Introduction
Heterogeneous nuclear ribonucleoprotein K (hnRNP K)
is a docking molecule that integrates nucleic acid-directed
processes with signal transduction pathways (1) and was initially identified as a member
of the hnRNP family in 1992 (2).
Over the past 20 years, clinical and basic science studies have
attempted to determine whether hnRNP K serves as an oncogene or
tumor suppressor gene; for example, numerous studies have reported
the oncogenic role of hnRNP K in gastric cancer (3), bladder cancer (4), colorectal cancer (5), malignant melanoma (6), hepatocellular carcinoma (7) and renal cell carcinoma (8). In contrast, Gallardo et al
(9) directly implicated hnRNP K as a
tumor suppressor in acute myeloid leukemias by generating an hnRNP
K haploinsufficient mouse model. In addition, Bastidas et al
(10) demonstrated that the
inactivation of hnRNP K was a potential driver in mycosis fungoides
development. However, there are currently conflicting results for
the same tumor. For example, previous studies have reported that
the hnRNP K protein was upregulated in gastric cell lines and
tissue microarrays, where it was associated with the poor survival
of patients with gastric cancer (3,11).
However, our previous study concluded that hnRNP K served a tumor
suppressive role in gastric cancer (12), which contradicts previous studies.
These findings are because hnRNP K has demonstrated the capacity to
regulate both tumor suppressive and oncogenic pathways, and both
the overexpression and knockdown result in cell proliferation and
apoptotic defects (13). Therefore,
it remains necessary to perform basic scientific studies to
determine the function of hnRNP K and its associated downstream
signaling pathways.
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most prevalent cancer, with a worldwide incidence of 600,000
cases reported annually (14).
Previous studies, including the tissue protein analysis of clinical
samples, have indicated that hnRNP K may be a biomarker for
predicting the potential for the malignant transformation of
precancerous lesions and the poor prognosis of patients with HNSCC
(15,16). However, to the best of our knowledge,
few studies have performed phenotypic experiments to verify the
genetic role of hnRNP K in HNSCC cell lines. In addition, the
underlying molecular mechanisms of hnRNP K in the development and
metastasis remain to be determined.
The present study aimed to investigate the roles of
hnRNP K in the proliferation and migration of HNSCC and to
determine the possible associated signaling pathways.
Materials and methods
Oncomine, UALCAN and TISIDB database
analysis
The Oncomine database (17,18)
(www.oncomine.org) was used to search for the fold
changes in hnRNP K expression levels in different types of tumor
using the following screening conditions: i) Gene name, hnRNP K;
ii) analysis type, cancer vs. normal; and iii) data type, mRNA. The
associations between hnRNP K expression levels and different
clinicopathological parameters of HNSCC were analyzed using the
UALCAN online database (http://ualcan.path.uab.edu/index.html). In the UALCAN
database, hnRNP K and HNSCC were used as the search criteria, and
the different clinical pathological parameters, such as sample
type, sex, age, ethnicity, tumor grade, individual cancer stages,
nodal metastasis status and HPV status, were selected and the
corresponding figures were downloaded. In the tumor-immune system
interactions database (TISIDB; http://cis.hku.hk/TISIDB), HNRNPK was used as the gene
symbol, with the search terms ‘clinical’ and ‘head and neck
squamous cell carcinoma’ as the cancer type to generate survival
curve.
Antibodies and chemical reagents
FBS, DMEM and minimum essential medium (MEM) were
purchased from Gibco; Thermo Fisher Scientific, Inc. The anti-hnRNP
K primary antibody (cat. no. ab52600) was purchased from Abcam,
while anti-β-Catenin (cat. no. 8480), anti-disheveled (Dvl)2 (cat.
no. 3224), anti-c-Jun (cat. no. 9165), anti-Met (cat. no. 8198),
anti-Cyclin-D1 (cat. no. 2978), anti-c-Myc (cat. no. 5605),
anti-matrix metalloproteinase (MMP)7 (cat. no. 3801) and
anti-β-actin (cat. no. 3700) primary antibodies were purchased from
Cell Signaling Technology, Inc. Moreover, anti-rabbit IgG,
HRP-linked antibody (cat. no. 7074) and anti-mouse IgG, HRP-linked
antibody (cat. no. 7076) were purchased from Cell Signaling
Technology, Inc. To confirm the outer β-catenin gene was expressed
and the plasmid transfection was successful, flag expression level
was analyzed using anti-flag (cat. no. M185-3L; Medical &
Biological Laboratories).
Cell culture and transfection
CAL-27 (human tongue squamous cell carcinoma) and
WI-38 (human embryo lung fibroblast) cell lines were cultured in
DMEM, while the FaDu (human pharyngeal squamous cell carcinoma)
cell line was cultured in MEM. All cells were purchased from
National Infrastructure of Cell Line Resource (http://www.cellresource.cn/), and the cells were
supplemented with 10% FBS and 100 U/ml penicillin and streptomycin,
and maintained in a humidified atmosphere with 5% CO2 at
37°C. The short hairpin RNA (shRNA/sh) sequence targeting hnRNP K
(shhnRNP K; 5′-GTGCTGATATTGAAACAAT-3′) (GV248-hnRNP K) and the
negative control (NC) lentivirus expressing green fluorescence
protein (shNC; 5′-TTCTCCGAACGTGTCACGT-3′) (GV248-NC) were provided
by Shanghai GeneChem Co., Ltd. The shhnRNP K and shNC (MOI=20) were
transfected into CAL-27 cells (5×105 cells per well)
using polybrene according to the manufacturer's protocol. The
duration of transfection was 12 h at 37°C followed by changing the
fresh medium. Transfected cells were used for subsequent
experiments after 72 h.
As the frozen shRNA after resuscitation was not
sufficient to perform experiments, siRNA was used to verify the
results of microarray results in order to save time. Small
interfering RNA (siRNA/si) targeting hnRNP K (sihnRNP K;
5′-GAGCUUCGAUCAAAAUUGATT-3′) and a non-specific siNC siRNA
(5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Guangzhou RiboBio
Co., Ltd. The target sequences in the two groups were identical to
those previously described (12).
The overexpression plasmid of β-Catenin (GV362-β-Catenin) and the
plasmid-NC (empty vector, GV362-NC) were provided by Shanghai
GeneChem Co., Ltd. The transfections were performed with
Lipofectamine 2000, and cells with sihnRNP K and siNC (30 pmol)
were harvested following 48 h of transfection at 37°C for further
experimentation.
Western blotting
Total protein was extracted from CAL-27, FaDu and
WI-38 cells using cold RIPA lysis buffer (Sigma-Aldrich; Merck
KGaA). Total protein was quantified using a bicinchoninic acid
protein assay kit (Pierce; Thermo Fisher Scientific, Inc.), the
protein samples were boiled with 2X SDS-PAGE protein loading buffer
(Beyotime Institute of Biotechnology) for 15 min and then the cell
lysates (20 µg per lane) were separated via 10% SDS-PAGE. The
separated proteins were subsequently transferred onto
nitrocellulose filter membranes (Pall Corporation) and blocked with
5% non-fat dry milk in Tris-buffered saline with 0.1% Tween-20 at
room temperature for 1 h. The membranes were then incubated with
the following primary antibodies overnight at 4°C: Anti-hnRNP K
primary antibody (1:10,000, cat. no. 8480; Abcam), anti-β-Catenin
(1:1,000, cat. no. 8480; Cell Signaling Technology), anti-Dvl2
(1:1,000, cat. no. 3224; Cell Signaling Technology), anti-c-Jun
(1:1,000, cat. no. 9165; Cell Signaling Technology), anti-Met
(1:1,000, cat. no. 8198; Cell Signaling Technology), anti-Cyclin-D1
(1:1,000, cat. no. 2978; Cell Signaling Technology), anti-c-Myc
(1:1,000, cat. no. 5605; Cell Signaling Technology), anti-MMP7
(1:1,000, cat. no. 3801; Cell Signaling Technology), anti-Flag
(1:1,000, M185-3L; Medical & Biological Laboratories) and
anti-β-actin (1:1,000, cat. no. 3700; Cell Signaling Technology).
Following the primary antibody incubation, the membranes were
incubated with anti-rabbit IgG, HRP-linked antibody (1:5,000; cat.
no. 7074) or anti-mouse IgG, HRP-linked antibody (1:5,000; cat. no.
7076) which was purchased from Cell Signaling Technology, Inc., for
1 h at room temperature. An enhanced chemiluminescence kit (cat.
no. 32106; Thermo Fisher Scientific, Inc.) was used to visualize
the membrane. β-actin was used as the loading control.
Densitometric analysis was performed using ImageJ software (version
1.8.0; National Institutes of Health).
Patient studies
A total of 20 paired tumor and adjacent normal
tissues (17 males and 3 females; median age, 63; age range, 44–84
years) were collected from patients with HNSCC at Beijing Tongren
Hospital (Beijing, China) between March 2018 and March 2019. The
patients had no history of radiotherapy or chemotherapy prior to
surgery. Written, informed consent was obtained from all patients
and the use of patient specimens for the experiments was approved
by the Ethics Committee of Beijing Tongren Hospital, Capital
Medical University (Beijing, China).
Immunohistochemistry
The patient tissue were fixed with 10% formalin at
room temperature for 24 h. Briefly, 5-µm sections of
paraffin-embedded tissue were deparaffinized in xylene and
rehydrated in a descending series of ethanol at room temperature.
The deparaffinized sections were incubated with 3%
H2O2 at room temperature for 15 min to
inhibit the endogenous peroxidase activity. The sections were then
boiled in antigen retrieval solution for 8 min and blocked with 10%
goat serum (cat. no. ZLI-9017; Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd.) for 30 min at 37°C. Subsequently, the
tissue sections were incubated with an anti-hnRNP K monoclonal
antibody (1:250) overnight at 4°C, followed by incubation with
anti-rabbit IgG, HRP-linked secondary antibody (1:5,000; cat. no.
7074) which was purchased from Cell Signaling Technology, Inc., for
1 h at room temperature. Fresh 3,3′-diaminobenzidine (DAB substrate
solution:DAB concentrate, 20:1) was added to the sections for a
color reaction at room temperature, and then, the samples were
counterstained with hematoxylin for 10 sec at room temperature.
Finally, the sections were dehydrated with an ascending series of
ethanol and cleared with xylene at room temperature, sealed with a
coverslip and visualized under a Zeiss Axio Imager Z2 Upright light
microscope (magnification, ×20; Zeiss AG).
Absolute cell count assays
CAL-27 cells were seeded into 96-well plates at a
density of 1,000 cells/well with three replicates per condition.
The plates were incubated at 37°C in a humidified incubator with 5%
CO2 for 24, 48, 72, 96 or 120 h, and the cell number was
checked by cell counter every 24 h.
Cell Counting Kit (CCK)-8 assay
CAL-27 cells were seeded into 96-well plates at a
density of 800 cells/well with three replicates per condition. The
plates were incubated at 37°C in a humidified incubator with 5%
CO2 for 24, 48, 72, 96 or 120 h and the cell viability
was analyzed every 24 h. Briefly, every 24 h, 100 µl serum-free
DMEM containing 10 µl CCK-8 reagent (Dojindo Molecular
Technologies, Inc.) was added to each well and incubated for 2 h
according to the manufacturer's protocol. The absorbance was
measured for 450 nm using microplate reader at room temperature.
Data were analyzed from ≥3 independent experiments.
Colony formation assay
CAL-27 cells were seeded into 6-well culture plates
(Corning Inc.) at a density of 1,000 cells/well, with each
condition set up in triplicate wells. The cells were cultured for
10 days at 37°C constant temperature CO2 incubator to
induce colony formation (>50 cells per colony). Following the
incubation, the cells were fixed with 4% paraformaldehyde at room
temperature for 30 min, and then stained with 1% crystal violet at
room temperature for 30 min. The colonies were counted and images
were captured using a Zeiss Axio Imager Z2 light microscope.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
The EdU incorporation assay was performed with
transfected CAL-27 cells using the Cell Light EdU
Apollo® 567 In Vitro Imaging kit (Guangzhou
RiboBio Co., Ltd.). For each group, 1×104 cells/well
were seeded into 24-well plates and incubated overnight at 37°C
constant temperature CO2 incubator. The EdU assay was
performed according to the manufacturer's protocol; however, the
cell nuclei were stained with DAPI instead of Hoechst 33342 for 30
min at room temperature, and the fixative used was 4%
paraformaldehyde for 30 min at room temperature. Five randomly
selected fields of view of EdU-positive cells were visualized using
Leica DMi8 fluorescence microscope (magnification, ×20). Image
analysis was performed using ImageJ software (version 1.8.0;
National Institutes of Health). Data were analyzed from ≥3
independent experiments.
Wound healing assay
A total of 1×106 CAL-27 cells/well were
plated into 6-well plates and cultured with complete medium in an
incubator at 37°C overnight until 100% confluence. Subsequently, a
linear scratch wound was generated in the cell monolayer using a
200-µl pipette tip. After washing with PBS 3 times, the cells were
cultured with serum-free DMEM at 37°C with 5% CO2. At 0,
12 and 24 h after the scratch was made, images were captured using
a Zeiss Axio Imager Z2 light microscope (magnification, ×10). The
wound area (%) was analyzed using ImageJ software (National
Institute of Health).
Transwell assay
CAL-27 cells were transfected with shhnRNP K and
shNC as described above. A total of 5×104 cells/well
were resuspended in 200 µl DMEM without FBS and seeded into the
upper compartments of Boyden chambers (Falcon; Corning Inc.) The
lower chamber was filled with 700 µl DMEM containing 10% FBS.
Following 12 h of incubation at 37°C, the non-migratory cells
remaining in the upper chamber were removed with cotton swabs and
the migratory cells in the lower chamber were washed with PBS three
times, fixed with 4% paraformaldehyde at room temperature for 30
min and stained with crystal purple for 15 min at room temperature.
The stained cells were visualized in five randomly selected fields
using a Zeiss Axio Imager Z2 light microscope (magnification,
×100).
Mouse xenograft model
Animal experiments were approved by the
Institutional Animal Care and Use Committee of the Institute of
Laboratory Animal Sciences, Peking Union Medical College (approval
no. GR-16002; Beijing, China). The nude mice were housed in
pathogen-free environment with 12-h light/dark cycle, controlled
humidity (50%) and temperature (26°C) and had free access to food
and water.
Equal numbers of shhnRNP K-and shNC-transfected
cells (5×106 cells) which were resuspended with PBS were
subcutaneously injected into the axilla of the left forelimb of 12
male BALB/c nude mice (six mice/group; age: 6-weeks-old; weight: 21
g; HFK Bioscience, http://www.hfkbio.com/). The tumor volume
(mm3) was determined every 2 days from the 5th day after
injection to the study endpoint using the following formula: Tumor
volume = (a × b2)/2, where ‘a’ was the longest tumor
diameter and ‘b’ was the shortest tumor diameter. The maximum tumor
volume of the study was 603.7 mm3. All animals were
sacrificed 4 weeks later by exposure to CO2 (25 l/min)
at a chamber displacement rate of 30% and subsequent cervical
dislocation, and the tumors were excised. Death was verified by
complete cessation of breathing, the lack of a heartbeat and
dilated pupils.
Microarray analysis and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cultured shhnRNP K and
shNC cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Gene expression profiles were analyzed by Shanghai OE Biotechnology
Corporation (https://www.oebiotech.com/). The hybridized signals
were assessed with Agilent SurePrint G3 Human gene expression v3
microarrays (Agilent Technologies, Inc.) for the downregulated
genes. A fold change threshold of ≤-2 and P≤0.05 were considered to
indicate a statistical significance. The differentially expressed
genes were then subjected to functional term enrichment analysis
using the Gene Ontology (GO) database (19) and signaling pathway enrichment
analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG)
database (20) using GeneSpring GX
software (version 14.9; Agilent Technologies, Inc.).
RT-qPCR was performed as previously described
(12). The reverse transcription
protocol was: 42°C for 60 min and 70°C for 5 min. The protocol of
PCR was as follows: 95°C, 2 min; 95°C, 10 sec; 60°C, 30 sec for 40
cycles in total. The primers used for the qPCR are provided in
Table I and the expression levels
were quantified using the 2−ΔΔCq method (21).
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5→3) |
|---|
| Heterogeneous
nuclear | F:
AGACCTGGAGACCGTTAC |
| ribonucleoprotein
K | R:
ATAAGCCATCTGCCATTC |
| β-Catenin | F:
GCGCCATTTTAAGCCTCTCG |
|
| R:
AAATACCCTCAGGGGAACAGG |
| Disheveled 2 | F:
CCTCCATCCTTCCACCCTAAT |
|
| R:
CATGCTCACTGCTGTCTCTCC |
| c-Jun | F:
TGAGTGACCGCGACTTTTCA |
|
| R:
TTTCTCTAAGAGCGCACGCA |
| Met | F:
CGACAGCTGACTTGCTGAGA |
|
| R:
AGGTTTATCTTTCGGTGCCCA |
| Cyclin-D1 | F:
GATCAAGTGTGACCCGGACT |
|
| R:
CTTGGGGTCCATGTTCTGCT |
| c-Myc | F:
TACAACACCCGAGCAAGGAC |
|
| R:
CGGGAGGCTGGTTTTCCA |
| Matrix | F:
GTCTCTGGACGGCAGCTATG |
| metalloproteinase
7 | R:
GATAGTCCTGAGCCTGTTCCC |
| GAPDH | F:
CATCAAGAAGGTGGTGAAGCAG |
|
| R:
CGTCAAAGGTGGAGGAGTGG |
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 5.0; GraphPad Software, Inc.) and data are
presented as the mean ± SD from three independent experiments.
Group comparisons between two groups were performed using an
unpaired two-tailed Student's t-test. Multiple group comparisons
were performed using a one-way ANOVA followed by a Bonferroni's
post hoc test. The overall survival analysis for low and high
expression levels of hnRNP K was performed using the Kaplan Meier
method and a log-rank test was used to determine the P-value.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Aberrant expression levels of hnRNP K
in HNSCC tissues and cells
To obtain an initial insight into hnRNP K expression
patterns, data mining using the Oncomine and UALCAN databases was
performed and the results are presented in Figs. 1A and S1. Compared with the normal tissues, the
mRNA expression levels of hnRNP K were upregulated in brain and CNS
cancer, cervical cancer, melanoma, myeloma and head and neck cancer
(Fig. 1A), and the expression levels
of hnRNP K in primary tumor of HNSCC was more upregulated than
normal head and neck tissues. Furthermore, hnRNP K expression
levels were positively associated with the grade, human
papillomavirus status and lymph node metastasis of patients with
HNSCC (Fig. S1E, G and H), while no
significant difference were recorded between the expression levels
of hnRNP K and the sex, age, ethnicity or tumor stage of the
patients with HNSCC (Fig. S1B-D and
F). In addition, data mining in the TISIDB revealed that high
expression levels of hnRNP K were associated with the significantly
poorer survival of patients with HNSCC compared with patients with
low expression levels (Fig. 1B).
Furthermore, the expression levels of hnRNP K were determined in
two HNSCC cell lines (CAL-27 and FaDu) and human embryonic lung
cells (WI-38) using RT-qPCR and western blotting analysis. The
results indicated that hnRNP K expression levels were upregulated
in CAL-27 and FaDu cells compared with the WI-38 cells (Fig. 1C and D). Next, 20 pairs of
paraffin-embedded cancerous and adjacent normal tissue samples from
patients with HNSCC were analyzed using immunohistochemistry. It
was revealed that hnRNP K was mainly expressed in the nuclei of
tumor tissue compared with the adjacent normal tissue, the
expression levels of hnRNP K in the cancer specimens were markedly
increased (Fig. 1E). Collectively,
these data indicated that hnRNP K may have an important role in the
development of HNSCC.
hnRNP K silencing reduces the
viability, proliferation and migration of CAL-27 cells
To investigate the function of hnRNP K in HNSCC
cells, shRNA was used to establish a stable hnRNP K-knockdown
CAL-27 cell line. Both hnRNP K mRNA and protein expression levels
in the shhnRNP K-transfected cells were downregulated compared with
the shNC-transfected cells (Fig. 2A and
B). The results of the CCK-8 and absolute cell count assays
indicated that the knockdown of hnRNP K significantly inhibited
CAL-27 cell viability and proliferation compared with the
shNC-transfected cells following 120 h of incubation (Fig. 2C and D). Furthermore, the results of
the colony formation assay revealed that the hnRNP K-knockdown
CAL-27 cell line formed significantly fewer and smaller clones
compared with the clones formed in the shNC group (Fig. 2E and F). The EdU incorporation assay
also demonstrated that hnRNP K silencing resulted in a markedly
reduced percentage of CAL-27 cells compared with the shNC group
(Fig. 2G). In addition, the hnRNP K
knockdown significantly inhibited the migratory ability of CAL-27
cells compared with the shNC group (Fig.
2H-K). Taken together, the present results indicated that hnRNP
K may promote the cell viability, proliferation and migration of
CAL-27 cells.
 | Figure 2.hnRNP K knockdown inhibits CAL-27
cell viability, proliferation and migration. The transfection
efficiency of hnRNP K knockdown using shRNA was analyzed using (A)
western blotting and (B) reverse transcription-quantitative PCR.
CAL-27 cell viability was determining using a (C) Cell Counting
Kit-8 assay and (D) an absolute count assay following the knockdown
of hnRNP K. (E) Representative images of the colony formation assay
used to evaluate the proliferation of CAL-27 cells after knocking
down the expression of hnRNP K. (F) Semi-quantification of the
results of the colony formation assay presented in part (E). (G) An
EdU incorporation assay was used to indicate the percentage of
proliferated CAL-27 cells following hnRNP K knockdown. (H) Wound
healing assay was used to determine the migratory ability of CAL-27
cells following the knockdown of hnRNP K (magnification, ×10). (I)
Semi-quantification of the wound healing assay results form part
(H). (J) hnRNP K knockdown decreased the cell migration of CAL-27
cells, as determined using a Transwell assay. Magnification, ×10.
(K) Semi-quantification of the number of migratory cells from part
(J). Error bars represent the mean ± SD of three independent
experiments, except for in part (K), where they represent the mean
± SD of five randomly selected fields of view. *P<0.05,
**P<0.01, ***P<0.001 vs. shNC. hnRNP K, heterogeneous nuclear
ribonucleoprotein K; sh/shRNA, short hairpin RNA; NC, negative
control; OD, optical density; EdU, 5-Ethynyl-2′-deoxyuridine. |
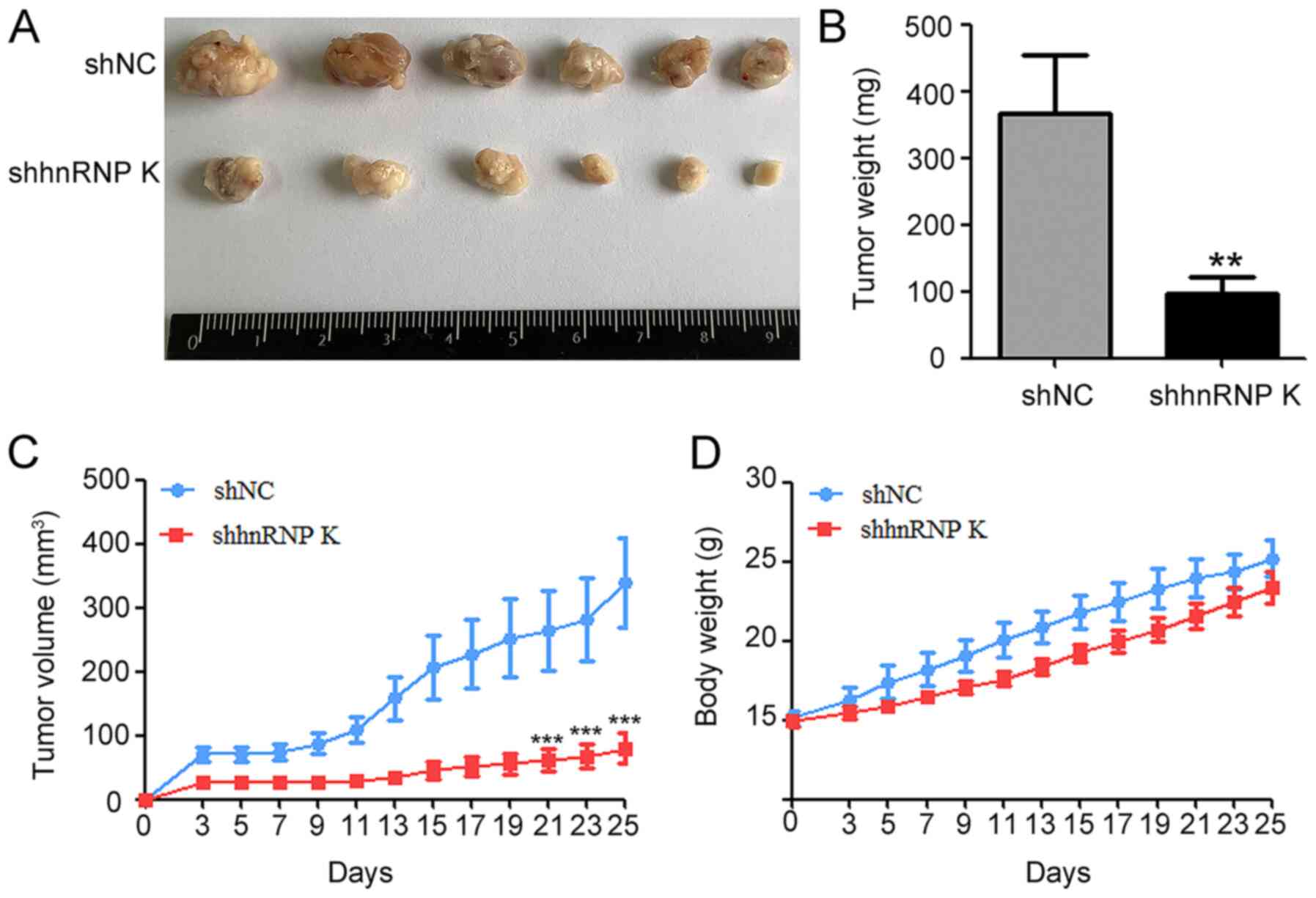
hnRNP K knockdown effectively
suppresses CAL-27 growth in vivo
To verify the oncogenic role of hnRNP K in CAL-27
cells in vivo, shhnRNP K- or shNC-transfected CAL-27 cells
were inoculated into BALB/c nude mice. Tumors derived from the
shhnRNP K cells exhibited slower growth compared with the tumors
derived from the shNC group (Fig.
3A-C). The differences in the tumor volume and weight between
the two groups were significant, which was shown as the tumor
weight and volume significantly decreased in the shhnRNP K group
compared with the shNC group (Fig.
3A-C), while there were no significant differences recorded in
the body weight of the mice between the two groups (Fig. 3D). These results indicated that hnRNP
K may promote the tumor formation by CAL-27 in vivo.
hnRNP K inhibits the activation of the
Wnt/β-Catenin signaling pathway
To determine the molecular mechanisms underlying the
phenotypic changes in hnRNP K-knockdown HNSCC cells, mRNA
microarray analysis was performed. GO functional term enrichment
analysis of the significantly differentially expressed genes of
shhnRNP K knockdown cells compared with the NC was used to identify
the role of differentially expressed hnRNP K. The results
identified that ‘Cell adhesion’ was the most highly enriched
biological process of the downregulated genes, which suggested that
hnRNP K may be an important modulator of HNSCC metastasis, while
receptor agonist activity was the most highly enriched molecular
function and extracellular region was the most highly enriched
cellular component (Fig. 4A).
Furthermore, the downregulated genes were also relevant to the
‘Response to virus’ and ‘Defense response to virus’, suggesting
that hnRNP K may be involved in the immune response (Fig.4A).
 | Figure 4.GO functional term and KEGG signaling
pathway enrichment analyses of hnRNP K-associated genes. (A) GO
annotation of downregulated mRNAs with the top 10 enrichment scores
in the categories of biological process, cellular components and
molecular functions. (B) KEGG signaling pathway enrichment analysis
of downregulated mRNAs with the top 30 enrichment scores. mRNA and
protein expression levels of hnRNP K, β-Catenin, Dvl2, c-Jun, Met,
Cyclin-D1, c-Myc and MMP7 were analyzed by (C) reverse
transcription-quantitative PCR and (D) western blotting,
respectively, following the knockdown of hnRNP K with siRNA in
CAL-27 cells. (E) Semi-quantification of the expression levels
presented in part (D). *P<0.05, **P<0.01, ***P<0.001 vs.
shNC. (F) Transfection efficiency of β-Catenin overexpression
plasmid was analyzed using western blotting. (G) Western blotting
analysis of flag, β-catenin, hnRNP K, c-Jun and c-Myc following the
inhibition of hnRNP K using siRNA and the plasmid-mediated
overexpression of β-Catenin in CAL-27 cells. (H) RT-qPCR analysis
of flag, β-catenin, hnRNP K, c-Jun and c-Myc following the
inhibition of hnRNP K using siRNA and the plasmid-mediated
overexpression of β-Catenin in CAL-27 cells. *P<0.05,
**P<0.01, ***P<0.001. GO, Gene Ontology, KEGG, Kyoto
Encyclopedia of Genes and Genomes; hnRNP K, heterogeneous nuclear
ribonucleoprotein K; Dvl2, disheveled 2; MMP7, matrix
metalloproteinase 7; si/siRNA, small interfering RNA; NC, negative
control. |
KEGG signaling pathway enrichment analysis of the
significantly differentially expressed genes was used to elucidate
the pathways and molecular interactions of hnRNP K in HNSCC. The
top 30 pathways associated with the downregulated mRNAs are listed
in Fig. 4B. The ‘Wnt signaling
pathway’ was the top pathway enriched by the downregulated genes,
which implied that the Wnt signaling pathway may be a target for
hnRNP K (Fig. 4B). Next, CAL-27
cells were transfected with sihnRNP K and siNC, and RT-qPCR
(Fig. 4C) and western blotting
analysis (Fig. 4D and E) revealed
that hnRNP K, β-catenin, Dvl2, c-Jun, Met, Cyclin-D1, c-Myc and
MMP7 expression levels were significantly downregulated in the
sihnRNP K-transfected cells compared with the siNC-transfected
cells. To determine if the β-Catenin pathway was involved in hnRNP
K regulating phenotype of HNSCC, β-Catenin was overexpressed with
plasmid (Fig. 4F). Following the
simultaneous transfection with si-hnRNP K and a β-Catenin
overexpression plasmid, the expression levels of β-Catenin, c-Myc
and c-Jun were significantly restored compared with the cells
transfected with the si-hnRNP K only (Fig. 4G and H).
These results indicated that the Wnt/β-Catenin
signaling pathway may be involved in the hnRNP K-driven inhibition
of proliferation and migration in HNSCC.
Discussion
The overexpression or amplification of oncogenes,
mutations and the de novo promoter methylation of tumor suppressor
genes have frequently been detected in HNSCC (22). hnRNP K is a member of the hnRNP
family; the hnRNP family comprises 19 hnRNP genes termed hnRNP A1
through to U (23). It is well
established that hnRNP K is overexpressed in various types of
cancer, such as hepatocellular carcinoma, bladder, pancreatic and
lung cancer (24–27). Wiesmann et al (28) proved that the downregulated
expression levels of hnRNP K had the potential to improve the
success rate of HNSCC radiotherapy. However, to the best of our
knowledge, previous studies on hnRNP K in HNSCC did not perform any
molecular investigations directly demonstrating the exact function
of hnRNP K and only a few studies have explored the downstream
regulatory mechanisms of hnRNP K (9,10,29) In
the present study, functional molecular studies were performed to
identify the exact roles of hnRNP K in HNSCC and the downstream
mechanisms involved.
The Oncomine and UALCAN databases were used to
confirm the upregulation of hnRNP K expression levels in HNSCC,
which indicated that hnRNP K may serve as an oncogene in the
development of HNSCC. Furthermore, the present study also revealed
that hnRNP K expression levels were significantly upregulated in
HNSCC tissues and cell lines, which was consistent with the results
from the Oncomine and UALCAN databases and previous studies
(15,16). Further data mining in the TISIDB
database suggested that the expression levels of hnRNP K were
negatively associated with the overall survival of patients with
HNSCC, which suggested that the hnRNP K mRNA expression levels may
serve as a potential biomarker to evaluate the prognosis of
patients with HNSCC.
In order to investigate the function of hnRNP K in
tumor development, hnRNP K was silenced in HNSCC cells, and the
subsequent in vitro and in vivo assays performed
indicated that hnRNP K knockdown attenuated the malignant and
migration properties of the cells. Similarly, Gao et al
(30) revealed that normal NIH3T3
mouse embryonic fibroblasts overexpressing hnRNP K were
characterized by enhanced malignant properties both in vitro
and in vivo. Thus, the present results further confirmed the
oncogenic activity of hnRNP K in promoting cell proliferation and
migration in HNSCC.
Although accumulating evidence has indicated that
hnRNP K has an important role in oncogenesis and metastasis, the
mechanisms remain unclear and controversial (31–33).
Therefore, to clarify the potential molecular
mechanisms of the regulation of hnRNP K, gene expression microarray
analysis was performed on hnRNP K-knockdown cells and the
corresponding NCs. KEGG signaling pathway enrichment analysis
revealed that the differentially expressed genes between the
shhnRNP K and shNC groups were enriched in the ‘Wnt signaling
pathway’, which indicated hnRNP K place an important role in HNSCC
development through Wnt signaling pathway. The knockdown of hnRNP K
in HNSCC cells blocked the Wnt signaling pathway, which was
demonstrated through the downregulated expression levels of
β-Catenin, Dvl2, c-Jun, Met, Cyclin-D1, c-Myc and MMP7; however,
following the simultaneous transfection with sihnRNP K and
β-Catenin overexpression plasmid, the expression levels of
β-Catenin, c-Myc and c-Jun were almost completely restored compared
with the sihnRNP K knockdown cells.
Previous studies have proved that the Wnt/β-Catenin
signaling pathway had a critical role in the development and
metastasis of HNSCC (34,35). β-Catenin is the central effector of
the Wnt signaling pathway, and aberrant β-Catenin expression was
identified to contribute to HNSCC therapy resistance and disease
relapse (36). In addition,
Cyclin-D1 is a target downstream effector activated by β-Catenin in
the Wnt signaling pathway, and the overexpression of cyclin D1 was
discovered to promote the proliferation and metastatic potential in
HNSCC (37–39). Furthermore, when the Wnt/β-Catenin
signaling pathway is activated, specific target oncogenes,
including c-Jun, MMP7 and c-Myc, were found to also be aberrantly
activated (40).
MMPs are a group of enzymes that cleave
extracellular matrix components, growth factors and cell-surface
receptors (41); they are deeply
involved in the process of metastasis and they were reported to be
overexpressed in patients with head and neck cancer with metastasis
compared with patients without metastasis (42). Previous studies have suggested that
MMP7 may promote tumor development and metastasis in HNSCC
(42–44). Thus, it was hypothesized that hnRNP K
may positively regulate the Wnt signaling pathway during the
proliferation and metastasis of HNSCC. To the best of our
knowledge, the present study was the first to suggest that hnRNP K
may regulate the development and metastasis of HNSCC through the
Wnt signaling pathway. Therefore, the suppression of the
Wnt/β-Catenin signaling pathway through hnRNP K may represent a
promising approach for the treatment of HNSCC.
Although this research has made progress in our
understanding, there are some limitations. Firstly, no normal head
and neck cell lines were available to use as the control cell line,
thus the present study used human embryo lung cells (WI-38). This
was considered to be acceptable, as the Human Protein Atlas network
(https://www.proteinatlas.org) indicated
that the expression levels of hnRNP K are conserved in all cell
lines. Secondly, the present study identified that hnRNP K
knockdown significantly impaired the cell proliferation and
migration of HNSCC cell lines; however, it did not investigate
whether hnRNP K overexpression had the opposite effects. Thus, this
will be investigated in future studies, if possible.
In summary, the present study observed that the
knockdown of hnRNP K markedly attenuated the viability,
proliferation and migration of HNSCC cells. In addition, the
knockdown of hnRNP K led to the downregulation of the protein
expression levels of β-Catenin, Dvl2, c-Jun, Met, Cyclin-D1, c-Myc
and MMP7 in HNSCC cells, which confirmed that the Wnt/β-Catenin
signaling pathway may be involved in the hnRNP K-driven inhibition
of proliferation and migration in HNSCC. These results may provide
a novel therapeutic biomarker for the treatment of HNSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research was funded by the National
Natural Science Foundation of China (grant no. 8160111965) and the
CAMS Innovation Fund for Medical Sciences (CIFMS) (grant no.
2016-I2M-3-019).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RG and ZH conceived and designed the study, and
critically revised the manuscript. HL performed all experiments and
drafted the manuscript. XY, ML, WZ, GZ, XZ, LC and WL helped
perform experiments and helped analyze some data of the study. XC
participated in initial design of the study and provided the
samples in the study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Animal experiments were approved by the
Institutional Animal Care and Use Committee of the Institute of
Laboratory Animal Sciences, Peking Union Medical College (approval
no. GR-16002; Beijing, China). Written, informed consent was
obtained from all patients and the use of patient specimens for the
experiments were approved by the Ethics Committee of Beijing
Tongren Hospital, Capital Medical University (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barboro P, Ferrari N and Balbi C: Emerging
roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in
cancer progression. Cancer Lett. 352:152–159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matunis MJ, Michael WM and Dreyfuss G:
Characterization and primary structure of the poly(C)-binding
heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell
Biol. 12:164–171. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang R, Zeng Y, Xu H, Chen Z, Xiang M, Fu
Y, Yin Y, Zhong J, Zeng M, Wang P, et al: Heterogeneous nuclear
ribonucleoprotein K is overexpressed and associated with poor
prognosis in gastric cancer. Oncol Rep. 36:929–935. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen X, Gu P, Xie R, Han J, Liu H, Wang B,
Xie W, Xie W, Zhong G, Chen C, et al: Heterogeneous nuclear
ribonucleoprotein K is associated with poor prognosis and regulates
proliferation and apoptosis in bladder cancer. J Cell Mol Med.
21:1266–1279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carpenter B, McKay M, Dundas SR, Lawrie
LC, Telfer C and Murray GI: Heterogeneous nuclear ribonucleoprotein
K is over expressed, aberrantly localised and is associated with
poor prognosis in colorectal cancer. Br J Cancer. 95:921–927. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wen F, Shen A, Shanas R, Bhattacharyya A,
Lian F, Hostetter G and Shi J: Higher expression of the
heterogeneous nuclear ribonucleoprotein k in melanoma. Ann Surg
Oncol. 17:2619–2627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo Y, Zhao J, Bi J, Wu Q, Wang X and Lai
Q: Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a tissue
biomarker for detection of early hepatocellular carcinoma in
patients with cirrhosis. J Hematol Oncol. 5:372012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Otoshi T, Tanaka T, Morimoto K and
Nakatani T: Cytoplasmic accumulation of heterogeneous nuclear
ribonucleoprotein K strongly promotes tumor invasion in renal cell
carcinoma cells. PLoS One. 10:e01457692015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gallardo M, Lee HJ, Zhang X, Bueso-Ramos
C, Pageon LR, McArthur M, Multani A, Nazha A, Manshouri T,
Parker-Thornburg J, et al: hnRNP K is a haploinsufficient tumor
suppressor that regulates proliferation and differentiation
programs in hematologic malignancies. Cancer Cell. 28:486–499.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bastidas Torres AN, Cats D, Mei H, Szuhai
K, Willemze R, Vermeer MH and Tensen CP: Genomic analysis reveals
recurrent deletion of JAK-STAT signaling inhibitors HNRNPK and
SOCS1 in mycosis fungoides. Genes Chromosomes Cancer. 57:653–664.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y, Jin X, Tian T and Yu DH:
Expression of hnRNPK in gastric carcinoma and its relationship with
Helicobacter pylori L-form infection. Zhonghua Zhong Liu Za
Zhi. 33:759–763. 2011.PubMed/NCBI
|
|
12
|
Huang H, Han Y, Yang X, Li M, Zhu R, Hu J,
Zhang X, Wei R, Li K and Gao R: HNRNPK inhibits gastric cancer cell
proliferation through p53/p21/CCND1 pathway. Oncotarget.
8:103364–103374. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gallardo M, Hornbaker MJ, Zhang X, Hu P,
Bueso-Ramos C and Post SM: Aberrant hnRNP K expression: All roads
lead to cancer. Cell Cycle. 15:1552–1557. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matta A, Tripathi SC, DeSouza LV, Grigull
J, Kaur J, Chauhan SS, Srivastava A, Thakar A, Shukla NK, Duggal R,
et al: Heterogeneous ribonucleoprotein K is a marker of oral
leukoplakia and correlates with poor prognosis of squamous cell
carcinoma. Int J Cancer. 125:1398–1406. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu CS, Chang KP, Chen LC, Chen CC, Liang
Y, Hseuh C and Chang YS: Heterogeneous ribonucleoprotein K and
thymidine phosphorylase are independent prognostic and therapeutic
markers for oral squamous cell carcinoma. Oral Oncol. 48:516–522.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Estilo CL, O-charoenrat P, Talbot S, Socci
ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y,
Boyle JO, et al: Oral tongue cancer gene expression profiling:
Identification of novel potential prognosticators by
oligonucleotide microarray analysis. BMC Cancer. 9:112009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ginos MA, Page GP, Michalowicz BS, Patel
KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL and Gaffney PM:
Identification of a gene expression signature associated with
recurrent disease in squamous cell carcinoma of the head and neck.
Cancer Res. 64:55–63. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al The gene ontology consortium, : Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mao L, Hong WK and Papadimitrakopoulou VA:
Focus on head and neck cancer. Cancer Cell. 5:311–316. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carpenter B, MacKay C, Alnabulsi A, MacKay
M, Telfer C, Melvin WT and Murray GI: The roles of heterogeneous
nuclear ribonucleoproteins in tumour development and progression.
Biochim Biophys Acta. 1765:85–100. 2006.PubMed/NCBI
|
|
24
|
Shu H, Hu J and Deng H: miR-1249-3p
accelerates the malignancy phenotype of hepatocellular carcinoma by
directly targeting HNRNPK. Mol Genet Genomic Med. 7:e008672019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aboushousha T, Hammam O, Helal N and El
Dahshan S: Impact of Cyclin D1 and heterogeneous nuclear
ribonucleoprotein-K (HnRNP-K) on urinary bladder carcinogenesis.
Asian Pac J Cancer Prev. 19:513–519. 2018.PubMed/NCBI
|
|
26
|
He D, Huang C, Zhou Q, Liu D, Xiong L,
Xiang H, Ma G and Zhang Z: HnRNPK/miR-223/FBXW7 feedback cascade
promotes pancreatic cancer cell growth and invasion. Oncotarget.
8:20165–20178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang F, Li W, Chen Y, Wang D, Han J and
Liu D: Downregulation of hnRNP K by RNAi inhibits growth of human
lung carcinoma cells. Oncol Lett. 7:1073–1077. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wiesmann N, Strozynski J, Beck C,
Zimmermann N, Mendler S, Gieringer R, Schmidtmann I and Brieger J:
Knockdown of hnRNPK leads to increased DNA damage after irradiation
and reduces survival of tumor cells. Carcinogenesis. 38:321–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu L, Luo C, Luo Y, Chen L, Liu Y, Wang
Y, Han J, Zhang Y, Wei N, Xie Z, et al: MRPL33 and its splicing
regulator hnRNPK are required for mitochondria function and
implicated in tumor progression. Oncogene. 37:86–94. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao R, Yu Y, Inoue A, Widodo N, Kaul SC
and Wadhwa R: Heterogeneous nuclear ribonucleoprotein K (hnRNP-K)
promotes tumor metastasis by induction of genes involved in
extracellular matrix, cell movement, and angiogenesis. J Biol Chem.
288:15046–15056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hope NR and Murray GI: The expression
profile of RNA-binding proteins in primary and metastatic
colorectal cancer: Relationship of heterogeneous nuclear
ribonucleoproteins with prognosis. Hum Pathol. 42:393–402. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du Q, Wang L, Zhu H, Zhang S, Xu L, Zheng
W and Liu X: The role of heterogeneous nuclear ribonucleoprotein K
in the progression of chronic myeloid leukemia. Med Oncol.
27:673–679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Inoue A, Sawata SY, Taira K and Wadhwa R:
Loss-of-function screening by randomized intracellular antibodies:
Identification of hnRNP-K as a potential target for metastasis.
Proc Natl Acad Sci USA. 104:8983–8988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang F, Zeng Q, Yu G, Li S and Wang CY:
Wnt/beta-catenin signaling inhibits death receptor-mediated
apoptosis and promotes invasive growth of HNSCC. Cell Signal.
18:679–687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Aminuddin A and Ng PY: Promising Druggable
target in head and neck squamous cell carcinoma: Wnt Signaling.
Front Pharmacol. 7:2442016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Roy S, Kar M, Roy S, Saha A, Padhi S and
Banerjee B: Role of β-catenin in cisplatin resistance, relapse and
prognosis of head and neck squamous cell carcinoma. Cell Oncol
(Dordr). 41:185–200. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sales KU, Giudice FS, Castilho RM, Salles
FT, Squarize CH, Abrahao AC and Pinto DS Jr: Cyclin D1-induced
proliferation is independent of beta-catenin in head and neck
cancer. Oral Dis. 20:e42–e48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hanken H, Gröbe A, Cachovan G, Smeets R,
Simon R, Sauter G, Heiland M and Blessmann M: CCND1 amplification
and cyclin D1 immunohistochemical expression in head and neck
squamous cell carcinomas. Clin Oral Investig. 18:269–276. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang B, Liu W, Li L, Lu J, Liu M, Sun Y
and Jin D: KAI1/CD82 and cyclin D1 as biomarkers of invasion,
metastasis and prognosis of laryngeal squamous cell carcinoma. Int
J Clin Exp Pathol. 6:1060–1067. 2013.PubMed/NCBI
|
|
40
|
Liang S, Zhang S, Wang P, Yang C, Shang C,
Yang J and Wang J: LncRNA, TUG1 regulates the oral squamous cell
carcinoma progression possibly via interacting with Wnt/β-catenin
signaling. Gene. 608:49–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gonzalez-Avila G, Sommer B, Mendoza-Posada
DA, Ramos C, Garcia-Hernandez AA and Falfan-Valencia R: Matrix
metalloproteinases participation in the metastatic process and
their diagnostic and therapeutic applications in cancer. Crit Rev
Oncol Hematol. 137:57–83. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rosenthal EL and Matrisian LM: Matrix
metalloproteases in head and neck cancer. Head Neck. 28:639–648.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fingleton B, Vargo-Gogola T, Crawford HC
and Matrisian LM: Matrilysin [MMP-7] expression selects for cells
with reduced sensitivity to apoptosis. Neoplasia. 3:459–468. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vento SI, Jouhi L, Mohamed H, Haglund C,
Mäkitie AA, Atula T, Hagström J and Mäkinen LK: MMP-7 expression
may influence the rate of distant recurrences and disease-specific
survival in HPV-positive oropharyngeal squamous cell carcinoma.
Virchows Arch. 472:975–981. 2018. View Article : Google Scholar : PubMed/NCBI
|