Introduction
Pancreatic ductal adenocarcinoma (PDAC) is an
aggressive and lethal neoplasm, with a 5-year survival rate <5%
in the United States (1,2). The typical pathological characteristics
of pancreatic cancer are rapid invasion into the surrounding
vascular, perineural and lymphatic systems, and frequent metastasis
to distant organs (3). These
features lead to this disease being frequently diagnosed at an
advanced stage, precluding the possibility of curative surgical
resection (2,3). Therefore, there is a critical need to
develop effective therapies to prevent the invasion and metastasis
of PDAC.
In PDAC, tumor cells are surrounded by a hyaluronan
(HA)-rich stroma, also known as desmoplasia (3–5). HA is a
large linear glycosaminoglycan of up to
106−107 Da in its native form that is
produced by hyaluronan synthases (HASs) and degraded into smaller
fragments by hyaluronidases (HYALs) (6). The high accumulation of HA occurs in
numerous types of cancer, including PDAC, and HA serves a critical
role in various cellular processes, including cell proliferation,
migration, invasion, metastasis, angiogenesis and resistance to
chemotherapeutic agents (4–6). Membrane-bound or free extracellular HA
binds to CD44 and the receptor for HA-mediated motility (RHAMM),
which alters the cellular physiology (6). The interaction between HA and these two
receptors induces CD44 receptor clustering and activates
intracellular RHAMM-regulated MAPK, leading to ERK phosphorylation
and activation of the transcription effectors activator protein-1
and NFκB, resulting in cell migration (6).
High CD44 expression is associated with a poor
prognosis in patients with PDAC, and its knockdown inhibits the
migration of PDAC cells (7). In our
previous study, it was demonstrated that RHAMM is upregulated in
PDAC cells and tissues, and is associated with a poor prognosis in
patients with PDAC following surgical resection (8). Previously, RHAMM has been used as a
promising target for antibody therapy to inhibit the extracellular
function of RHAMM on the surface of cancer cells (9). Therefore, the present study
hypothesized that the migration of PDAC cells may decrease
following inhibition of RHAMM. To test this hypothesis, small
interfering (si)RNA knockdown was used to determine the effects of
RHAMM on the migratory ability of PDAC cells.
Materials and methods
Cell lines and reagents
A total of eight PDAC cell lines were used in the
present study. BxPC-3, Panc-1, Capan-2, CFPAC1 and AsPC-1 were
purchased from American Type Culture Collection, NOR-P1 from the
RIKEN BioResource Center, and SUIT-2 and KP-2 from the Japanese
Collection of Research Bioresources Cell Bank. The characteristics
of the eight PDAC cell lines are shown in Table I. These cell lines had recently been
obtained when the experiments were performed and their identities
were routinely monitored by short tandem repeat profiling. Primary
fibroblasts (sk-f2) derived from PDAC tissues were donated by
Kyushu University (Fukuoka, Japan). PDAC cell lines and sk-f2 cells
were maintained in RPMI-1640 medium supplemented with 10% FBS and
1% streptomycin and penicillin (all Thermo Fisher Scientific, Inc.)
in a 5% CO2 incubator at 37°C. 4-methylumbelliferone
(4-MU) and 12-O-tetradecanoyl-phorbol-13-acetate (TPA) were
purchased from Sigma-Aldrich (Merck KGaA).
 | Table I.Characteristics of eight pancreatic
ductal adenocarcinoma cell lines. |
Table I.
Characteristics of eight pancreatic
ductal adenocarcinoma cell lines.
| Cell line | Derivation | Metastasis | Proliferation,
h |
Differentiation |
|---|
| BxPC-3 | Primary tumor | No | 48-60 | Moderate to
poor |
| PANC-1 | Primary tumor | Yes | 52 | Poor |
| NOR-P1 | Liver
metastasis | Yes | 16 | Poor |
| SUIT-2 | Liver
metastasis | Yes | 38.2 | Moderate |
| KP-2 | Primary tumor | No | 48 | Moderate |
| Capan-2 | Primary tumor | No | 96 | Well |
| CFPAC-1 | Liver
metastasis | Yes | 31 | Well |
| AsPC-1 | Ascites | Yes | 38-40 | Poor |
Tissue samples
Patients aged ≥18 years old with histologically
confirmed PDAC diagnosed and a Karnofsky Performance Score ≥80 were
enrolled. The main exclusion criteria were as follows: i) Central
nervous system metastasis; and ii) other malignant tumors. Surgical
tissue specimens (matched pairs of primary pancreatic tumor and
adjacent non-tumor tissues, 1 cm away from the tumor tissue)
collected from 20 patients with PDAC (11 men and 9 women; age
range, 34–82 years; median age, 64 years) between August 2012 and
December 2014 at the School of Medicine of the University of
Occupational and Environmental Health (Kitakyushu, Japan) were
evaluated by an experienced pathologist. All samples were collected
and handled in an identical manner. Tissue aliquots were placed in
RNAlater® Stabilization Solution (Ambion; Thermo Fisher
Scientific, Inc.) at −80°C until further use. The present study was
approved by the Ethics Committee of the University of Occupational
and Environmental Health (Kitakyushu, Japan), and written informed
consent was obtained from all patients who approved the use of
their tissues for unspecified research purposes.
Trypan blue dye exclusion (TBDE) assay
of cell viability
The effects of RHAMM siRNA (100 nM), 4-MU (1,000 µM)
and TPA (100 ng/ml) on cell viability were analyzed using TBDE
assays as measurements of cytotoxicity. PDAC cells
(~2×105) were cultured at 37°C in the medium. The
untreated and RHAMM siRNA-treated cells were harvested after 24 h
of transfection, while the untreated, 4-MU- and TPA-treated cells
were harvested after 48 h of treatment. Subsequently, 4% trypan
blue was added at room temperature for 3 min, and the cells were
counted using a LUNA™ automated cell counter (Logos Biosystems,
Inc.), according to the manufacturer's protocol. Cytotoxicity was
determined from the number of viable cells (no color) in treated
samples as a percentage of that in the untreated control.
siRNA targeting RHAMM
siRNA for RHAMM (ON-TARGETplus Human HMMR
siRNA-SMARTpool L-010409-00-0005) and negative control siRNA
(siGENOME Non-Targeting siRNA Pool D-001206-13-05) were purchased
from Cytiva. All PDAC cell lines were transfected with 100 nM siRNA
using DhermaFECT 1 Transfection Reagent (Cytiva) for 24 h at 37°C,
according to the manufacturer's protocol. The target sequences
were: GUUAACAGCCAGUGAGAUA [molecular weight (MW), 13,414.8 g/mol];
GGACUAAUGAACUACUAAA (MW, 13,384.8 g/mol); GAUACUACCUUGCCUGCUU (MW,
13,429.9 g/mol); CAAGUGGCGUCUCCUCUAU (MW, 13,444.8 g/mol).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from all cells (untreated and
treated) and tissues (matched pairs of primary pancreatic tumor and
adjacent non-tumor tissues) using the RNeasy® Mini kit
(Qiagen, Inc.). Single-stranded cDNA was synthesized from 1 µg
total RNA using the SuperScript® VILO™ MasterMix
(Invitrogen; Thermo Fisher Scientific, Inc.) for 10 min at 25°C, 1
h at 42°C and 5 min at 85°C. mRNA expression analyses of RHAMM
(Hs00234864_m1), HAS1 (Hs00758053_m1), HAS2 (Hs00193435_m1), HAS3
(Hs00193436_m1), HYAL1 (Hs00201046_m1), HYAL2 (Hs01117343_g1),
HYAL3 (Hs00185910_m1) and the housekeeping gene GAPDH
(Hs02758991_g1), used as a control, were performed using
TaqMan® Fast Universal PCR Master Mix, TaqMan Gene
Expression Assays and Step One Plus real-time PCR instrument (all
Applied Biosystems; Thermo Fisher Scientific, Inc) at room
temperature for 45 min. The thermocycling conditions used for qPCR
consisted of an initial denaturation step for 20 sec at 95°C,
followed by 40 cycles of denaturation for 1 sec at 95°C, annealing
for 20 sec at 60°C and extension for 30 sec at 72°C. The relative
quantification was performed using the comparative
2−ΔΔCq method (10).
Migration assay
After 24 h of treatment with siRNAs, the cells were
harvested and counted, and ~2×104 cells were seeded in
100 µl medium with 1% FBS in the top chamber with an uncoated
membrane (24-well Transwell insert; 8-µm pore size; BD
Biosciences). A total of 900 µl medium containing 10% FBS was used
as a chemoattractant in the lower chamber, and the cells were
allowed to migrate at 37°C with 5% CO2 for 48 h.
Non-migrating cells on the upper surface of the membrane were
removed with a cotton swab. Migrating cells that penetrated onto
the lower surface of the membrane were fixed with 70% methanol for
10 min, stained with hematoxylin for 10 min and eosin for 5 min,
all at room temperature, and air-dried. The number of migrating
cells was counted in six randomly selected fields (magnification,
×400) using a light microscope.
ELISA for determination of HA
concentration
The untreated, RHAMM siRNA-, 4-MU- and TPA-treated
cells were cultured in the medium at 37°C for 48 h, while the
monoculture and co-culture cells were cultured in the medium at
37°C for 72 h, after which the medium was collected and clarified
by centrifugation at room temperature (1,000 × g for 15 min). The
supernatant fraction was aliquoted and stored at −80°C until
further use. HA concentrations in cell culture medium and thawed
pancreatic tissue extracts were determined using an HA ELISA kit
(cat. no. DHYALO; R&D Systems, Inc.) as previously described
(11).
SUIT-2-based models
A total of ~4×105 SUIT-2 cells were
maintained in 2 ml serum-free medium at 37°C for 12 h.
Subsequently, the medium was replaced with fresh medium containing
1% FBS with or without 4-MU (1,000 µM) or TPA (100 ng/ml) at 37°C
for 48 h. SUIT-2 cells (2×105 /ml) in 1 ml serum-free
medium were seeded in the upper chamber (High Density, Translucent
PET Membrane 6-well insert, 0.4-µm pore size; BD Biosciences) and 3
ml serum-free medium was added into the lower chamber as
monoculture. Simultaneously, sk-f2 cells (1×105/ml) in 3
ml serum-free medium were seeded in the lower chamber for
co-culture with SUIT-2 cells at 37°C for 72 h. For all samples, the
supernatant was collected and clarified via centrifugation at room
temperature (1,000 × g for 15 min), and RNA was extracted as
previously described (8).
Statistical analysis
Data are presented as the mean ± SD (n=3 or n=6).
All statistical analyses were performed using SPSS statistical
software (v21.0; IBM Corp.). Differences in RHAMM mRNA expression,
migrating cell number and HA production between untreated and
treated groups were compared using an independent-samples t-test.
Differences in HAS1-3 and HYAL1-3 mRNA expression between untreated
and treated groups were compared using an independent-samples
t-test (for individual cell lines) and the Mann-Whitney U test
(when combining the expression levels of all cell lines).
Spearman's rank correlation coefficient was used to assess the
correlation between RHAMM mRNA expression and HA concentration in
PDAC cell lines and tissues, and the NOR-P1 sample was used as a
reference. P<0.05 was considered to indicate a statistically
significant difference.
Results
Effects of RHAMM-knockdown on
migration and HA production of PDAC cells
siRNA was used to knock down RHAMM expression in
eight PDAC cell lines for 24 h, and the collected cells
(2×105 /ml) were seeded in 2 ml medium with 1% FBS for
48 h. RT-qPCR revealed that transfection of PDAC cell lines with
100 nM siRNA targeting RHAMM resulted in 18–86% knockdown of
RHAMM mRNA expression. The transfection of five PDAC cell
lines (Panc-1, NOR-P1, SUIT-2, KP-2 and Capan-2) with siRNA
targeting RHAMM resulted in a significant decrease in RHAMM mRNA
expression compared with control siRNA-treated cell lines (Fig. 1). RHAMM-knockdown had no significant
effects on cell viability, as assessed using a TBDE test (Fig. S1).
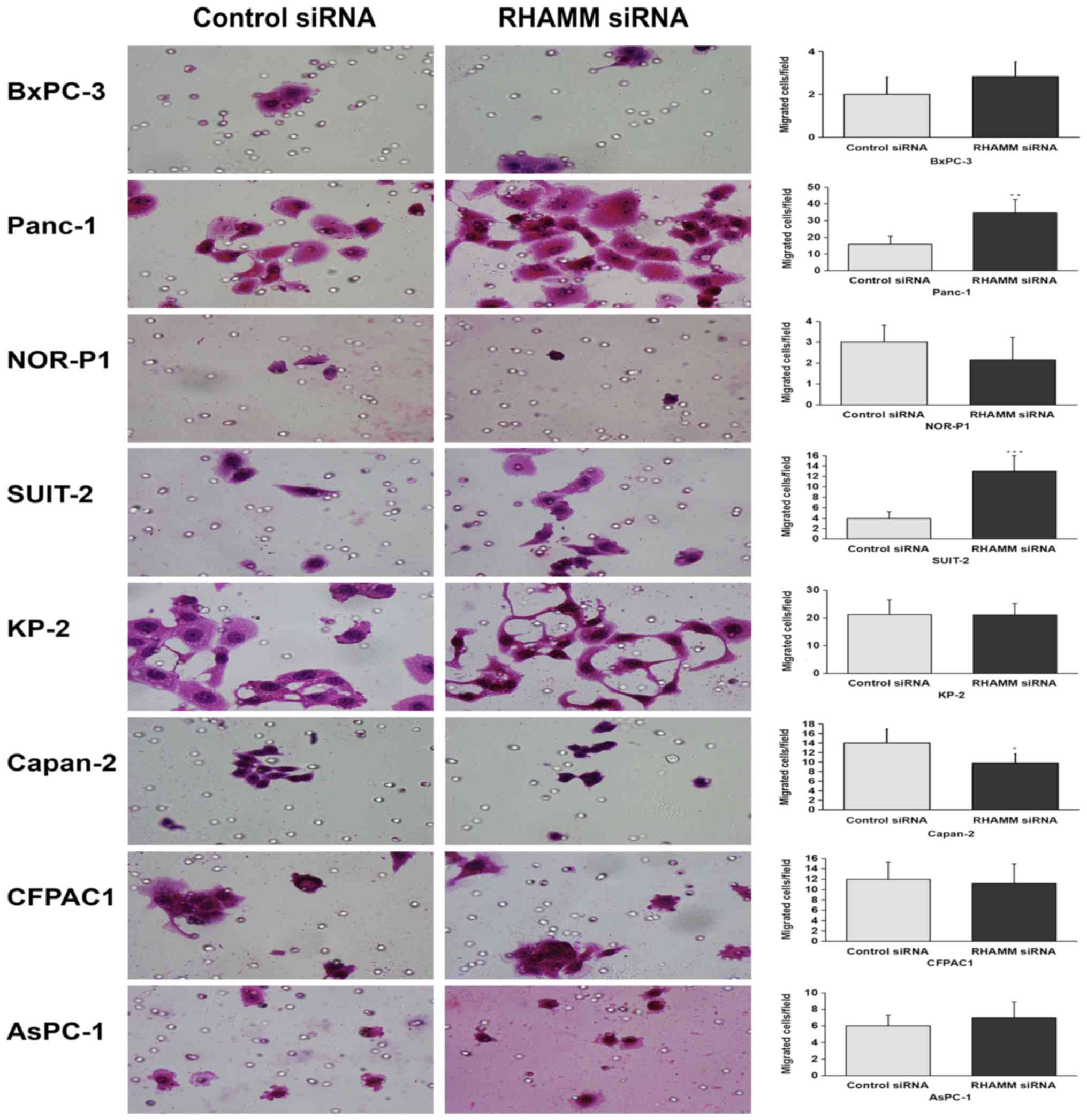
In response to RHAMM-knockdown, the number of
migrating cells was significantly increased in two cell lines
(Panc-1 and SUIT-2) and significantly decreased in Capan-2 cells,
while no significant difference was observed in the remaining cell
lines (NOR-P1, BxPC-3, AsPC-1, CFPAC1 and KP-2; Fig. 2) compared with control siRNA-treated
groups. Furthermore, HA production was significantly increased in
two cell lines (Panc-1 and SUIT-2) and significantly decreased in
three cell lines (NOR-P1, CFPAC1 and Capan-2), while no significant
differences were observed in KP-2, BxPC-3 and AsPC-1 cell lines
compared with control-siRNA treated groups (Fig. 3).
To elucidate the mechanism underlying HA modulation
via RHAMM-knockdown, the mRNA expression levels of HA-synthesizing
enzymes, HAS1-3 (Figs. 4–6), and HA-degrading enzymes, HYAL1-3
(Figs. 7–9), in PDAC cells were analyzed using
RT-qPCR. RHAMM-knockdown led to a significantly increased
production of HAS2 mRNA expression in six cell lines (BxPC-3,
Panc-1, SUIT-2, KP-2, CFPAC1 and AsPC-1; Fig. 5), HYAL1 mRNA expression in five cell
lines (Panc-1, NOR-P1, Capan-2, CFPAC1 and AsPC-1; Fig. 7), HYAL2 mRNA expression in three cell
lines (NOR-P1, Capan-2 and AsPC-1; Fig.
8) and HYAL3 mRNA expression in two cell lines (Panc-1 and
AsPC-1; Fig. 9), compared with the
control siRNA-treated groups. As aforementioned, RHAMM-knockdown
led to increased production of HA in two cell lines (Panc-1 and
SUIT-2), which may have been induced by a significant increase in
combined HAS2 mRNA expression compared with the control
siRNA-treated groups (Fig. 5), and
decreased production of HA in three cell lines (NOR-P1, CFPAC1 and
Capan-2), which may have been induced by significantly increased
combined HYAL1 mRNA expression compared with the control
siRNA-treated groups (Fig. 7).
Correlation between RHAMM mRNA
expression and HA concentration
The present study hypothesized that there may be a
correlation between RHAMM mRNA expression and HA concentration. To
test this, RHAMM mRNA expression and HA concentration were examined
in eight PDAC cell lines and tissues from 20 patients with PDAC.
Fig. 10A shows a significant
linear negative correlation between RHAMM mRNA expression and
HA concentration in PDAC cell lines (r=−0.738; P=0.037).
Additionally, Fig. 10B shows a
significant linear negative correlation between RHAMM mRNA
expression and HA concentration in PDAC tissues (r=−0.759;
P<0.0001).
SUIT-2 cells were selected to further confirm the
negative correlation between RHAMM mRNA expression and HA
concentration, since this cell line exhibits low mRNA expression
levels of RHAMM (8) and high HA
production (12). HA concentration
was significantly decreased (P<0.0001) and RHAMM mRNA expression
levels were significantly increased (P=0.006) when SUIT-2 cells
were treated with an HA inhibitor (4-MU; 1,000 µM) compared with
untreated cells (Fig. 11A). By
contrast, HA concentration was significantly increased (P=0.012)
and RHAMM mRNA expression levels were significantly decreased
(P=0.049) when SUIT-2 cells were treated with an HA stimulator
(TPA; 100 ng/ml) compared with untreated cells (Fig. 11B). 4-MU and TPA had no significant
effects on SUIT-2 cell viability (data not shown) when
applied at the same concentrations used in a previous study
(11). Additionally, HA
concentration was significantly increased (P<0.0001) and RHAMM
mRNA expression levels were significantly decreased (P<0.0001)
when SUIT-2 cells were co-cultured with primary fibroblasts
compared with monoculture (Fig.
11C).
Discussion
In the present study, the effects of RHAMM on PDAC
cell migration were investigated using RHAMM-knockdown. In contrast
to our hypothesis that cell migration would be inhibited,
RHAMM-knockdown resulted in a significantly increased number of
migrating cells in two cell lines (Panc-1 and SUIT-2) and a
significantly decreased number of migrating cells in Capan-2 cells,
while no significant differences were observed in the remaining
cell lines (NOR-P1, BxPC-3, AsPC-1, CFPAC1 and KP-2) compared with
untreated groups. Several human PDAC cell lines (PaTu 8902, PaTu
II, PaTu 8988s, PaTu 8988t and HPAF) synthesize and secrete HA, and
in human primary pancreatic carcinomas, the highest level of HA is
detected between the tumor mass and normal tissue (13). This suggests that HA may promote
tumor invasion (14–17). In our previous study (12), it was demonstrated that HA
significantly increases PDAC cell migration compared with untreated
groups. Therefore, it was suspected that RHAMM-knockdown may lead
to increased HA production, resulting in the aforementioned
phenomena. In the present study, ELISA and RT-qPCR assays suggested
that HA production in two cell lines (Panc-1 and SUIT-2) may have
been increased in a manner dependent on increased HAS2 mRNA
expression, while HA production in three other cell lines (NOR-P1,
CFPAC1 and Capan-2) may have been decreased in a manner dependent
on increased HYAL1 mRNA expression. Therefore, there is reason to
hypothesize that RHAMM may regulate PDAC cell migration via changes
in HA production.
The RHAMM gene is localized on human chromosome band
5q33.2 and contains 18 exons (18).
RHAMM mRNA expression is controlled by its promoter at the
transcriptional level in a cell cycle-dependent manner, and RHAMM
protein levels follow mRNA expression levels in the early phases of
the cell cycle (18). However, RHAMM
protein levels peak during the S phase and decrease before the
highest RHAMM mRNA expression level is reached at the
G2/M phase (18).
Furthermore, RHAMM mRNA and protein levels are suppressed by p53
(18,19). Lin et al (20) demonstrated that the androgen receptor
regulates RHAMM expression. In the present study, RHAMM mRNA
expression and HA concentration were measured in PDAC cell lines,
tissues and SUIT-2-based models, revealing that RHAMM mRNA
expression was significantly negatively correlated with HA
concentration. To the best of our knowledge, the present study was
the first to demonstrate that HA may be negatively correlated with
RHAMM mRNA expression.
The present findings suggest a complex mechanism
regulating RHAMM mRNA expression and HA synthesis in PDAC cells at
the genetic level. There may be a negative correlation between
RHAMM mRNA expression and HA production in a subset of PDAC cell
lines. Future studies should confirm whether there is a negative
correlation between RHAMM expression and HA production in PDAC cell
lines and tissues at the protein level, and explore its mechanism
in depth in order to provide a stronger scientific basis for the
evaluation of the risk and benefit of a therapeutic strategy
targeting RHAMM.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Medical and
Health Science and Technology Innovation Foundation of Jinshan
District of Shanghai (grant no. 2018-3-23), the Intramural Funding
from Shanghai Public Health Clinical Center (grant no.
KY-GW-2018-03), the Shanghai Municipal Committee for Health and
Family Planning (grant no. 201740194), the Clinical Science and
Technology Innovation Foundation of Shanghai Shenkang Hospital
Development Center (grant no. SHDC12018112) and the National 13th
Five-Year Grand Program on Key Infectious Disease Control (grant
no. 2018ZX10302103-003).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
NS, YZ and XC conceived the experimental design. XC,
SW, HY and HT performed the experiments. XC, SW, GS, LW, JZ, LS, HL
and SR analyzed the data. XC, SW, NS and YZ wrote the paper. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the University of Occupational and Environmental
Health (Kitakyushu, Japan), and written informed consent was
obtained from all patients who approved the use of their tissues
for unspecified research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RHAMM
|
receptor for hyaluronic acid-mediated
motility
|
|
HA
|
hyaluronic acid
|
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
HAS
|
HA synthase
|
|
HYAL
|
hyaluronidases
|
|
HMW-HA
|
high-molecular weight HA
|
|
4-MU
|
4-methylumbelliferone
|
|
TPA
|
12-O-tetradecanoyl-phorbol-13-acetate
|
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bardeesy N and DePinho RA: Pancreatic
cancer biology and genetics. Nat Rev Cancer. 2:897–909. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mahadevan D and Von Hoff DD: Tumor-stroma
interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther.
6:1186–1197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Erkan M, Reiser-Erkan C, Michalski CW and
Kleeff J: Tumor microenvironment and progression of pancreatic
cancer. Exp Oncol. 32:128–131. 2010.PubMed/NCBI
|
|
6
|
Schwertfeger KL, Cowman MK, Telmer PG,
Turley EA and McCarthy JB: Hyaluronan, inflammation, and breast
cancer progression. Front Immunol. 6:2362015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li XP, Zhang XW, Zheng LZ and Guo WJ:
Expression of CD44 in pancreatic cancer and its significance. Int J
Clin Exp Pathol. 8:6724–6731. 2015.PubMed/NCBI
|
|
8
|
Cheng XB, Sato N, Kohi S, Koga A and
Hirata K: Receptor for hyaluronic acid-mediated motility is
associated with poor survival in pancreatic ductal adenocarcinoma.
J Cancer. 6:1093–1098. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gust KM, Hofer MD, Perner SR, Kim R,
Chinnaiyan AM, Varambally S, Moller P, Rinnab L, Rubin MA, Greiner
J, et al: RHAMM (CD168) is overexpressed at the protein level and
may constitute an immunogenic antigen in advanced prostate cancer
disease. Neoplasia. 11:956–963. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng XB, Sato N, Kohi S and Yamaguchi K:
Prognostic impact of hyaluronan and its regulators in pancreatic
ductal adenocarcinoma. PLoS One. 8:e807652013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng XB, Kohi S, Koga A, Hirata K and
Sato N: Hyaluronan stimulates pancreatic cancer cell motility.
Oncotarget. 7:4829–4840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mahlbacher V, Sewing A, Elsasser HP and
Kern HF: Hyaluronan is a secretory product of human pancreatic
adenocarcinoma cells. Eur J Cell Biol. 58:28–34. 1992.PubMed/NCBI
|
|
14
|
Fries H, Elsasser HP, Mahlbacher V,
Neumann K and Kern HF: Localisation of hyaluronate (HA) in primary
tumors and nude mouse xenografts of human pancreatic carcinomas
using a biotinylated HA-binding protein. Virchows Arch. 424:7–12.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McBride WH and Bard JB:
Hyaluronidase-sensitive halos around adherent cells. Their role in
blocking lymphocyte-mediated cytolysis. J Exp Med. 149:507–515.
1979. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suzuki Y, Nishida Y, Naruse T, Gemba T and
Ishiguro N: Pericellular matrix formation alters the efficiency of
intracellular uptake of oligonucleotides in osteosarcoma cells. J
Surg Res. 152:148–156. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Toole BP: Hyaluronan-CD44 interactions in
cancer: Paradoxes and possibilities. Clin Cancer Res. 15:7462–7468.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sohr S and Engeland K: RHAMM is
differentially expressed in the cell cycle and downregulated by the
tumor suppressor p53. Cell cycle. 7:3448–3460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Godar S and Weinberg RA: Filling the
mosaic of p53 actions: P53 represses RHAMM expression. Cell cycle.
7:34792008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin SL, Chang D, Chiang A and Ying SY:
Androgen receptor regulates CD168 expression and signaling in
prostate cancer. Carcinogenesis. 29:282–290. 2008. View Article : Google Scholar : PubMed/NCBI
|