Introduction
Acute lymphoblastic leukemia (ALL) is a type of
malignancy that develops in the thymus from an immature thymocyte
(1). ALL is caused by the
accumulation of genetic and epigenetic aberrations, which results
in altered cell differentiation, proliferation, apoptosis and
self-renewal capacity (1). T-cell
ALL (T-ALL) accounts for ~15% of all cases of ALL in children and
25% of cases in adults (2,3). Studies in the past several decades have
identified a considerable number of genetic factors that are
involved in the development and progression of T-ALL (4,5).
However, the molecular signaling pathways involved in the
progression of ALL are not well understood.
Long non-coding RNAs (lncRNAs) are sequences of RNA
>200 nucleotides long involved in regulating a range of
physiological and pathophysiological processes (6). Altered expression of various lncRNAs
has been demonstrated to promote cancer development, and some
differentially expressed lncRNAs may possess clinical potential in
treating patients with cancer (7,8). lncRNA
associated with poor prognosis of hepatocellular carcinoma (AWPPH)
is a recently identified oncogenic lncRNA in liver and bladder
cancer (9,10). In liver cancer, lncRNA AWPPH
expression is increased and promotes cancer progression by
interacting with Y-box binding protein 1 (9). In bladder cancer, lncRNA AWPPH is
involved in the regulation of cancer progression by regulating the
activities of the SMAD family member 4 via the enhancer of zeste 2
polycomb repressive complex 2 subunit (10). lncRNA AWPPH expression was
additionally upregulated in glioma based on our preliminary
transcriptome analysis data (data not shown). In the present study,
it was demonstrated that lncRNA AWPPH may regulate the cancerous
behaviors of cells in pediatric T-ALL by interacting with the
Rho-associated protein kinase 2 (ROCK2), a well-characterized
oncogene which is expressed in different types of cancer (11).
Materials and methods
Human materials and cell lines
Bone marrow containing malignant cells was obtained
from 32 patients with pediatric T-ALL and 32 age- and sex-matched
healthy volunteers. Patients were admitted to The First Clinical
Hospital Affiliated to Harbin Medical University (Heilongjiang,
China) between June 2014 and July 2018. The inclusion criteria for
recruitment were: i) Patients were diagnosed with ALL for the first
time; and ii) patients had otherwise normal function of the major
organs. The exclusion criteria were: i) Patients had received
treatment for ALL in the past 3 months; ii) other clinical
disorders were observed; and iii) patients failed to cooperate with
the researchers. The ALL group included 19 males and 13 females
(age range, 7–14 years; mean age, 10.8±1.9 years). In the control
group, there were 18 males and 14 females (age range, 7–14 years;
mean age, 10.6±1.7 years). The present study was approved by the
Ethics Committee of The First Clinical Hospital Affiliated to
Harbin Medical University. All participants' guardians signed
informed consent.
Cells of Loucy (cat. no. CRL-2629) were obtained
from the American Type Culture Collection. Cells were cultured in
RPMI-1640 medium (cat. no. 30–2001; American Type Culture
Collection) supplemented with 10% FBS (Sigma-Aldrich; Merck KGaA)
and Penicillin-Streptomycin (100 U/ml; Sigma-Aldrich; Merck KGaA)
at 37°C and 5% CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
The GenElute™ Total RNA Purification kit
(Sigma-Aldrich; Merck KGaA) was used for RNA extraction from bone
marrow samples and in vitro cultured cells, followed by
preparation of cDNA samples using AMV Reverse Transcriptase (VWR
International) with the following thermocycling conditions: 25°C
for 5 min, 53°C for 20 min and 75°C for 10 min. All PCR mixtures
were prepared using the SuperScript III Platinum One-Step qRT-PCR
kit (Thermo Fisher Scientific, Inc.). The ABI 7500 system was used
to perform all PCR reactions with the following conditions: 1 min
at 95°C, followed by 40 cycles of 15 sec at 95°C and 40 sec at
57.5°C. Ct values were normalized using the
2−ΔΔCq method (12).
Primer sequences were as follows: lncRNA AWPPH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′; ROCK2 forward,
5′-GTGTCGGCTCCTCTGATCTC-3′ and reverse, 5′-GGCATGTCTGGATGACCTCT-3′;
and GAPDH (reference gene) forward, 5′-CTGGATGGTCGCTGCTTTTTA-3′ and
reverse, 5′-AGGGGGATGAGTCGTGATTT-3′.
Vectors and cell transfection
pcDNA3.1 vectors expressing ROCK2 and lncRNA AWPPH
were designed and prepared by Sangon Biotech Co., Ltd., along with
empty vectors. Cells were cultivated overnight to reach 70–80%
confluence before transfection. Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform
cell transfection using 15 nM vectors. Transfection with empty
vectors was used as the negative control (NC). Treatment with the
Lipofectamine® 2000 reagent alone was also used as
another control (C). Cells were cultivated in fresh medium for 48 h
prior to the subsequent assays.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was measured using the CCK-8
assay (Sigma-Aldrich; Merck KGaA) at 24 h after transfection,
according to the manufacturer's instructions. Briefly,
3×103 cells/well were added to a 96-well plate, followed
by the addition of 10 µl CCK-8 solution every 24 h up to 96 h.
Following incubation at 37°C for 4 h, a microplate reader (Bio-Rad
Laboratories, Inc.) was used to measure the absorbance at 450
nm.
Cell apoptosis assay
Cell apoptosis was analyzed using a cell apoptosis
assay 24 h after transfection. Cells were centrifuged at room
temperature at 1,500 × g for 10 min to remove the supernatant.
Cells were washed with PBS and counted. Subsequently,
1×106 cells were stained with 500 µl binding buffer, 5
µl FITC-labeled Annexin V and 5 µl propidium iodide solution. After
incubation in the dark for 10 min, apoptotic cells were detected
using the FACSCalibur flow cytometry system (BD Biosciences). Data
was analyzed using Flowing Software version 2.5 (Turku
Bioscience).
Western blot analysis
Total protein was extracted from in vitro
cultivated Loucy cells at 48 h after transfection using a Total
Protein Extraction kit (cat. no. NBP2-37853; Novus Biologicals,
LLC). BCA assay (Invitrogen; Thermo Fisher Scientific, Inc.) was
performed to measure protein concentration. Following protein
denaturation (5 min in boiling water), electrophoresis was
performed (30 µg per lane) using 10% SDS-PAGE, and PVDF membranes
were used for gel transfer. Following incubation with 5% skimmed
milk for 2 h at 22°C, membranes were incubated with ROCK2
(dilution, 1:2,000; cat. no. ab71598; Abcam) and GAPDH (dilution,
1:1,000; cat. no. ab9485; Abcam) rabbit anti-human primary
antibodies for 18 h at 4°C, followed by incubation with
immunoglobulin G-horseradish peroxidase goat anti-rabbit secondary
antibody (dilution, 1:1,000; cat. no. MBS435036; MyBioSource, Inc.)
for 2 h at room temperature. Subsequently, membranes were incubated
with ECL reagent (Sigma-Aldrich; Merck KGaA) to detect signals.
Data were analyzed using ImageJ v1.46 (National Institutes of
Health).
Statistical analysis
GraphPad Prism 6 software (GraphPad Software, Inc.)
was used for data analysis. Data are presented as the mean ±
standard deviation of three biological replicates. Differences
between two groups were determined using an unpaired Student's
t-test. Differences among three or more groups were analyzed using
one-way ANOVA with Tukey's post hoc test. Correlations between
lncRNA AWPPH and ROCK2 expression were determined using Pearson's
correlation coefficient. Diagnostic values of lncRNA AWPPH and
ROCK2 for T-ALL were determined using receiver operating
characteristic (ROC) curve analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
lncRNA AWPPH and ROCK2 mRNA levels are
upregulated in patients with pediatric T-ALL
lncRNA AWPPH and ROCK2 mRNA expression in bone
marrow was detected by RT-qPCR. Compared with the control group,
lncRNA AWPPH (Fig. 1A) and ROCK2
mRNA (Fig. 1B) expression was
significantly upregulated in patients with T-ALL. In addition,
western blotting results revealed that ROCK2 protein levels were
higher in patients (n=4) than in controls (n=2) (P<0.05;
Fig. 1C). It is worth noting that
due to a lack of quality protein samples, the number of assayed
patients was limited.
lncRNA AWPPH and ROCK2 upregulation
distinguishes patients with T-ALL from healthy controls
The diagnostic values of lncRNA AWPPH and ROCK2 for
T-ALL were analyzed by ROC curve analysis. In the present analysis,
true positive cases were patients with T-ALL, while true negative
cases were healthy controls. For lncRNA AWPPH, the area under the
curve (AUC) was 0.8954 (95% CI, 0.8168–0.9740; standard error,
0.04008; Fig. 2A). For ROCK2 mRNA,
the AUC was 0.9194 (95% CI, 0.8537–0.9851; standard error, 0.03351;
Fig. 2B).
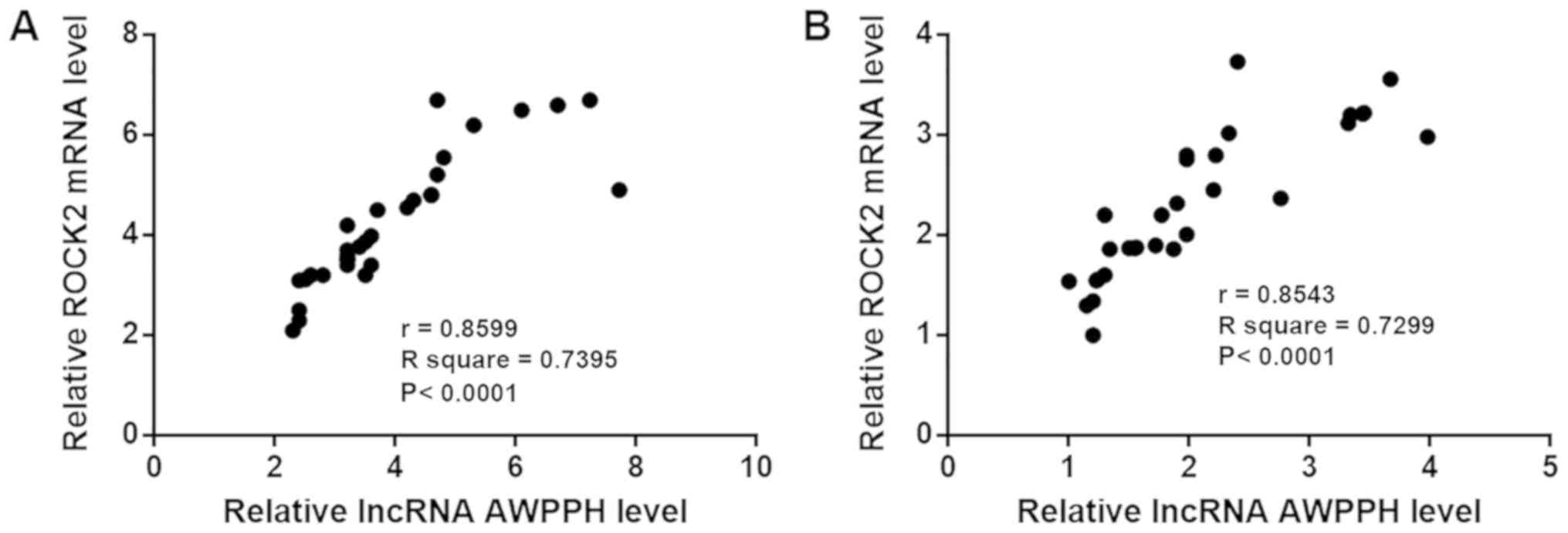
lncRNA AWPPH and ROCK2 expression are
positively correlated
The correlation between lncRNA AWPPH and ROCK2
expression was analyzed using Pearson's correlation coefficient. As
shown in Fig. 3, the expression
levels of lncRNA AWPPH and ROCK2 were positively correlated in
patients with T-ALL (Fig. 3A) and in
healthy controls (Fig. 3B).
lncRNA AWPPH and ROCK2 regulate each
other in T-ALL cells
Expression levels of lncRNA AWPPH and ROCK2 were
detected 24 h after transfection. As shown in Fig. 4A, overexpression of lncRNA AWPPH and
ROCK2 was achieved in the human T-ALL Loucy cell line (P<0.05;
Fig. 4A). Compared with the C and NC
groups, overexpression of ROCK2 significantly upregulated lncRNA
AWPPH expression (P<0.05; Fig.
4B). In addition, overexpression of lncRNA AWPPH upregulated
ROCK2 expression at both the mRNA and protein levels (P<0.05;
Fig. 4C).
lncRNA AWPPH and ROCK2 overexpression
regulate T-ALL cell behaviors
Cell proliferation and apoptosis were analyzed by
CCK-8 and cell apoptosis assays, respectively, at 24 h after
transfection. Compared with the C and NC groups, overexpression of
either lncRNA AWPPH or ROCK2 significantly promoted proliferation
(Fig. 5A) and inhibited apoptosis
(Fig. 5B) of Loucy cells
(P<0.05).
Discussion
lncRNA AWPPH is a recently identified oncogenic
lncRNA in liver cancer and bladder cancer (9,10). To
the best of our knowledge, the involvement of lncRNA AWPPH in other
human diseases remains unknown. The present study revealed that the
expression levels of lncRNA AWPPH and ROCK2 were upregulated in
pediatric T-ALL. In addition, lncRNA AWPPH and ROCK2 overexpression
upregulated each other's expression, suggesting that they may
promote the development and progression of T-ALL.
Previous studies have demonstrated that the
development of T-ALL is accompanied by changes in the expression
patterns of a large set of lncRNAs (13,14), and
that the differential expression of lncRNAs defines the subtypes of
this disease (13). The present
study reported the upregulation of lncRNA AWPPH in pediatric T-ALL.
lncRNA AWPPH has been associated with the regulation of cancer cell
proliferation (9,10). Consistently, the present study
revealed that upregulation of lncRNA AWPPH significantly promoted
the proliferation of T-ALL cells. Additionally, the present
findings improved the understanding of the function of lncRNA
AWPPH, suggesting that lncRNA AWPPH may be an inhibitor of cancer
cell apoptosis in T-ALL.
ROCK2 is a type of serine/threonine kinase that is
involved in the regulation of smooth muscle contraction,
cytokinesis, focal adhesion and formation of actin stress fibers
(15). ROCK2 is frequently
upregulated in cancer development, and inhibition of ROCK2
expression can improve cancer treatment (11,15–17). In
the present study, ROCK2 expression was significantly upregulated
in patients with T-ALL compared with in healthy controls. ROCK2
upregulation distinguished patients with T-ALL from healthy
controls.
It is well-known that ROCK2 may exert its biological
functions by interacting with lncRNAs (18). Notably, in the present study lncRNA
AWPPH and ROCK2 upregulated each other's expression in T-ALL cells.
However, the molecular mechanism underlying the interaction between
lncRNA AWPPH and ROCK2 remains unknown. It was hypothesized that
lncRNA AWPPH and ROCK2 may directly interact with each other, or
that their interaction may be regulated by some non-pathological
mediators, since the expression levels of lncRNA AWPPH and ROCK2
were positively correlated in both patients and healthy controls.
In addition, AWPPH and ROCK2 did not affect each other's expression
in silencing assays (data not shown). Therefore, AWPPH and ROCK2
may interact with each other in a unidirectional way.
However, only one cell line was used in the present
study. Therefore, future studies should include multiple cell lines
to further verify the conclusions made in the present study. Future
studies should focus on the identification of pathological factors
that mediate the interaction between lncRNA AWPPH and ROCK2.
In conclusion, lncRNA AWPPH and ROCK2 were
upregulated in pediatric T-ALL. lncRNA AWPPH and ROCK2 may
upregulate each other's expression to promote the development of
this disease.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XL designed the experiments. XL and FS performed the
experiments. HS analyzed data. XL drafted the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Clinical Hospital Affiliated to Harbin
Medical University (Harbin, China). All participants' guardians
signed informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ribera JM, Hentrich M and Barta SK: Acute
lymphoblastic leukemia//HIV-associated hematological malignancies.
Springer. (Cham). 145–151. 2016.PubMed/NCBI
|
|
2
|
Chiaretti S and Foà R: T-cell acute
lymphoblastic leukemia. Haematologica. 94:160–162. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karrman K and Johansson B: Pediatric
T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer.
56:89–116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grabher C, von Boehmer H and Look AT:
Notch 1 activation in the molecular pathogenesis of T-cell acute
lymphoblastic leukaemia. Nat Rev Cancer. 6:347–359. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang J, Ding L, Holmfeldt L, Wu G,
Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et
al: The genetic basis of early T-cell precursor acute lymphoblastic
leukaemia. Nature. 481:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao X, Liu Y and Yu S: Long noncoding RNA
AWPPH promotes hepatocellular carcinoma progression through YBX1
and serves as a prognostic biomarker. Biochim Biophys Acta Mol
Basis Dis. 1863:1805–1816. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu F, Zhang X, Yu Q, Han G, Diao F, Wu C
and Zhang Y: LncRNA AWPPH inhibits SMAD4 via EZH2 to regulate
bladder cancer progression. J Cell Biochem. 119:4496–4505. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rath N and Olson MF: Rho-associated
kinases in tumorigenesis: Re-considering ROCK inhibition for cancer
therapy. EMBO Rep. 13:900–908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2011.
View Article : Google Scholar
|
|
13
|
Wallaert A, Durinck K, Van Loocke W, Van
de Walle I, Matthijssens F, Volders PJ, Avila Cobos F, Rombaut D,
Rondou P, Mestdagh P, et al: Long noncoding RNA signatures define
oncogenic subtypes in T-cell acute lymphoblastic leukemia.
Leukemia. 30:1927–1930. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ngoc PC, Tan SH, Tan TK, Chan MM, Li Z,
Yeoh AEJ, Tenen DG and Sanda T: Identification of novel lncRNAs
regulated by the TAL1 complex in T-cell acute lymphoblastic
leukemia. Leukemia. 32:2138–2151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nakagawa O, Fujisawa K, Ishizaki T, Saito
Y, Nakao K and Narumiya S: ROCK-I and ROCK-II, two isoforms of
Rho-associated coiled-coil forming protein serine/threonine kinase
in mice. FEBS Lett. 392:189–193. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vigil D, Kim TY, Plachco A, Garton AJ,
Castaldo L, Pachter JA, Dong H, Chen X, Tokar B, Campbell SL and
Der CJ: ROCK1 and ROCK2 are required for non-small cell lung cancer
anchorage-independent growth and invasion. Cancer. 72:5338–5347.
2012.
|
|
17
|
Wang L, Hou G, Xue L, Li J, Wei P and Xu
P: Autocrine motility factor receptor signaling pathway promotes
cell invasion via activation of ROCK-2 in esophageal squamous cell
cancer cells. Cancer Invest. 28:993–1003. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi S, Peng Q, Shao X, Xie J, Lin S, Zhang
T, Li Q, Li X and Lin Y: Self-assembled tetrahedral DNA
nanostructures promote adipose-derived stem cell migration via
lncRNA XLOC 010623 and RHOA/ROCK2 signal pathway. ACS Appl Mater
Interfaces. 8:19353–19363. 2016. View Article : Google Scholar : PubMed/NCBI
|