% cell viability, B is the lower
concentration considering the same two values, C is the inhibition
rate determined for B, and D is the inhibition rate determined for
A (20,21).
Statistical analysis
Statistical analysis was carried out using the JMP
Pro software (version 14; SAS Institute, Inc.). The experiments
were repeated three times, and the data are presented as the mean ±
SD. Comparisons between two groups were analyzed using F-test
followed by unpaired Student's t-test. Multi-group comparisons were
analyzed using one-way ANOVA and Tukey's post hoc test, and
Kruskal-Wallis test was used for comparison between groups divided
by TNM staging. P<0.05 was considered to indicate a
statistically significant difference.
Results
SMYD2 is overexpressed in OCCC
cells
The expression of several histone methyltransferases
reported to be overexpressed in other types of cancer (7,13,20–25)
were measured using RT-qPCR in order to evaluate their role in OCCC
(Fig. 1). SMYD2 mRNA expression was
significantly increased in OCCC tissues, compared with normal
ovarian tissue (P=0.023). However, the expression of the eight
remaining histone methyltransferases was similar in OCCC and normal
ovarian tissue. The expression of SMYD2 in OCCC cell lines is also
shown in Fig. S1. Only OVISE cells
exhibited significantly higher SMYD2 expression compared with the
other cell lines, but there was no difference in SMYD2 expression
among the other cell lines.
 | Figure 1.SMYD2 expression in OCCC. (A) Histone
methyltransferase mRNA levels in OCCC clinical samples and normal
ovarian samples. The data are displayed as box plots with the
median, the average (represented by Xs), the maximum and minimum,
the interquartile ranges and the outliers (represented by circles).
(B) SMYD2 expression levels in individual study participants. The
data is displayed as the mean ± SD. *P<0.05. N.S., not
significant; OCCC, ovarian clear cell carcinoma; EZH2, enhancer of
zeste 2 polycomb repressive complex 2 subunit; SMYD, SET and MYND
domain containing 2; SETD, SET domain containing lysine
methyltransferase; SUV39H2, suppressor of variegation 3–9 homolog
2; EHMT2, euchromatic histone lysine methyltransferase 2. |
The association between SMYD2 expression and age or
TNM stage was also evaluated (Table
SIV). Tumor tissues were divided according to SMYD2 expression,
with expression ≥ median levels considered ‘high’, and expression
< median classified as ‘low’. The average age in both groups was
53 years. Moreover, SMYD2 expression was not associated with TNM
stage. Thus, SMYD2 expression was not associated with these
clinical variables.
SMYD2 promotes OCCC cell
proliferation
The effect of SMYD2 silencing on cell proliferation
was evaluated in the OVTOKO and OVISE cell lines, which have been
previously used as representative OCCC cell lines (22,23),
using two specific siRNAs against SMYD2. OVTOKO and OVISE cells
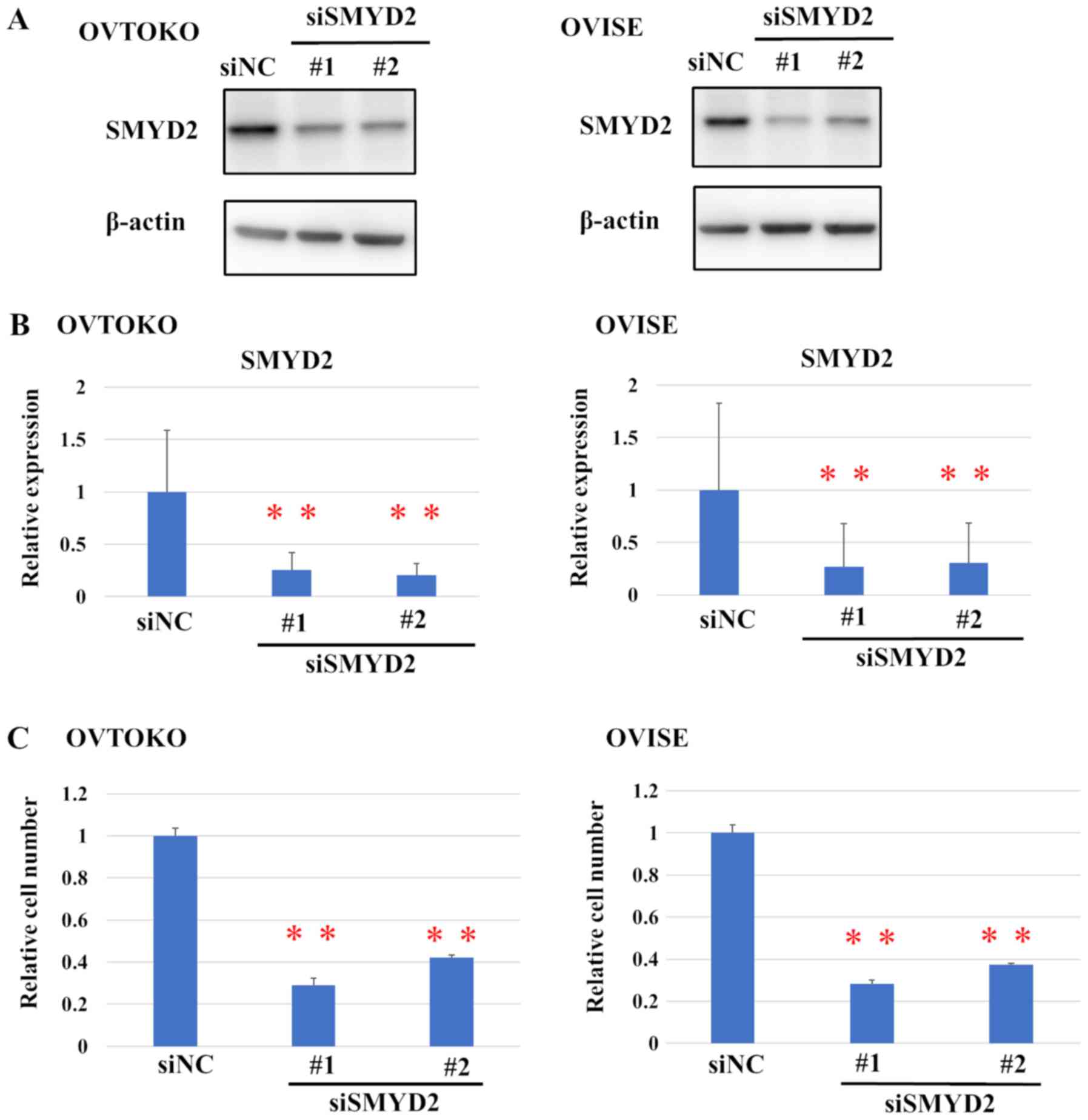
were transfected with siSMYD2#1, siSMYD2#2 or siNC SMYD2 expression
was evaluated using western blot analysis (Fig. 2A). Transfection with either siMYD2#1
or #2 led to a significant reduction in the relative protein levels
of SMYD2, compared with siNC (Fig.
2B).
Moreover, SMYD2 knockdown also significantly
affected the viability of OCCC cells (Fig. 2C). Furthermore, the effect of a
SMYD2-selective inhibitor, LLY-507, was also evaluated. Although
the dose-response curve did not follow a strictly sigmoid trend,
the viability of the OVTOKO, TOV-21G and OVMANA cell lines
predominantly increased with the concentration of LLY-507
(IC50, 0.95, 1.20 and 1.17 µM, respectively; Fig. 3).
Suppression of SMYD2 induces apoptosis
in OCCC cells
To examine the mechanism through which SMYD2
modulates the proliferation of OCCC cell lines, the levels of the
apoptosis marker PARP were determined by western blot analysis
(Fig. 4A). Cleaved PARP was detected
in the SMYD2-knockdown group in OVTOKO and OVISE cell lines.
Moreover, SMYD2 knockdown also significantly increased the
proportion of OVTOKO and OVISE cells in the sub-G1 phase
of the cell cycle, compared with siNC, suggesting that SMYD2
silencing induced apoptosis (Figs.
4B and S2).
Discussion
SMYD2, which belongs to the group of transcriptional
regulators containing SET and MYND domains, is responsible for
complex transcriptional regulation through histone methylation and
non-histone protein methylation (24,25).
Previous studies have suggested that SMYD2 is upregulated in
several types of cancer, including pediatric acute lymphoblastic
leukemia, gastric cancer and breast cancer (26–28).
Additionally, previous studies have highlighted the role of SMYD2
in the regulation of cancer cell growth (13,17). For
example, SMYD2 is upregulated in gastric cancer and has important
effects on cell proliferation (26).
In the field of gynecologic cancer, an association between poor
prognosis of cervical cancer and SMYD2 upregulation has been
reported (29). Additionally, our
previous study indicated that SMYD2 knockdown could suppress HGSOC
cell proliferation and induce apoptosis (18). The findings of the present study are
consistent with these previous reports. Higher SMYD2 expression was
observed in OCCC tumor samples, compared with normal ovarian
tissue. Furthermore, SMYD2 silencing inhibited OCCC cell
proliferation and induced apoptosis. A SMYD2 selective inhibitor,
LLY-507, also attenuated the proliferation of three OCCC cell
lines.
It has been demonstrated that SMYD2 could attenuate
the tumor suppressive function of p53 through methylation of Lys370
in the H1299 cell line, which is a human non-small cell lung
carcinoma cell line (12). However,
distinct mechanisms may account for HGSOC and OCCC cell apoptosis
following SMYD2 knockdown. As >90% of HGSOC cells have p53
mutations (18), it is possible that
SMYD2 promotes apoptosis independently of p53 methylation. However,
because p53 mutations are relatively infrequent in OCCC (30), SMYD2 knockdown could induce apoptosis
by suppressing p53 methylation. Moreover, other mechanisms might
underlie the role of SMYD2 in carcinogenesis in OCCC, as SMDY2 can
methylate several proteins other than histones (13–15). For
instance, SMYD2 can methylate RB1 at Lys810, thereby modulating the
cell cycle in bladder carcinoma (13). Furthermore, it has been reported that
SMYD2 may affect the promotion of colorectal cancer metastasis by
suppressing APC2 and activating the Wnt/β-catenin signaling pathway
(31). Additionally, it has been
reported that SMYD2-mediated EML4-ALK methylation affects signaling
pathways and proliferation of non-small cell lung cancer cells
(16).
A previous study has indicated that LLY-507 induces
apoptosis and suppresses HGSOC cell proliferation (18). Moreover, SMYD2 inhibitors have a
long-term inhibitory effect on the proliferation of HGSOC cells in
colony-formation assays (18).
Another previous study has also indicated that LLY-507 suppresses
the proliferation of esophageal, liver and breast cancer cells
(32). The sensitivity to LLY-507
did not increase in a time-dependent manner in liver cancer cells,
but breast cancer cells were 5 times more sensitive to LLY-507
after 7 days of treatment compared with after 3–4 days of
treatment, suggesting that proliferation of breast cancer cells may
be mediated, at least in part, by a SMYD2-dependent epigenetic
mechanism (32). Consistent with
these previous findings, the present study indicated that LLY-507
could inhibit the proliferation of OCCC cell lines. However, the
antitumor effect of LLY-507 may differ in OCCC cell lines compared
with HGSOC cells. Patients with OCCC rarely present p53 mutations,
whereas >90% of patients with HGSOC display p53 mutations
(33). Therefore, it is speculated
that the cytostatic effect of LLY-507 may be associated with the
p53 signaling pathway in OCCC and alternative pathways, such as
through histone modification in HGSOC. However, further studies are
required to confirm this hypothesis. Moreover, there was no
association between the cytostatic effect induced by SMYD2
suppression and its expression in OCCC cell lines.
The present study has several limitations. As all
experiments were carried out in vitro, extensive in
vivo validation is required to ascertain whether SMYD2 might
serve as a potential therapeutic target in OCCC. Moreover, the
mechanism of action underlying the effect of SMYD2 on cell
proliferation and apoptosis, including the potential role of p53
methylation, has not been addressed. Lastly, endometriosis is
reportedly a precursor of OCCC (34), and therefore analysis of SMYD2
expression in OCCC should be compared with normal ovarian tissues
as well as with tissues of endometriosis in the future.
Nevertheless, the present findings suggest that SMYD2 increases the
proliferation of OCCC cells in vitro, suggesting a potential
therapeutic avenue for SMYD2 inhibition in the treatment of
OCCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was financially supported by a
Grant-in-Aid for Scientific Research (grant nos. 18K09249, 15K10705
and 17K11268) and a Grant-in-Aid for Research Activity Start-up
(grant no. 15H06173) from The Ministry of Education, Culture,
Sports, Science and Technology of Japan. This research was also
supported by the Project for Cancer Research and Therapeutic
Evolution from The Japan Agency for Medical Research and
Development (grant no. 17K11268).
Availability of data and materials
All data generated and analyzed during this study
are included in this published article.
Authors' contributions
MK, KS and KO conceived and designed the study. MK,
KS, RH and SK designed the experiments. All experiments were
performed by MK. The data were analyzed and interpreted by SO, AKu,
AKa, HH, YK, TK, MS, AT, MT, YM, TT, KN, OH, YO and TF. MK and KS
prepared the manuscript and figures. MK, KS, KO, RH, SK, TT, YO and
TF reviewed and revised the manuscript for important intellectual
content. Technical and material support was provided by SO, AKu, HH
and YK. All the authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Human Genome,
Gene Analysis Research Ethics Committee at the University of Tokyo.
Written informed consent was obtained from the patients for the
research use of the tumor specimens and their clinical data as a
whole.
Patient consent for publication
Not applicable.
Competing interests
KS has a research grant from Daiichi-Sankyo Co.,
Ltd. KO has a research grant from Daiichi-Sankyo Co., Ltd. and
AstraZeneca Plc. and received a lecture fee from Chugai
Pharmaceutical Co., Ltd. and AstraZeneca Plc. All other authors
declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
OCCC
|
ovarian clear cell carcinoma
|
|
siRNA
|
small interfering RNA
|
|
NC
|
negative control
|
|
SMYD2
|
SET and MYND domain containing 2
|
References
|
1
|
Serov SF, Scully R and Sobin LH:
Histologic typing of ovarian tumors. Geneva: World Health
Organization; 1973
|
|
2
|
Mabuchi S, Sugiyama T and Kimura T: Clear
cell carcinoma of the ovary: Molecular insights and future
therapeutic perspectives. J Gynecol Oncol. 27:e312016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crotzer DR, Sun CC, Coleman RL, Wolf JK,
Levenback CF and Gershenson DM: Lack of effective systemic therapy
for recurrent clear cell carcinoma of the ovary. Gynecol Oncol.
105:404–408. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takano M, Sugiyama T, Yaegashi N, Sakuma
M, Suzuki M, Saga Y, Kuzuya K, Kigawa J, Shimada M, Tsuda H, et al:
Low response rate of second-line chemotherapy for recurrent or
refractory clear cell carcinoma of the ovary: A retrospective Japan
Clear Cell Carcinoma Study. Int J Gynecol Cancer. 18:937–942. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Varier RA and Timmers HT: Histone lysine
methylation and demethylation pathways in cancer. Biochim Biophys
Acta. 1815:75–89. 2011.PubMed/NCBI
|
|
6
|
Hamamoto R, Saloura V and Nakamura Y:
Critical roles of non-histone protein lysine methylation in human
tumorigenesis. Nat Rev Cancer. 15:110–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sone K, Piao L, Nakakido M, Ueda K,
Jenuwein T, Nakamura Y and Hamamoto R: Critical role of lysine 134
methylation on histone H2AX for gamma-H2AX production and DNA
repair. Nat Commun. 5:56912014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bitler BG, Aird KM, Garipov A, Li H,
Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih IeM,
Conejo-Garcia JR, et al: Synthetic lethality by targeting EZH2
methyltransferase activity in ARID1A-mutated cancers. Nat Med.
21:231–238. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kojima M, Sone K, Oda K, Hamamoto R,
Kaneko S, Oki S, Kukita A, Machino H, Honjoh H, Kawata Y, et al:
The histone methyltransferase WHSC1 is regulated by EZH2 and is
important for ovarian clear cell carcinoma cell proliferation. BMC
Cancer. 19:4552019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oki S, Sone K, Oda K, Hamamoto R, Ikemura
M, Maeda D, Takeuchi M, Tanikawa M, Mori-Uchino M, Nagasaka K, et
al: Oncogenic histone methyltransferase EZH2: A novel prognostic
marker with therapeutic potential in endometrial cancer.
Oncotarget. 8:40402–40411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brown MA, Sims RJ III, Gottlieb PD and
Tucker PW: Identification and characterization of Smyd2: A split
SET/MYND domain-containing histone H3 lysine 36-specific
methyltransferase that interacts with the Sin3 histone deacetylase
complex. Mol Cancer. 5:262006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang J, Perez-Burgos L, Placek BJ,
Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T
and Berger SL: Repression of p53 activity by Smyd2-mediated
methylation. Nature. 444:629–632. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho HS, Hayami S, Toyokawa G, Maejima K,
Yamane Y, Suzuki T, Dohmae N, Kogure M, Kang D, Neal DE, et al: RB1
methylation by SMYD2 enhances cell cycle progression through an
increase of RB1 phosphorylation. Neoplasia. 14:476–486. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Piao L, Kang D, Suzuki T, Masuda A, Dohmae
N, Nakamura Y and Hamamoto R: The histone methyltransferase SMYD2
methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in
cancer cells. Neoplasia. 16:257–264, 264.e2. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hamamoto R, Toyokawa G, Nakakido M, Ueda K
and Nakamura Y: SMYD2-dependent HSP90 methylation promotes cancer
cell proliferation by regulating the chaperone complex formation.
Cancer Lett. 351:126–133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang R, Deng X, Yoshioka Y, Vougiouklakis
T, Park JH, Suzuki T, Dohmae N, Ueda K, Hamamoto R and Nakamura Y:
Effects of SMYD2-mediated EML4-ALK methylation on the signaling
pathway and growth in non-small-cell lung cancer cells. Cancer Sci.
108:1203–1209. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deng X, Hamamoto R, Vougiouklakis T, Wang
R, Yoshioka Y, Suzuki T, Dohmae N, Matsuo Y, Park JH and Nakamura
Y: Critical roles of SMYD2-mediated β-catenin methylation for
nuclear translocation and activation of Wnt signaling. Oncotarget.
8:55837–55847. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kukita A, Sone K, Oda K, Hamamoto R,
Kaneko S, Komatsu M, Wada M, Honjoh H, Kawata Y, Kojima M, et al:
Histone methyltransferase SMYD2 selective inhibitor LLY-507 in
combination with poly ADP ribose polymerase inhibitor has
therapeutic potential against high-grade serous ovarian carcinomas.
Biochem Biophys Res Commun. 513:340–346. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yokogi S, Tsubota T, Kanki K, Azumi J,
Itaba N, Oka H, Morimoto M, Ryoke K and Shiota G: Wnt/Beta-catenin
signal inhibitor HC-1 sensitizes oral squamous cell carcinoma cells
to 5-fluorouracil through reduction of CD44-positive population.
Yonago Acta Med. 59:93–99. 2016.PubMed/NCBI
|
|
21
|
Lyles RH, Poindexter C, Evans A, Brown M
and Cooper CR: Nonlinear model-based estimates of IC(50) for
studies involving continuous therapeutic dose-response data.
Contemp Clin Trials. 29:878–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Price C, Gill S, Ho ZV, Davidson SM,
Merkel E, McFarland JM, Leung L, Tang A, Kost-Alimova M, Tsherniak
A, et al: Genome-wide interrogation of human cancers identifies
EGLN1 dependency in clear cell ovarian cancers. Cancer Res.
79:2564–2579. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Anglesio MS, Wiegand KC, Melnyk N, Chow C,
Salamanca C, Prentice LM, Senz J, Yang W, Spillman MA, Cochrane DR,
et al: Type-specific cell line models for type-specific ovarian
cancer research. PLoS One. 8:e721622013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh PK: Histone methyl transferases: A
class of epigenetic opportunities to counter uncontrolled cell
proliferation. Eur J Med Chem. 166:351–368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abu-Farha M, Lambert JP, Al-Madhoun AS,
Elisma F, Skerjanc IS and Figeys D: The tale of two domains:
Proteomics and genomics analysis of SMYD2, a new histone
methyltransferase. Mol Cell Proteomics. 7:560–572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Komatsu S, Ichikawa D, Hirajima S, Nagata
H, Nishimura Y, Kawaguchi T, Miyamae M, Okajima W, Ohashi T,
Konishi H, et al: Overexpression of SMYD2 contributes to malignant
outcome in gastric cancer. Br J Cancer. 112:357–364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li LX, Zhou JX, Calvet JP, Godwin AK,
Jensen RA and Li X: Lysine methyltransferase SMYD2 promotes triple
negative breast cancer progression. Cell Death Dis. 9:3262018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sakamoto LH, Andrade RV, Felipe MS,
Motoyama AB and Pittella Silva F: SMYD2 is highly expressed in
pediatric acute lymphoblastic leukemia and constitutes a bad
prognostic factor. Leuk Res. 38:496–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun JJ, Li HL, Ma H, Shi Y, Yin LR and Guo
SJ: SMYD2 promotes cervical cancer growth by stimulating cell
proliferation. Cell Biosci. 9:752019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ho ES, Lai CR, Hsieh YT, Chen JT, Lin AJ,
Hung MH and Liu FS: p53 mutation is infrequent in clear cell
carcinoma of the ovary. Gynecol Oncol. 80:189–193. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meng F, Liu X, Lin C, Xu L, Liu J, Zhang
P, Zhang X, Song J, Yan Y, Ren Z and Zhang Y: SMYD2 suppresses APC2
expression to activate the Wnt/β-catenin pathway and promotes
epithelial-mesenchymal transition in colorectal cancer. Am J Cancer
Res. 10:997–1011. 2020.PubMed/NCBI
|
|
32
|
Nguyen H, Allali-Hassani A, Antonysamy S,
Chang S, Chen LH, Curtis C, Emtage S, Fan L, Gheyi T, Li F, et al:
LLY-507, a cell-active, potent, and selective inhibitor of
protein-lysine methyltransferase SMYD2. J Biol Chem.
290:13641–13653. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cole AJ, Dwight T, Gill AJ, Dickson KA,
Zhu Y, Clarkson A, Gard GB, Maidens J, Valmadre S, Clifton-Bligh R
and Marsh DJ: Assessing mutant p53 in primary high-grade serous
ovarian cancer using immunohistochemistry and massively parallel
sequencing. Sci Rep. 6:261912016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ishibashi H, Takano M, Miyamoto M, Soyama
H, Matsuura H, Aoyama T, Yoshikawa T, Kato K, Tsuda H and Furuya K:
Role of endometriosis as a prognostic factor for post-progression
survival in ovarian clear cell carcinoma. Mol Clin Oncol.
7:1027–1031. 2017.PubMed/NCBI
|