Introduction
Renal cell carcinoma (RCC) is a complex,
multifaceted malignant disease comprising different and specific
entities, with over two-thirds of all cases being clear cell RCC
(ccRCC) (1). Of all ccRCCs cases,
60–80% are considered to be associated with the biallelic
inactivation of the Von Hippel Lindau (VHL) gene, which causes
aberrant accumulation of hypoxia-inducible factor (HIF) (2). In addition, HIF is a critical component
of several proteins that constitute the cellular machinery for
adaption to hypoxia, which is the result of rapid cell
proliferation and poor vascularization of solid tumors (3). Although previous evidence from genetic
and biological studies indicates that hypoxia-associated pathways
play key roles in the development and progression of renal cancer
(4), the underlying molecular
mechanism is complex and remains elusive.
As a highly conserved developmental signaling
pathway, deregulated Notch signaling has been reported to be
associated with tumorigenesis. The core Notch signaling pathway is
composed of five ligands (Jagged-1 and −2 and δ-like-1, −3, and −4)
and four receptors (Notch1-4). The binding of a ligand with a Notch
receptor results in two proteolytic cleavages of the receptor,
which leads to the release of the Notch intracellular domain
(NICD). Subsequently, NICD translocates into the nucleus, where it
interacts with the DNA-binding factors [CBF1/RBP-J, Su (H), Rbpsuh
and Lag-1, CSL] to regulate the expression of downstream genes
(5,6). Clinical studies have demonstrated that
Notch1 signaling is activated in ccRCC and may be a useful
biomarker for prognosis (7,8), whereas treatment with Notch1 inhibitors
may inhibit the progression of ccRCC (9). While the Notch1 signaling pathway has
been demonstrated to be involved in the progression of ccRCC, less
is known on whether and how Notch regulates the cellular hypoxic
response, particularly Notch3, the structure and function of which
differs from those of the other three Notch receptors (10). In the present study, RO4929097, a
γ-secretase inhibitor that inhibits NICD expression, thereby
decreasing the expression of the downstream Notch targets (11), was used to alter NICD3 expression.
The present study aimed to investigate the effects of altered NICD3
expression on cell proliferation, cell cycle progression and HIF-2α
protein expression, which may prove to be of value in the treatment
of RCC.
Materials and methods
Cell lines and cell culture
The human ccRCC cell lines, 786-O and ACHN were
purchased from American Type Culture Collection and maintained in
RPMI-1640 medium or minimal essential medium (MEM) supplemented
with 10% fetal bovine serum (all purchased from HyClone; Cytiva),
respectively. 786-O and ACHN cells were treated with or without
RO4929097 (cat. no. HY-11102; MCE), and were cultured under
normoxic (21% O2, 37°C and 5% CO2) or hypoxic
(2% O2, 37°C and 5% CO2) conditions for 48 h,
or with 200 µM cobalt chloride (CoCl2) (cat. no. C7300;
Beijing Solarbio Science & Technology Co., Ltd.) under 21%
O2, 37°C and 5% CO2 condition for 48 h.
Cell Counting Kit-8 (CCK-8) assay
786-O and ACHN cells were seeded into 96-well plates
at a density of 10,000 cells/well and cultured under normoxic or
hypoxic conditions at 37°C for 48 h. Then, according to the
manufacturer's protocol, the cell proliferation rates of 786-O and
ACHN were subsequently analyzed via the CCK-8 assay (cat. no.
DJDB4000X; Dojindo Molecular Technologies, Inc.).
EdU assay
EdU incorporation assay was performed using the
Cell-Light™ EdU Apollo®488 In Vitro Flow
Cytometry kit (cat. no. C10310-3; Guangzhou Ribobio Co., Ltd.),
according to the manufacturer's protocol. Briefly, 786-O and ACHN
cells were respectively cultured in RPMI-1640 and MEM medium
supplemented with 10 µM EdU for 2 h at 37°C, and washed with cold
phosphate buffered saline (PBS) containing 1% bovine serum albumin
(Beijing Solarbio Science & Technology Co., Ltd.) for three
times. Cells were resuspended in 500 µl of 1X Apollo reaction
buffer and subsequently incubated at room temperature for 30 min.
786-O and ACHN cells were re-washed twice with PBS containing 0.5%
Triton X-100, stained with 1X Hoechst33342 reaction buffer for 5
min at room temperature, re-washed twice with PBS containing 0.5%
Triton X-100, and subsequently added to 500 µl PBS. Cells were
observed under an inverted immunofluorescence microscope at ×10
magnification [IX70/SPOT RT-KE (color); Olympus
Corporation/Diagnostic Instruments Inc.] and EdU-positive cells
were counted using ImageJ software (version 1.52; National
Institute of Health).
Colony formation assay
786-O and ACHN cells were trypsinized and seeded
into 6-well plates at a density of 500 cells/well. The RPMI-1640
and MEM medium with 10% fetal bovine serum were replaced with fresh
media every 48 h, and cells were cultured at 37°C under normoxic
and hypoxic conditions, respectively. After 10 days, the size of
colonies was observed in the control group (untreated cells). When
the colonies size reached size >50 cells, the medium was removed
and the formed colonies were stained with 10% methylene blue
(Beijing Solarbio Science & Technology Co., Ltd.) in 70%
ethanol at room temperature for 5 min. The staining solution was
removed and washed three times with PBS to remove background
staining. Triplicate wells were set up for each condition, with or
without RO4929097 under normoxic or hypoxic conditions, and cells
were observed under a light microscope at ×2 magnification
[SZX12/SPOT RT-KE (color); Olympus Corporation/Diagnostic
Instruments Inc.]. The integrated optical density (IOD) of each
well was analyzed using Image-Pro Plus 6.0 software (Media
Cybernetics, Inc.).
Cell cycle analysis
Cell lines 786-O and ACHN with or without RO4929097
in normoxia or hypoxia were collected and washed with PBS by
centrifugation at 60 × g for 5 min at 4°C, prior to fixation in 75%
alcohol overnight at −20°C. Cells were washed three times with cold
PBS and resuspended in 1 ml PBS containing 1% Triton X-100, 40 µg
propidium iodide and 100 µg RNase A (both from Sigma-Aldrich; Merck
KGaA), and incubated at 4°C for at least 30 min. The staining
solution was then removed and cells were washed twice with PBS.
Samples were resuspended in 0.5 ml PBS and analyzed for DNA
contents using a flow cytometer (FACSCalibur; BD Biosciences) and
ModFit LT software (version 3.3; FACSCalibur; BD Biosciences).
Vector construction and
transfection
The coding sequence of NICD3 was cloned using a pCLE
NICD3 construct (cat. no. 26894; Addgene, Inc.), amplified and
re-cloned into a p3×FLAG-CMV-14 vector (cat. no. E7908;
Sigma-Aldrich; Merck KGaA). The plasmid DNAs were
sequence-confirmed and named as p3×FLAG-CMV-14-NICD. For the
transfection experiments, according to the manufacturer's
instructions, 786-O and ACHN cells (2×105 cells/well)
were seeded in 6-well plates overnight, then transfected with
p3×FLaG-CMV-14 vector as the control or p3×FLaG-CMV-14-NICD vector
(2 µg/ml) using Lipofectamine® 3000 (cat. no. L3000015;
Thermo Fisher Scientific, Inc.). After 24 h post-transfection,
RO4929097 was added to cell culture wells. Subsequently, the cells
were cultured under hypoxic conditions for 48 h, and the cells were
harvested for further analysis.
Western blotting
786-O and ACHN cells were lysed using RIPA lysis
buffer (cat. no. P0013C; Beyotime Institute of Biotechnology). Cell
extracts were collected via centrifugation and protein
concentration was measured by BCA method. Subsequently, 50 µg
protein per lane was separated by SDS-PAGE on a 10% gel and
transferred to PVDF membranes. The membranes were blocked with 5%
non-fat milk for 2 h at room temperature, and then incubated with
the following primary antibodies: Rabbit polyclonal anti-HIF-2
(1:500; cat. no. NB100-122; Novus Biologicals, LLC), rabbit
polyclonal anti-Notch3 (1:500; cat. no. ab23426; Abcam), mouse
monoclonal anti-cyclinD1 (1:500; cat. no. 60186-1-Ig; ProteinTech
Group, Inc.), rabbit polyclonal anti-CDK4 (1:1,000; cat. no.
YT5198; http://www.immunoway.com/), rabbit
polyclonal anti-PCNA (1:1,000; cat. no. ab152112; Abcam) and mouse
monoclonal anti-β-actin (1:1,000; cat. no. AF0003; Beyotime
Institute of Biotechnology) at 4°C overnight. Following the primary
incubation, membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit (1:10,000) or anti-mouse IgG
(1:10,000) (cat. nos. A0208 and A0216, respectively; both from
Beyotime Institute of Biotechnology) secondary antibodies at room
temperature for 2 h. Protein bands were visualized using the
chemiluminescent reagent SuperSignal West Pico Chemiluminescent
Substrate (Thermo Fisher Scientific, Inc.). Densitometry values
were normalized to levels of β-actin. Quantitation analysis for
western blot were performed using ImageJ software (version 1.52;
National Institute of Health).
Statistical analysis
Statistical analysis was performed using (version
6.0; GraphPad Software, Inc.). All experiments were performed in
triplicate and data are presented as the mean ± standard deviation.
One-way analysis of variance, followed by Tukey's post hoc test was
used to compare differences between multiple groups. GraphPad Prism
6.0 was used to create the histograms. P<0.05 was considered to
indicate a statistically significant difference.
Results
RO4929097 dose-dependently inhibits
NICD3 expression and decreases ccRCC cell proliferation in normoxia
and hypoxia
Aberrant cell proliferation is the main biological
characteristic of cancer (12), and
is affected by the tumor microenvironment, including the
O2 concentration around cancer cells (13). In the present study, cells were
treated with or without RO4929097 for 48 h. Western blot analysis
demonstrated that NICD3 expression decreased with increasing
concentration of RO4929097 in 786-O and ACHN cells (Fig. 1A). Moreover, after the treatment with
20 µM RO4929097 for 48 h, not many dead cells were obviously
observed via invert-microscope, suggesting that the direct
cytotoxicity of RO4929097 to cells was low. The CCK-8 assay was
performed to determine whether inhibition of Notch3 decreased cell
proliferation. As presented in Fig.
1B, a significant increase in proliferation was induced by
hypoxia in the control groups of 786-O cells (P<0.01) but not
ACHN cells, and the increases weren't significant in the treatment
groups of two cell lines. However, the proliferation of 786-O and
ACHN cells was significantly inhibited by RO4929097 under normoxic
and hypoxic conditions (P<0.05 in 786-O in normoxia; P<0.01
in 786-O in hypoxia; P<0.01 in ACHN in normoxia or hypoxia;).
EdU incorporation assay was performed to validate the effect of
RO4929097 or hypoxia on cell proliferation. The results
demonstrated that the number of EdU-positive cells significantly
increased in response to hypoxia in the control groups and the
treatment groups of two cell lines (all P<0.01), whereas the
number of EdU-positive cells significantly decreased following
RO4929097 treatment in normoxia and hypoxia (all P<0.05)
(Fig. 1C).
 | Figure 1.Proliferation analysis of 786-O and
ACHN cells under normoxic or hypoxic conditions, with or without
RO4929097. (A) Western blot analysis of NICD3 protein expression in
786-O and ACHN cells treated with RO4929097. Left, representative
blot images. NICD3 bands, 90 kDa and N3ICD with the transmembrane
domain bands, >90 kDa. Right, histograms of the relative protein
expression. (B) Cell Counting Kit-8 assay was performed to assess
cell proliferation. (C) EdU incorporation assay was performed to
assess cell proliferation. Left, representative images of cells.
Right, histograms presenting the numbers of EdU-positive cells
counted using ImageJ software. (D) Analysis of cell proliferation
with the colony formation assay. Left, representative images of
cell colonies. Right, histograms of the IODs of methylene
blue-stained cells measured using Image-Pro Plus 6.0 software.
*P<0.05, **P<0.01. NICD3, Notch3 intracellular domain; EdU,
5-Ethynyl-2′-deoxyuridine; IOD, integrated optical density; ns, not
significant. |
The colony formation assay is considered the gold
standard for analyzing cell proliferation potential in cellular
systems in vitro (14,15). In
the present study, 786-O and ACHN cells were cultured under
normoxic or hypoxic conditions for 10 days. At the end of the
culture period, the cells derived from a single cell were separable
and did not form a dense clone. Thus, the sum of the IOD of cells
stained with methylene blue in each well was measured, rather than
the number of colonies. The results demonstrated that under
normoxic conditions, the sum of the IOD significantly decreased
following RO4929097 treatment in 786-O cells (P<0.01), but not
in ACHN cells. Furthermore, under hypoxic conditions, the sum of
the IOD significantly increased in both 786-O and ACHN cells
(P<0.05 in 786-O and P<0.01 in ACHN), the effects of which
were reversed following RO4929097 treatment (P<0.05 in 786-O and
P<0.01 in ACHN) (Fig. 1D).
Collectively, these results suggest that hypoxia promotes the
proliferation of ccRCC cells in vitro, and inhibiting NICD3
expression may repress the proliferation of ccRCC cells under both
normoxic and hypoxic conditions.
Downregulation of NICD3 inhibits RCC
cell cycle progression via CDK4 and cyclin D1, under both normoxic
and hypoxic conditions
Cell replication and division are tightly regulated
by the cell cycle, and dysregulation of the cell cycle leads to
aberrant cell proliferation in cancer (16). Cell cycle distribution was assessed
to determine the molecular mechanism underlying the regulatory
effect of Notch3 on the proliferation of ccRCC cells. As presented
in Fig. 2A and B, hypoxia decreased
the percentage of cells in the G1 phase compared with
that in normoxia in 786-O (P<0.01) but not ACHN, and RO4929097
treatment significantly increased the percentage of cells in the
G1 phase, under both normoxic (all P<0.05) and
hypoxic conditions (all P<0.01). Furthermore, the increased
range of the percentage of cells in the G1 phase induced
by RO4929097 in hypoxia was higher compared with that in
normoxia.
The biological properties modulated by hypoxic
signaling and Notch signaling pathways are due to the initial
expression levels of a set of downstream genes, such as cyclin D1,
CDK4, P21 and so on (2,17–21). For
example, hypoxic signaling and Notch signaling regulated the
migration and invasion of breast cancer via their regulation to the
targets, snail and slug (21). Thus,
the expression levels of cyclin D1 and CDK4, which regulate
G1-S transition (22,23),
were detected via western blotting. Western blot analysis
demonstrated that hypoxia upregulated the expression of cyclin D1
and CDK4 of the control groups (all P<0.01). After treatment
RO4929097, their expressions were downregulated in no matter
normoxia (all P<0.05) or hypoxia (all P<0.01) (Fig. 2C). Given that an increasing number of
cells in the G1 phase indicates low proliferation
(24,25), consistent with those of cell cycle
analysis,, the results suggested that downregulation of NICD3 may
decrease cell proliferation by regulating the cell cycle.
NICD3 overexpression rescues the
expression of HIF-2α, CDK4, cyclin D1 and PCNA, which are inhibited
by RO4929097 in hypoxia
A set of cellular responses, such as proliferation,
drug resistance and metastasis, caused by low oxygen levels are
mediated by HIFs (3,4,13). HIF-α
is an oxygen-dependent transcriptional activator, and three α
subunits (HIF-1α, HIF-2α and HIF-3α) have been identified (4). Several lines of investigation indicate
that HIF-2α, but not HIF-1α, is upregulated in ccRCC, which
promotes ccRCC procession (26,27). In
addition, HIF-1α is not expressed in 786-O cells, which lack HIF-1α
mRNA (28). Previous studies have
reported that HIFs interact with types of Notch in cancer, such as
in breast cancer and prostate cancer (29,30),
thus the effect of hypoxia on Notch3 expression and the effect of
NICD3 on HIF-2α expression were assessed in the present study.
First, the vector containing NICD3 coding sequence was constructed.
The results demonstrated that NICD3 expression significantly
increased in 786-O and ACHN cells following transfection, by 47.0
and 168.1%, respectively (Fig. 3A).
To determine the effect of hypoxia on Notch3, the expression levels
of the full length of Notch3 (Notch3FL), NICD3 with transmembrane
domain (TD+NICD3) and NICD3 were detected via western blotting. As
the anti-Notch3 antibody used in this present study could bind with
Human NOTCH3 aa 2300 to the C-terminus (C terminal), which
Notch3FL, TD+NICD3 and NICD3 all contain, the bands of different
molecular weights, 220kDa, more than 90kDa and 90kDa, relatively
represented Notch3FL, TD+NICD3 and NICD3. The results demonstrated
that the expression levels of Notch3FL, TD+NICD3 and NICD3 did not
change in 786-O and ACHN cells following exposure to 2%
O2 for 48 h (Fig. 3B).
The effects of CoCl2 and 2% O2 on HIF-2α
expression were subsequently assessed. As presented in Fig. 3C, 2% O2, but not
CoCl2, increased HIF-2α expression in 786-O cells
(P<0.01), whereas HIF-2α expression increased in ACNH cells
following treatment with both CoCl2 (P<0.05) and 2%
O2 (P<0.01), respectively. These results may be due
to the inactivation of VHL in 786-O cells (2,28). As
presented in Fig. 3D, HIF-2α
expression was significantly decreased by RO4929097 under 2%
O2 (all P<0.01), but not 21% O2 for 48 h,
the effects of which were reversed following overexpression of
NICD3 overexpression. Collectively, these results suggest that
NICD3 affects HIF-2α expression; however, hypoxia does not affect
Notch3 expression.
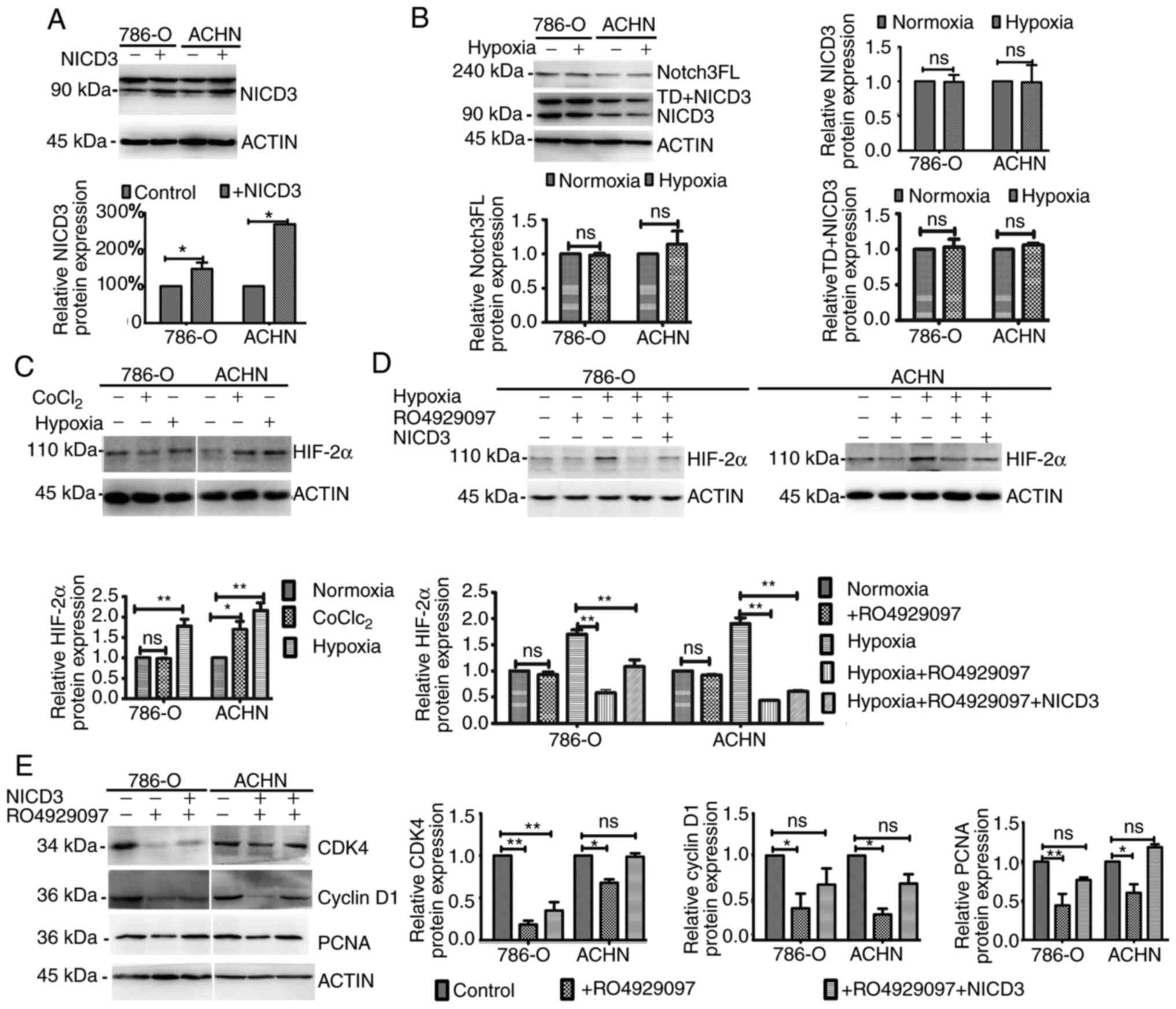 | Figure 3.Western blot analysis following
altered NICD3 expression. (A) NICD3 protein expression in 786-O and
ACHN cells transfected with or without the vector containing the
coding sequence of NICD3. Upper panels, representative blot images.
N3ICD bands, 90 kDa and TD+NICD3 bands, >90 kDa. Lower panels,
histograms of the relative protein expression. (B) Protein
expression levels of Notch3FL, TD+NICD3 and NICD3 in 786-O and ACHN
cells under 21% O2 and 2% O2 conditions.
Upper left, representative blot images. Upper right and lower
panels, histograms of the relative protein expression. (C) HIF-2α
expression in 786-O and ACHN cells treated with 200 µM
CoCl2 or 2% O2. Upper panels, representative
blot images. Lower panels, histograms of the relative protein
expression. (D) HIF-2α expression following altered NICD3
expression in normoxia or hypoxia. Upper panels, representative
blot images. Lower panels, histograms of the relative protein
expression. (E) CDK4, cyclin D1 and PCNA expression following
altered NICD3 expression in hypoxia. Left panels, representative
blot images. Right panels, histograms of the relative protein
expression. *P<0.05; **P<0.01. NICD3, Notch3 intracellular
domain; TD, transmembrane domain; FL, full length; HIF,
hypoxia-inducible factor; CDK, cyclin-dependent kinase; PCNA,
proliferating cell nuclear antigen; ns, non-significant. |
To further investigate the association between
hypoxia and Notch3 signaling, the effects of altered NICD3
expression on the cell cycle and proliferation-associated protein
expression in hypoxia were assessed in the present study. Western
blot analysis demonstrated that compared with the control groups,
the protein levels of CDK4, cyclin D1 and PCNA decreased in
RO4929097-treated 786-O and ACHN cells cultured under 2%
O2 (all P<0.05), whereas NICD3 overexpression rescued
the protein levels, suggesting that the hypoxia-induced protein
expression was regulated by NICD3 (Fig.
3E). Taken together, the results of the present study suggest
that regulation of NICD3 to these proteins is associated with its
regulation to HIF-2α.
Discussion
Notch family members are single-pass transmembrane
proteins that function both as cell surface receptors and nuclear
transcriptional regulators. The latter function is predominantly
mediated by NICD. Upon Notch binding to their ligands, the
extracellular subunit, NEC, dissociates from the transmembrane
subunit (5). NEC is subsequently
cleaved by a disintegrin, metalloprotease and γ-secretase, whereas
the NICD is released into the cytoplasm and translocates into the
nucleus to regulate the transcription of Notch target genes, which
subsequently regulates proliferation and differentiation (6,31). In
the present study, RO4929097 dose-dependently inhibited NICD3
expression in 787-O and ACHN cells, suggesting that RO4929097
downregulates Notch3 signaling.
Hypoxia and activation of HIFs have also been
described in the major RCC subtypes, and are closely associated
with ccRCC development and progression (4). In normoxia, HIF-α molecules are
subjected to a regulatory process involving the enzymatic
hypoxylation of conserved prolyl and asparaginyl residues, thus
leading to VHL protein-mediated ubiquitination and proteasomal
degradation (2). The HIF-specific
prolyl hydroxylases have an iron-binding core, which can be
replaced by CoCl2, resulting in the inhibition of HIF-1α
degradation; thus, CoCl2 is used to mimic chemical
hypoxia effects (32). However, to
the best of our knowledge, it has not yet been demonstrated that
the function of cells in response to hypoxia can completely be
recapitulated by CoCl2. In the present study,
CoCl2 did not affect HIF-2α protein expression in 786-O
cells, which lack VHL (28).
However, 2% O2 increased HIF-2α protein expression in
786-O and ACHN cells. These findings were consistent with the
results that demonstrated that 2% O2 promoted cell
proliferation and cell cycle progression in 786-O and ACHN cells,
suggesting that culturing cells in 2% O2 for 48 h did
induce the response of 786-O and ACHN cells to hypoxia in the
present study. In D324 medulloblastoma cells, CoCl2 does
not significantly alter HIF-2α expression (33). Previous studies have demonstrated
that hypoxia increases HIF-2α protein expression in 786-O cells
(34), and also increases cyclin D1
expression, which is a target gene of HIF-2α (17). The results of the present study
demonstrated that although the CoCl2 experimental group
exhibited different results from 2% O2, 2% O2
did affect ccRCC through its regulation of HIF-2α expression.
Multiple signaling pathways, including the Notch
signaling pathway, have been reported to affect the response to
hypoxia and interact with HIFs at several levels (29,35–37). In
the present study, although hypoxia did not affect Notch3
expression, NICD3 was demonstrated to regulate HIF-2α protein
expression, suggesting an association between Notch3 and HIF-2α in
ccRCCs. Furthermore, downregulation of NICD3 was demonstrated to
decrease the proliferation of 787-O and ACHN cells under both
normoxic and hypoxic conditions, suggesting the involvement of
Notch3 in the progression of ccRCC in normoxia, and the response of
ccRCC cells to hypoxia.
High cell proliferation is characterized by rapid
cell division and G1-S transition (16,24,25). A
previous study demonstrated that Notch3 affects cell cycle
progression (38). In esophageal
squamous cell carcinoma, inhibiting Notch3 induces G1
phase arrest and inhibits cell proliferation (39). In Notch-dependent T-cell lymphomas,
cyclin D3 and CDK4 are highly expressed, and γ-secretase inhibitor
induces G1 arrest in these cell lines (18). HIFs have also been reported to affect
cell cycle progression through transcriptional targets, such as
cyclin D1 and CDK inhibitors, p21 and p27 (2,19,20). In
the present study, RO4929097 treatment increased the number of
cells in the G1 phase by downregulating the expression
of cyclin D1 and CDK4 in normoxia and hypoxia, and the increased
range of the percentage of cells in the G1 phase induced
by RO4929097 in hypoxia was higher compared with that in normoxia.
Notably, overexpression of NICD3 reversed the RO4929097-induced
downregulated expression of cyclin D1 and CDK4 in hypoxia. Taken
together, these results suggest that Notch3 promotes cell cycle
progression of ccRCCs by regulating cyclin D1 and CDK4 expression,
and acts synergistically with HIFs to regulate cyclin D1 and CDK4
expression in hypoxia.
In conclusion, the results of the present study
demonstrated that Notch3 is closely associated with the cell
proliferation of ccRCC cells through its regulatory effects on the
cell cycle, and by acting synergistically with HIF-2α in hypoxia.
These findings provide novel insights into targeting Notch3 as a
promising therapeutic option in RCC. However, the exact molecular
mechanisms underlying the synergistical effect of Notch3 and HIF-2α
on ccRCC tumorigenesis and progression were not investigated in the
present study, and further investigations are required.
Acknowledgements
The authors would like to thank Dr Ying Zhang and Dr
Miao Yu (Center of Science Experiments, China Medical University,
Shenyang, China) for their technical assistance with the flow
cytometer.
Funding
The present study was funded by The Scientific
Research Fund of Liaoning Science and Technology Department,
Shenyang, China (grant no. 20180551143).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QH, CL and BW conceived and designed the present
study. QH, YF, FH, BL, JX, WS, DH and HK performed the experiments,
and QH drafted the initial manuscript, analyzed and interpreted the
data, and performed statistical analysis. CL and BW revised the
manuscript for intellectual content. All authors read and approved
the manuscript and agreed to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bergerot P, Lamb P, Wang E and Pal SK:
Cabozantinib in combination with immunotherapy for advanced renal
cell carcinoma and urothelial carcinoma: Rationale and clinical
evidence. Mol Cancer Ther. 18:2185–2193. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baba M, Hirai S, Yamada-Okabe H, Hamada K,
Tabuchi H, Kobayashi K, Kondo K, Yoshida M, Yamashita A, Kishida T,
et al: Loss of von hippel-lindau protein causes cell density
dependent deregulation of cyclinD1 expression through
hypoxia-inducible factor. Oncogene. 22:2728–2738. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rajendran JG, Mankoff DA, O'Sullivan F,
Peterson LM, Schwartz DL, Conrad EU, Spence AM, Muzi M, Farwell DG
and Krohn KA: Hypoxia and glucose metabolism in malignant tumors:
Evaluation by [18F]fluoromisonidazole and
[18F]fluorodeoxyglucose positron emission tomography
imaging. Clin Cancer Res. 10:2245–2252. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schödel J, Grampp S, Maher ER, Moch H,
Ratcliffe PJ, Russo P and Mole DR: Hypoxia, hypoxia-inducible
transcription factors, and renal cancer. Eur Urol. 69:646–657.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Artavanis-Tsakonas S, Matsuno K and
Fortini ME: Notch signaling. Science. 268:225–232. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhagat TD, Zou Y, Huang S, Park J, Palmer
MB, Hu C, Li W, Shenoy N, Giricz O, Choudhary G, et al: Notch
pathway is activated via genetic and epigenetic alterations and is
a therapeutic target in clear cell renal cancer. J Biol Chem.
292:837–846. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jędroszka D, Orzechowska M and Bednarek
AK: Predictive values of notch signalling in renal carcinoma. Arch
Med Sci. 13:1249–1254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fendler A, Bauer D, Busch J, Jung K,
Wulf-Goldenberg A, Kunz S, Song K, Myszczyszyn A, Elezkurtaj S,
Erguen B, et al: Inhibiting WNT and NOTCH in renal cancer stem
cells and the implications for human patients. Nat Commun.
11:9292020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hosseini-Alghaderi S and Baron M: Notch3
in development, health and disease. Biomolecules. 10:4852020.
View Article : Google Scholar
|
|
11
|
Olsauskas-Kuprys R, Zlobin A and Osipo C:
Gamma secretase inhibitors of notch signaling. Onco Targets Ther.
6:943–955. 2013.PubMed/NCBI
|
|
12
|
Sonnenschein C and Soto AM: Symposium on
the control of cell proliferation and cancer. Cancer Res.
49:61611989.PubMed/NCBI
|
|
13
|
Hubbi ME and Semenza GL: Regulation of
cell proliferation by hypoxia-inducible factors. Am J Physiol Cell
Physiol. 309:C775–C782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Katz D, Ito E, Lau KS, Mocanu JD,
Bastianutto C, Schimmer AD and Liu FF: Increased efficiency for
performing colony formation assays in 96-well plates: Novel
applications to combination therapies and high-throughput
screening. Biotechniques. 44:ix–xiv. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Braselmann H, Michna A, Heß J and Unger K:
CFAssay: Statistical analysis of the colony formation assay. Radiat
Oncol. 10:2232015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Raju RR, Lau KW, Tran MG, Sowter HM,
Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL and Ratcliffe
PJ: Contrasting properties of hypoxia-inducible Factor 1 (HIF-1)
and HIF-2 in von hippel-lindau-associated renal cell carcinoma. Mol
Cell Biol. 25:5675–5686. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Joshi I, Minter LM, Telfer J, Demarest RM,
Capobianco AJ, Aster JC, Sicinski P, Fauq A, Golde TE and Osborne
BA: Notch signaling mediates G1/S cell-cycle progression in T cells
via cyclin D3 and its dependent kinases. Blood. 113:1689–1698.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gardner LB, Li Q, Park MS, Flanagan WM,
Semenza GL and Dang CV: Hypoxia inhibits G1/S transition through
regulation of p27 expression. J Biol Chem. 276:7919–7926. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Green SL, Freiberg RA and Giaccia AJ:
P21(Cip1) and p27(Kip1) regulate cell cycle reentry after hypoxic
stress but are not necessary for hypoxia-induced arrest. Mol Cell
Biol. 21:1196–1206. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen J, Imanaka N, Chen J and Griffin JD:
Hypoxia potentiates notch signaling in breast cancer leading to
decreased E-cadherin expression and increased cell migration and
invasion. Br J Cancer. 102:351–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Michalides R: Prognosis for G1 cell-cycle
regulators: Useful for predicting course of disease and for
assessment of therapy in cancer. J Pathol. 188:341–343. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Milde-Langosch K and Riethdorf S: Role of
cell-cycle regulatory proteins in gynecological cancer. J Cell
Physiol. 196:224–244. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He L, Lu N, Dai Q, Zhao Y, Zhao L, Wang H,
Li Z, You Q and Guo Q: Wogonin induced G1 cell cycle arrest by
regulating wnt/β-catenin signaling pathway and inactivating CDK8 in
human colorectal cancer carcinoma cells. Toxicology. 312:36–47.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J, Li Q, Yuan J, Wang J, Chen Z, Liu
Z, Li Z, Lai Y, Gao J and Shen L: CDK4/6 inhibitor-SHR6390 exerts
potent antitumor activity in esophageal squamous cell carcinoma by
inhibiting phosphorylated Rb and inducing G1 cell cycle arrest. J
Transl Med. 15:1272017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salama R, Masson N, Simpson P, Sciesielski
LK, Sun M, Tian YM, Ratcliffe PJ and Mole DR: Heterogeneous effects
of direct hypoxia pathway activation in kidney cancer. PLoS One.
10:e01346452015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen C and Kaelin WG Jr: The VHL/HIF axis
in clear cell renal carcinoma. Semin Cancer Biol. 23:18–25. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shinojima T, Oya M, Takayanagi A, Mizuno
R, Shimizu N and Murai M: Renal cancer cells lacking hypoxia
inducible factor (HIF)-1α expression maintain vascular endothelial
growth factor expression through HIF-2α. Carcinogenesis.
28:529–536. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
De Francesco EM, Maggiolini M and Musti
AM: Crosstalk between notch, HIF-1α and GPER in breast cancer EMT.
Int J Mol Sci. 19:20112018. View Article : Google Scholar
|
|
30
|
Marignol L, Rivera-Figueroa K, Lynch T and
Hollywood D: Hypoxia, notch signalling, and prostate cancer. Nat
Rev Urol. 10:405–413. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kandasamy K, Mohan SS, Raju R,
Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD,
Mathivanan S, Pecquet C, et al: NetPath: A public resource of
curated signal transduction pathways. Genome Biol. 11:R32010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yuan Y, Hilliard G, Ferguson T and
Millhorn DE: Cobalt inhibits the interaction between
hypoxia-inducible factor-alpha and von hippel-lindau protein by
direct binding to hypoxia-inducible factor-alpha. J Biol Chem.
278:15911–15916. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mutvei AP, Landor SK, Fox R, Braune EB,
Tsoi YL, Phoon YP, Sahlgren C, Hartman J, Bergh J, Jin S, et al:
Notch signaling promotes a HIF2α-driven hypoxic response in
multiple tumor cell types. Oncogene. 37:6083–6095. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zimmer M, Ebert BL, Neil C, Brenner K,
Papaioannou I, Melas A, Tolliday N, Lamb J, Pantopoulos K, Golub T
and Iliopoulos O: Small molecule inhibitors of HIF-2a translation
link its 5′-UTR iron-responsive element (IRE) to oxygen sensing.
Mol Cell. 32:838–848. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kong LY, Xi Z, Ma WT, Yang FY, Niu LD and
Shi JH: Effects of notch signal on the expressions of HIF-α and
autophagy-related genes BECLIN1, LC3I, LC3II in oxygen-glucose
deprivation induced myocardial cell injury. Zhongguo Ying Yong
Sheng Li Xue Za Zhi. 35:165–168. 2019.(In Chinese). PubMed/NCBI
|
|
36
|
Yu N, Wu JL, Xiao J, Fan L, Chen SH and Li
W: HIF-1α regulates angiogenesis via notch1/STAT3/ETBR pathway in
trophoblastic cells. Cell Cycle. 18:3502–3512. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Moriyama H, Moriyama M, Ozawa T, Tsuruta
D, Iguchi T, Tamada S, Nakatani T, Nakagawa K and Hayakawa T: Notch
signaling enhances stemness by regulating metabolic pathways
through modifying p53, NF-κB, and HIF-1α. Stem Cells Dev.
27:935–947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dang TP: Notch, apoptosis and cancer. Adv
Exp Med Biol. 727:199–209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu Z, Ren Y, Zhang M, Fan T, Wang Y, Zhao
Q, Liu HM, Zhao W and Hou G: FLI-06 suppresses proliferation,
induces apoptosis and cell cycle arrest by targeting LSD1 and Notch
pathway in esophageal squamous cell carcinoma cells. Biomed
Pharmacother. 107:1370–1376. 2018. View Article : Google Scholar : PubMed/NCBI
|

















