Introduction
Colorectal cancer (CRC) is the third most common
cause of cancer-related mortality among both men and women
(1). In the past decade,
chemotherapy and molecular targeted treatment have improved the
overall survival (OS) in patients with metastatic CRC to ~30 months
(2). These drugs, however, have some
limitations, including drug resistance and side effects, so the
development of new therapeutic options to prevent metastatic spread
and eventually improve patient survival is necessary (3).
Cancer immunotherapy is considered as a major
scientific and medical breakthrough (4), and several immune checkpoint-directed
antibodies have increased the OS in patients with various cancers
and are approved by the Food and Drug Administration (5,6). For
example, PD-1 inhibitor nivolumab and pembrolizumab are used for
deficient mismatch repair (dMMR) or microsatellite-instability-high
(MSI-H) CRC world-wide (7,8).
However, immunotherapies, including immune
checkpoint inhibitors to proficient-MMR CRC, have not achieved
durable objective responses against CRC (9,10).
Improving the efficacy of immunotherapies requires two approaches.
One is the use of combination therapy to alter ‘cold tumors’ which
are characterized by the absence of T cell infiltration, to ‘hot
tumors’ characterized by the accumulation of proinflammatory
cytokines and T cell infiltration (5,11). The
other is the identification of biomarkers to select patients who
are expected to respond well to immunotherapy.
The authors of the present study previously reported
phase I and II studies in which five epitope peptides were
administered to advanced-stage CRC patients (12,13). In
these studies, a low neutrophil/lymphocyte ratio and a low plasma
interleukin (IL)-6 level were the potential markers for improved
survival time of vaccinated patients (14,15).
Furthermore, it was also shown that several miRNAs and the
integrity of plasma cell-free DNA were predictive biomarkers for
active immunotherapy using epitope peptides (15–18).
This study aimed to identify novel predictive
biomarkers to select patients who are highly responsive to
immunotherapy to improve the efficacy of immunotherapy. To this
end, a comprehensive analysis of proteins in tumor tissues was
performed and sialic acid-binding immunoglobulin type lectin
(Siglec)-7 was identified as a potential predictive biomarker for
immunotherapy.
Siglecs are a family of transmembrane receptors
predominantly found in both innate and adaptive immune cells,
involved in distinguishing between self and non-self-cells by
recognizing sialic acids at the cellular surface (19,20).
Siglec-7, the seventh member of the Siglec family, is mainly
expressed on natural killer (NK) cells, monocytes, macrophages, and
a minor subset of CD8+ T cells (21,22), and
acts as an inhibitory receptor. The cytoplasmic portion of Siglec-7
contains immune receptor tyrosine-based inhibition motifs (ITIMs),
which provide inhibitory signals by recruiting the
SH2-domain-containing tyrosine phosphatase (SHP)-1 and SHP2
(22). SHP1 and SHP2 inhibit NK cell
activation pathways such as the NKG2D pathway, suppressing NK cell
cytotoxicity to tumor cells (23).
However, it has never been evaluated for its possible role in
cancer immunotherapy. In the present study, Siglec-7 was evaluated
for its potential role as a novel biomarker for active
immunotherapy.
Materials and methods
Summary of the phase II study
To assess the clinical benefits of cancer
vaccination treatment, a phase II study was conducted using five
human leukocyte antigen (HLA)-A*24:02-restricted peptides,
including kinase of the outer chloroplast membrane 1 (KOC1)
(24), translocase of outer
mitochondrial membrane 34 (TOMM34) (25), ring finger protein 43 (RNF43)
(26), vascular endothelial growth
factor receptor (VEGFR) 1 and 2 (27,28).
This phase II study was a non-randomized, HLA-A status double-blind
study. The detailed protocol of this phase II study was previously
described (13). Briefly, the
therapy consisted of a cocktail of five therapeutic epitope
peptides in addition to oxaliplatin-containing chemotherapy. The
cocktail containing 3 mg of each of the five peptides was mixed
with 1.5 ml of incomplete Freund's adjuvant and administered
subcutaneously into the thigh or axilla regions every week for 13
weeks, followed by the vaccination once every 2 weeks. Patients ≥20
years old with histologically confirmed advanced CRC who were
chemotherapy-naïve, who had adequate functions of critical organs,
and had a life expectancy of ≥3 months were eligible. Between
February 2009 and November 2012, 96 chemotherapy-naïve CRC patients
were enrolled with masked HLA-A*24:02 status.
Sample collection
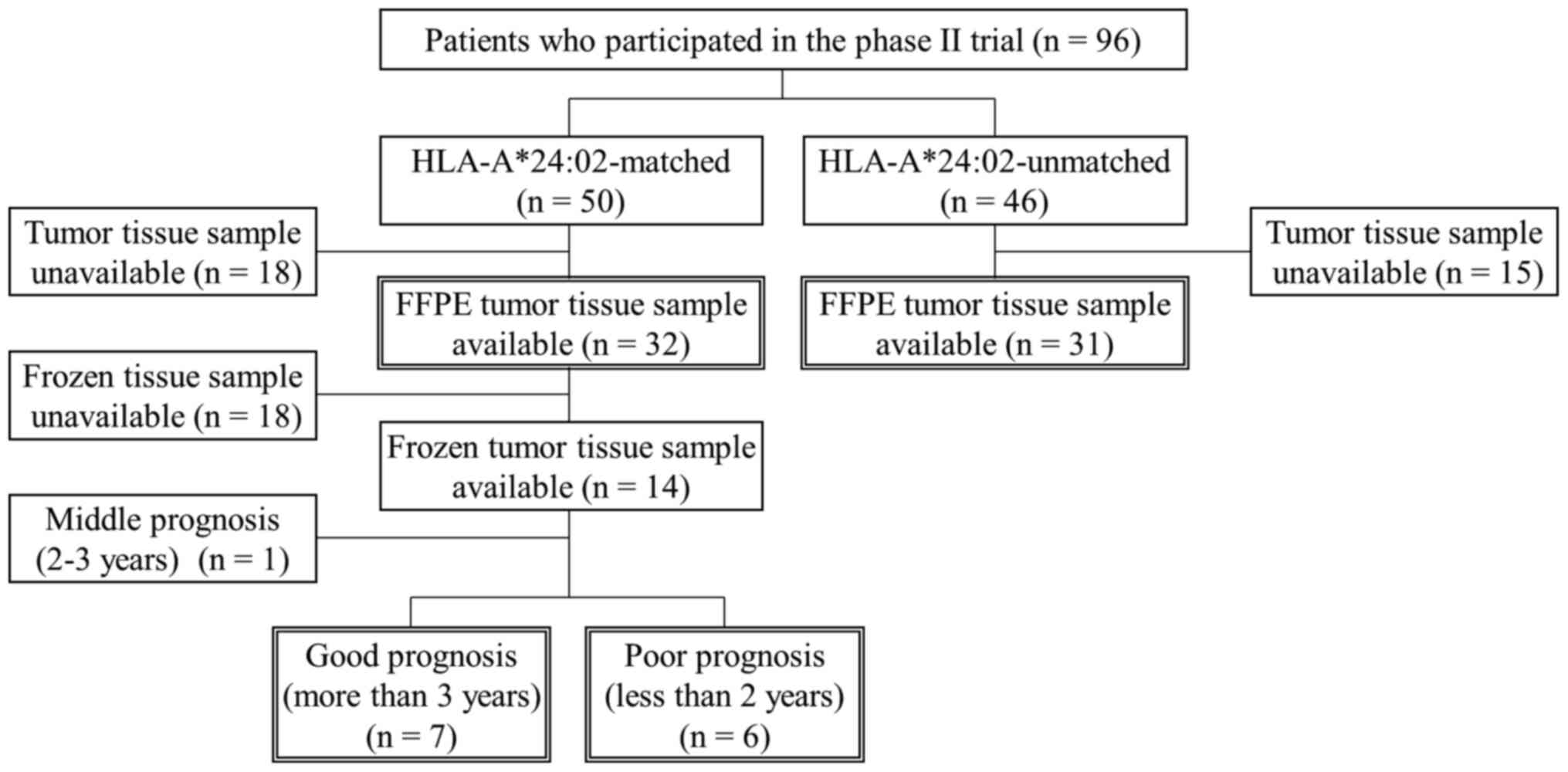
From the 96 patients who were enrolled in the phase
II trial (50 were HLA-A*24:02-matched and 46 were unmatched), 63
formalin-fixed paraffin-embedded (FFPE) tissue samples of primary
CRC were obtained (32 were HLA-A*24:02-matched and 31 were
unmatched) (Fig. 1). In 14 of the 32
HLA-A*24:02-matched patients, fresh tissues were also snap-frozen
in liquid nitrogen and preserved at −80°C until further
examination. Primary CRC tissues were obtained by surgery prior to
the vaccine treatment at Yamaguchi University Hospital and
affiliated hospitals. All samples were obtained with the patients'
written informed consent. This study was conducted according to the
Declaration of Helsinki and was approved by the Institutional
Ethics Review Boards of Yamaguchi University (approval no. H20-102;
Clinical Trials Registry: UMIN000001791).
Comprehensive proteomic analysis of
tumor tissue
A comprehensive analysis of the protein levels in
tumor tissue lysate was performed using the SOMAscan (SomaLogic,
Inc.) to quantify 1,129 biologically relevant proteins as
previously described (29). Frozen
CRC tissue samples were available from patients who survived for
either more than three years or less than two years (Fig. 1). According to the manufacturer's
protocol (SomaLogic, Inc.), the total protein of the frozen CRC
tissue sample was extracted with lysis buffer T-PER Tissue Protein
Extraction Reagent (Thermo Fisher Scientific, Inc.) supplemented
with Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific,
Inc.) through a Qiagen TissueLyser (Qiagen). Samples were sent to
SomaLogic and analyzed using the SOMAscan assay. In this assay,
protein signals were converted to nucleotide signals using
chemically modified nucleotides so that quantification could be
done using relative fluorescence signal on microarrays. For this
reason, SOMAscan measurements were presented as relative
fluorescence units (RFUs).
Western blot analysis
Western blot analysis was performed as previously
described (30), using the same
extracts as those used in the comprehensive analysis of SOMAscan.
Briefly, protein samples (10 µg) were separated on 10% SDS-PAGE and
transferred onto a PVDF membrane (Bio-Rad Laboratories, Inc.).
Membranes were blocked by pre-incubation with 3% skim milk for 30
min at room temperature and then were incubated with anti-Siglec-7
antibody (ProteinTech Group, Inc.) at 4ºC overnight. After washing
3 times with Tris-buffered saline with Tween-20 (TBST) buffer, the
membranes were incubated with the corresponding secondary antibody
for 1 h at room temperature. Immunoreactions were detected using an
enhanced chemiluminescence (ECL) western blotting detection system
and an Amersham Imager 600 (GE Healthcare Life Sciences).
Densitometry analysis was performed using ImageJ software (National
Institutes of Health) (31). Since
the protein levels of VCP, one of the housekeeping proteins, are
more stable compared to other housekeeping proteins, such as
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and actin, VCP was
chosen as the loading control (30,32,33).
Immunohistochemistry
Immunohistochemistry was carried out on 4-µm-thick
FFPE sections. For staining Siglec-7, sections were deparaffinized
through xylene and graded alcohols, and antigen retrieval was
performed in 10 mM Tris-EDTA buffer pH 9.0 (Dako) in a microwave at
95°C for 20 min. Endogenous peroxidase activity in the sections was
blocked with 3% hydrogen peroxidase for 20 min, and nonspecific
protein binding was blocked with Protein Block Serum-Free (Dako)
for 10 min. The staining procedures were performed in a Dako
Autostainer (Dako) according to the manufacturer's protocol.
Sections were incubated with an anti-Siglec-7 antibody (rabbit
polyclonal, 13939-1-AP, ProteinTech Group, Inc.; dilution 1:800) at
room temperature for 1 h. After washing 3 times with
phosphate-buffered saline (PBS), the sections were incubated with
the corresponding secondary antibody for 30 min. The reactions were
visualized with 3,3′-diaminobenzidine chromogen (DAB; Dako) and
counterstained with Mayer's hematoxylin. Images were acquired using
the All-in-one fluorescence microscope BZ-X710 (Keyence).
Considering the important role of tumor-infiltrating
lymphocytes (TILs) for the CRC prognosis, the densities of
CD3+, CD4+, CD8+, and forkhead box
P3 (FOXP3)+ T cells in CRC tissues were also examined.
Immunohistochemistry for TILs was performed as previously described
(34,35). Briefly, using the Ventana Discovery
XT staining system (Ventana), the sections were incubated with
anti-CD3 antibody (mouse monoclonal, 518110079; Ventana), anti-CD4
(mouse monoclonal, 518108816; Ventana), anti-CD8 (mouse monoclonal,
IR623; Dako; dilution 1:50), and anti-FOXP3 (mouse monoclonal,
ab20034; Abcam; dilution 1:100). The microscopic images were
acquired using a high-resolution digital slide scanner
NanoZoomer-XR C12000 (Hamamatsu Photonics).
Immunofluorescence
Immunofluorescence was carried out on 4-µm-thick
FFPE sections the same way as immunohistochemistry. Sections were
deparaffinized and antigen retrieval was performed in 10 mM
Tris-EDTA buffer pH 9.0 (Dako) in a microwave at 95°C for 20 min.
Nonspecific protein binding was blocked with Protein Block
Serum-Free (Dako) for 10 min. Sections were incubated with an
antibody mixture (1:800 diluted anti-Siglec-7 antibody, and 1:400
diluted anti-CD68 antibody; mouse monoclonal, Ab783; Abcam) at 4°C
overnight. The next day, after washing 3 times with PBS, sections
were incubated with secondary antibody mixture (1:1,000 diluted
anti-mouse Alexa Fluor 568 and 1:1,000 diluted anti-rabbit Alexa
Fluor 488; Thermo Fisher Scientific) for 60 min at room
temperature. Slides were counterstained with DAPI blue to visualize
nuclei. All staining procedures were performed manually, and
stained sections were visualized and photographed using the
All-in-one fluorescence microscope BZ-X710 (KEYENCE; magnification,
×200). From each section, 10 fields near the center of the tumor
with the highest density of Siglec-7-positive cells and
CD68-positive cells were manually selected by observers. Images
were analyzed with an algorithm for positive pixel count using
ImageJ software (NIH) to quantify the expression levels of Siglec-7
and CD68. The threshold intensity was set at 40 for Siglec-7 and
CD68 staining. The results were presented as a percent of the total
positive area to the area of the examined fields.
Measurement of TILs
Based on the immunohistochemistry for TILs, the
number of TILs was measured as previously described (34,35).
Briefly, intratumoral-infiltrating CD3+,
CD4+, CD8+ and FOXP3+ cells were
defined as TILs and their numbers were measured. Those found in the
peritumoral stroma and extratumoral lymphoid structures were
excluded from this analysis. A computerized image analysis system
Tissue Studio (Definiens) was used to score all tumor lesions. The
numbers of TILs were recorded in square millimeters as the mean
number of positive cells per tumor tissue unit.
Statistical analysis
In comprehensive protein analysis, differential
expression of proteins was detected using the log2 and
Fisher ratio using Microsoft Excel 2010 (Microsoft Corporation)
(36). The log2 ratio for
a protein k was calculated according to the following
formula:
log2 ratio=log2(x¯k(good prognosis group)x¯k(poor prognosis group)),
where x¯k is
the kth protein of the sample mean of the good or
poor prognosis group. The Fisher ratio F for a protein
k was calculated using the following formula:
formula: F(k)=[x¯k(good prognosis group)-x¯k(poor prognosis group)]212[sk2(good prognosis group)+sk2(poor prognosis group)],
where sk2 is
the kth protein of the sample variance of the
good or poor prognosis group.
Differences between the two groups were estimated
using the Welch's t-test, which was selected for this study because
recent statistical recommendations and simulation studies suggest
using this test under either homoscedasticity or heteroscedasticity
conditions (37). The categorical
variables were compared using the χ2 or Fisher's exact
tests. The strength of a correlation between two groups was
assessed by the Spearman's rank correlation coefficient. The
optimal cut-off values of the expression levels of Siglec-7, CD3,
CD4, CD8, and FOXP3 were determined using either the median value
or the time-dependent receiver operating characteristic (ROC) curve
analysis using the Kaplan-Meier (KM) estimation method and Youden's
index (sensitivity + specificity - 1) (38). The survival curves were estimated
using the KM method and tested using the log-rank test. All
statistical analyses were performed using R language for 64-bit
Windows (version 3.6.1, R Development Core Team). P<0.05 was
considered to indicate a statistically significant difference.
Results
Selection of candidate protein to
predict the efficacy of vaccination
Comprehensive analysis of the expression profiles of
1,129 proteins in 13 frozen CRC tissue samples from
HLA-A*24:02-matched patients was performed. The patients were
divided into good and poor prognosis groups; in 7 cases with good
prognosis, the patients had OS of 3 years or more and in 6 cases
with poor prognosis, the patients had OS of 2 years or less.
Comparing the protein expression levels of the two groups, 23
proteins satisfied the absolute log2 ratio ≥1 and the
Fisher ratio ≥1. Of the 23 proteins, Table I shows the 10 proteins with the
highest Fisher ratio. The expression level of Sonic hedgehog (SHH)
in the good prognosis group was significantly higher than that in
the poor prognosis group (P=0.022). In contrast, the expression
levels of Siglec-7 and fibronectin were significantly higher in the
poor prognosis group than those in the good prognosis group
(P=0.016 and 0.025, respectively). Among them, Siglec-7 was
selected as a candidate protein because of the lowest P-value.
 | Table I.Predictive markers from comprehensive
proteomic analysis of tumor tissue. |
Table I.
Predictive markers from comprehensive
proteomic analysis of tumor tissue.
|
|
| Good prognosis
(n=7) | Poor prognosis
(n=6) |
|
| Welchs t-test |
|---|
|
|
|
|
|
|
|
|
|---|
| Rank | Target protein | Mean | SD | Mean | SD | |Log2
ratio| | Fisher ratio | P-value |
|---|
| 1 | Sonic Hedgehog | 492.5 | 226.9 | 230.3 | 82.7 | 1.1 | 3.66 | 0.022 |
| 2 | ICOSLG | 14492.5 | 14465.9 | 41296.1 | 34872.0 | 1.5 | 2.29 | 0.089 |
| 3 | Lysozyme | 9204.3 | 5769.5 | 19217.0 | 12240.3 | 1.1 | 2.20 | 0.079 |
| 4 | Siglec-7 | 934.8 | 511.9 | 2272.1 | 1121.8 | 1.3 | 2.00 | 0.016 |
| 5 | Siglec-9 | 416.4 | 167.9 | 1158.8 | 1044.2 | 1.5 | 1.62 | 0.089 |
| 6 | Fibronectin | 4794.1 | 3336.5 | 13751.4 | 8483.4 | 1.5 | 1.60 | 0.025 |
| 7 | FCGR3B | 1494.4 | 827.5 | 3934.3 | 3370.6 | 1.4 | 1.42 | 0.089 |
| 8 | TIMP1 | 6566.0 | 4235.2 | 14537.2 | 11608.0 | 1.1 | 1.32 | 0.117 |
| 9 | LBP | 3525.2 | 2064.2 | 12051.2 | 14610.8 | 1.8 | 1.22 | 0.152 |
| 10 | C1q | 14432.5 | 10855.0 | 32501.8 | 22469.4 | 1.2 | 1.20 | 0.085 |
Confirmation of candidate protein
expression using western blot analysis
To validate the results obtained in comprehensive
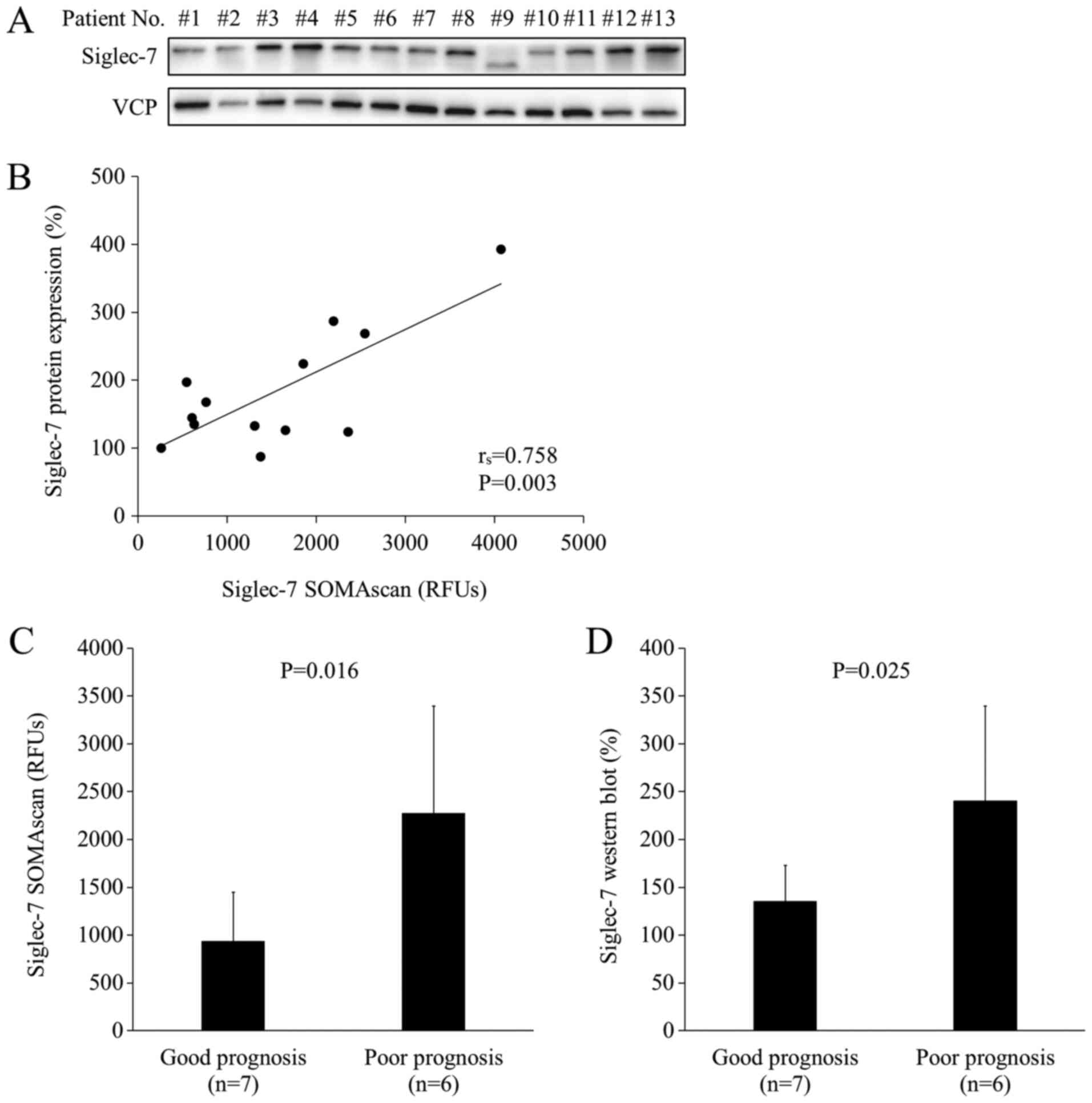
analysis of SOMAscan, western blot analysis was performed using the
same 13 samples as the ones used in SOMAscan analysis (Fig. 2A). As shown in Fig. 2A, the protein band in lane #9 was
lower than those in the other lanes. Siglec-7 has three isoforms,
and the shorter isoform may have been highly expressed in the tumor
tissue of patient #9 compared with the other patients. The levels
of Siglec-7 showed a positive correlation between SOMAscan
measurements and western blot measurements (rs=0.758, P=0.00268;
Fig. 2B). The mean expression levels
of Siglec-7 were significantly higher in the poor prognosis group
based on both SOMAscan analysis (P=0.016) and in western blot
analysis (P=0.025) (Fig. 2C and D).
These results indicated that the levels of Siglec-7 protein in the
CRC tissue were significantly higher in patients with poor
prognosis than in those with good prognosis in HLA-A*24:02-matched
cohort.
Localization of Siglec-7 in tumor
tissue
To identify the localization of Siglec-7 in CRC
tissue, immunohistochemistry and immunofluorescence were performed.
Immunohistochemistry showed that Siglec-7 was expressed in stromal
cells located between or around tumor cells (Fig. 3A). Immunofluorescence showed that
Siglec-7 was expressed in stromal cells which also expressed CD68
(Fig. 3B). These results indicated
that Siglec-7 was expressed in intratumoral macrophages.
 | Figure 3.Siglec-7 expression in tumor
microenvironment and its correlation with patient prognosis. (A)
Stromal cells in the tumor area expressed Siglec-7 (indicated by
arrows), whereas tumor cells did not (indicated by arrowheads).
They were defined based on their locations and morphological
findings. Scale bar, 100 µm. (B) Representative immunofluorescence
images of Siglec-7 (green), CD68 (red), and nuclei (blue) in CRC
tissue. Almost all of the Siglec-7+ cells expressed CD68
simultaneously, whereas there were some cells expressing only CD68.
The upper row represents a tumor with high expression level of
Siglec-7, and the lower one shows a tumor with low expression.
Scale bar, 5 µm. (C) Overall survival in patients with high levels
of Siglec-7 expression as detected with immunofluorescence images
was significantly (P=0.017) shorter than that in patients with low
levels of Siglec-7 expression in HLA-A*24:02-matched group. (D)
There was no significant difference in overall survival between
patients with high and low levels of Siglec-7 expression in
HLA-A*24:02-unmatched patients. Siglec-7, sialic acid-binding
immunoglobulin-like lectin 7; HLA, human leukocyte antigen; CRC,
colorectal cancer; RFUs, relative fluorescence units. |
Validation of Siglec-7 as a predictive
biomarker of vaccination
The levels of Siglec-7 expression in 63 CRC tissue
samples from 32 HLA-A*24:02-matched patients and 31
HLA-A*24:02-unmatched patients were examined using
immunofluorescence (Fig. 1; Table II). The levels of Siglec-7
expression ranged from 0.00001 to 7.81% (median, 0.0279%), and from
0.0400 to 0.457% (median, 0.120%) in HLA-A*24:02-matched and
-unmatched patients, respectively. The comprehensive proteomic
analysis in the present study was based on the survival of stage IV
patients. Since the median OS among stage IV CRC patients is
approximately 3 years, the optimal cut-off value was determined
using ROC curve analysis at 36 months. This analysis was performed
in HLA-A*24:02-matched patients because HLA-restricted peptides
vaccines are theoretically effective for these patients. The
cut-off value was presented as a percentage of the total positive
area of Siglec-7 to the area of the examined fields. A percent of
0.213 was selected as the cut-off value for Siglec-7 expression. In
the HLA-A*24:02-matched patients, the OS in patients with high
Siglec-7 expression was significantly shorter than that in patients
with low Siglec-7 expression (P=0.017; Fig. 3C). In contrast, in the
HLA-A*24:02-unmatched patients, there was no significant difference
in OS between patients with high or low Siglec-7 expression
(P=0.910; Fig. 3D). In patients with
low Siglec-7 expression, there was a significant difference in OS
between HLA-A*24:02-matched and -unmatched patients (P=0.041;
Fig. S1A), whereas there was no
significant difference in patients with high Siglec-7 expression
(P=0.179; Fig. S1B). The levels of
Siglec-7 expression in tumor tissue were correlated with that of
CD68 (rs=0.786, P<0.001; Fig.
S2A). However, there was no significant difference in OS
between patients with high and low levels of CD68 expression in
HLA-A*24:02-matched patients (P=0.528; Fig. S2B). These results indicated that
Siglec-7 expression in tumor microenvironment might be a predictive
biomarker of the efficacy of cancer vaccine therapy.
 | Table II.Characteristics of patients in the
phase II study whose tissues were analyzed by
immunofluorescence. |
Table II.
Characteristics of patients in the
phase II study whose tissues were analyzed by
immunofluorescence.
|
| HLA-A*24:02 |
|
|---|
|
|
|
|
|---|
|
Characteristics | Matched (n=32) | Unmatched
(n=31) | P-value |
|---|
| Age |
|
Mean | 67.9 | 64.3 | 0.069 |
|
Range | 47-82 | 47-77 |
|
| Sex |
|
Male | 13 | 18 | 0.211 |
|
Female | 19 | 13 |
|
| Unresectable
site |
|
Liver | 18 | 24 | 0.300 |
|
Lung | 11 | 9 |
|
|
Dissemination | 3 | 2 |
|
|
Bone | 0 | 2 |
|
| Lymph
node | 3 | 9 |
|
|
Other | 3 | 1 |
|
| Number of
metastatic organs |
|
One | 26 | 19 | 0.068 |
|
Two | 6 | 8 |
|
|
Three | 0 | 4 |
|
| Location of
tumor |
|
Colon | 22 | 24 | 0.572 |
|
Rectum | 10 | 7 |
|
Relationship of TIL infiltration and
prognosis with Siglec-7 expression
Because TILs have been reported as biomarkers for
CRC, they were analyzed using immunohistochemistry in CRC tissue
samples from 32 HLA-A*24:02-matched patients, the same as those
used for Siglec-7 analysis (Fig.
S3). Using ROC curve analysis at 36 months, the optimal cut-off
values were determined as 440.1, 133.8, 52.6 and 17.8 for
CD3+, CD4+, CD8+ and
FOXP3+ cell densities, respectively. There was no
significant difference in OS between patients with high and low
numbers of TILs including CD4+, CD8+ and
FOXP3+ T cells (P=0.319, 0.605 and 0.242, respectively;
Fig. 4), although there was a trend
for better OS in patients with high infiltration of CD3+
lymphocytes (P=0.065; Fig. 4A).
Next, the correlation between Siglec-7 expression and TILs in CRC
tissues was examined. There were no significant associations
between the levels of Siglec-7 expression detected in
immunofluorescence and the numbers of CD3+,
CD4+, CD8+ and FOXP3+ T cells in
immunohistochemistry (P=0.565, 0.154, 0.982 and 0.676,
respectively; Fig. 5). These
findings indicated that lymphocytes and monocytes/macrophage
infiltration might be independent.
Discussion
The purpose of the present study was to explore
proteins as novel biomarkers to predict the efficacy of
immunotherapy before treatment. First, it was demonstrated that
high levels of Siglec-7 expression in tumor tissues were associated
with shorter OS in patients treated with peptide vaccines for
metastatic CRC. Second, it was shown that Siglec-7 was expressed in
macrophages in CRC tissue. Further, there was no significant
correlation between the level of Siglec-7 expression and the number
of TILs in CRC tissue. These results indicated that high levels of
Siglec-7 expression in intratumoral macrophages can be a negative
biomarker of the vaccine treatment efficacy against metastatic CRC.
To our knowledge, this is the first report showing the relationship
between Siglec-7 and CRC prognosis.
In the comprehensive proteomic analysis, the good
and poor prognosis groups showed significant differences in
expression levels of Siglec-7, SHH, and fibronectin. SHH is a
ligand for the Hedgehog signaling pathway, which is critical for
embryonic development and carcinogenesis (39). Although increased expression of SHH
has been associated with poor prognosis in patients with various
malignancies, including CRC (40,41), the
present study obtained opposite results in this aspect.
Furthermore, fibronectin is a ligand for many members of the
integrin receptor family and it is involved in cell adhesion,
migration, growth, and differentiation (42). Because the relationship between
fibronectin and CRC has been already reported (43,44), it
was difficult to find additional roles for this protein as a
biomarker in cancer vaccination against CRC. For these reasons, SHH
and fibronectin were excluded as candidates for predictive
biomarkers.
Low levels of Siglec-7 expression in tumor tissue
was associated with better prognosis in HLA-A*24:02-matched
patients, but not in the unmatched patients. HLA-restricted epitope
peptides show theoretical antitumoral effects only in HLA-matched
patients. And only HLA-A*24:02-matched patients were considered to
be treated with vaccines in the present study. Therefore, the
resulting difference in OS based on Siglec-7 expression was only in
HLA-A*24:02-matched patients, indicating that Siglec-7 was not a
prognostic marker for CRC but a predictive biomarker for cancer
vaccination.
Siglec-7, a member of the CD33-related Siglecs, is
mainly expressed in NK cells and monocytes/macrophages (22). The distribution of
Siglec-7+ cells has been reported to differ between
peripheral blood and colonic lamina propria (45). In the peripheral blood, 75% of
Siglec-7+ cells were NK cells and 8% were monocytes. In
colonic lamina propria, in contrast, 76% of Siglec-7+
cells were monocyte/macrophage lineages and 4% were NK cells. In
this study, Siglec-7 was observed mostly in CD68+ cells
in CRC tissue, thereby it was suggested that intratumoral
macrophages expressed Siglec-7. The role of Siglec-7 in macrophage
has been poorly explored, whereas Siglec-9, another CD33-related
Siglec that shares 84% sequence homology with Siglec-7, was
reported to play an inhibitory role in macrophages (46). Specifically, Siglec-9 mediated
reduction in proinflammatory cytokine tumor necrosis factor (TNF)-α
production and potent increment in anti-inflammatory cytokine IL-10
production via ITIMs (47).
Therefore, it was hypothesized that Siglec-7-expressing macrophages
may mediate the reduction in secretion of proinflammatory cytokine
TNF-α and increase in secretion of anti-inflammatory cytokine
IL-10, resulting in immunosuppression of the tumor
microenvironment. MSI status, another factor related to the tumor
microenvironment, was also analyzed in the present study, and only
one patient had MSI-high CRC (data not shown). Although the level
of Siglec-7 expression was low in the MSI-high CRC, the
relationship between Siglec-7 expression in CRC tissue and MSI
status was not analyzed because it was statistically
inappropriate.
Cancer vaccination shows antitumoral effects by
introducing tumor antigen-specific cytotoxic T lymphocytes (CTLs).
Described as the cancer-immunity cycle (48), injected HLA-restricted epitope
peptides are captured and presented to T cells by dendritic cells
via HLA molecules. Then, activated tumor antigen-specific CTLs
infiltrate the tumor, recognizing and killing target cancer cells.
However, CTLs may have their function inhibited by PD-L1 and
immunosuppressive mediators such as IL-10 and transforming growth
factor-β in the tumor microenvironment (49,50).
Siglec-7 may pose an obstacle to CTLs by mediating
immunosuppression of tumor microenvironment via regulation of TNF-α
and IL-10 secretions, resulting in suppressed efficacy of vaccine
treatment against metastatic CRC. These mechanisms may explain the
association between high levels of Siglec-7 expression in
intratumoral macrophages and poor prognosis in HLA-A*24:02-matched
patients.
TILs, especially CD3+ and
CD8+ T cells, are prognostic biomarkers for CRC
(28,51). For instance, a scoring system based
on CD3+ and CD8+ T cells densities within the
tumor and its invasive margin, the immunoscore, was demonstrated to
be a strong prognostic factor for CRC patients (52,53). In
the present study, Siglec-7 expression was not associated with
CD3+, CD4+, CD8+ and
FOXP3+ T cells. It was suggested that Siglec-7 was an
independent biomarker from TILs. The analysis of Siglec-7 might
have led to these results by assessing macrophages rather than
lymphocytes in the tumor microenvironment.
The present study, however, had several
limitations. The first one is the small number of patients enrolled
in this study. Second, multivariate analysis, including
clinicopathological factors to adjust for confounding factors, was
not performed because it was statistically inappropriate due to the
small number of patients. The third limitation concerns the lack of
mechanistic studies. Nonetheless, understanding the functions of
Siglec-7 in the tumor environment might lead to novel
immunotherapeutic strategies such as the alteration of cold tumor
to hot tumor. For example, because Siglecs are endocytic receptors
suitable for drug delivery, the alteration may be achieved by
administering a Siglec-7-specific antibody conjugated to toxins or
chemotherapeutic agents to deplete Siglec-7-expressing macrophages
(54). Finally, the relationship
between Siglec-7 expression and other immunologically important
molecules including PD-1, PD-L1 and HLA expressions were not
evaluated.
In conclusion, Siglec-7 expression in macrophages
in tumor tissue might be a novel predictive biomarker for the
efficacy of immunotherapy against metastatic CRC. Further studies
are needed to confirm the utility of Siglec-7 as a predictive
biomarker and to analyze the role of Siglec-7 in the tumor
microenvironment.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Hiroko
Takenouchi (Department of Translational Research and Developmental
Therapeutics against Cancer, Yamaguchi University School of
Medicine) for her technical support.
Funding
This study was performed as a research program of
the Project for Development of Innovative Research on Cancer
Therapeutics (P-DIRECT; grant no. 11039020) and The Japan Agency
for Medical Research and Development (AMED; grant no.
15cm0106085h0005). This study was supported in part by a grant for
Leading Advanced Projects for Medical Innovation (LEAP; grant no.
16am0001006h0003) from the Japan Agency for Medical Research and
Development.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
KY, SH and HN designed the study. KY, NS, MX, NF,
RT, SY, STo, SM, HM, YT, SK, YS, YW, MI, STa, TI, TU, YHo, HK, TF
and YK contributed to patient recruitment and collection of data,
and analysis and interpretation of data. KY, YN, HO and YHa
performed the statistical analysis. KY, SH and HN wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was carried out according to the
Declaration of Helsinki on experimentation on human subjects and
was approved by the Institutional Ethics Review Boards of Yamaguchi
University (approval number: H20-102; Clinical Trials Registry:
UMIN000001791). Written informed consent for participation in this
study was obtained from each patient.
Patient consent for publication
Written informed consent for publication was
obtained from each patient at the time of enrollment.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Loupakis F, Cremolini C, Masi G, Lonardi
S, Zagonel V, Salvatore L, Cortesi E, Tomasello G, Ronzoni M, Spadi
R, et al: Initial therapy with FOLFOXIRI and bevacizumab for
metastatic colorectal cancer. N Engl J Med. 371:1609–1618. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sanchez-Castañón M, Er TK, Bujanda L and
Herreros-Villanueva M: Immunotherapy in colorectal cancer: What
have we learned so far? Clin Chim Acta. 460:78–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Myint ZW and Goel G: Role of modern
immunotherapy in gastrointestinal malignancies: A review of current
clinical progress. J Hematol Oncol. 10:862017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hazama S, Tamada K, Yamaguchi Y, Kawakami
Y and Nagano H: Current status of immunotherapy against
gastrointestinal cancers and its biomarkers: Perspective for
precision immunotherapy. Ann Gastroenterol Surg. 2:289–303. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kono K: Advances in cancer immunotherapy
for gastroenterological malignancy. Ann Gastroenterol Surg.
2:244–245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Le DT, Durham JN, Smith KN, Wang H,
Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et
al: Mismatch repair deficiency predicts response of solid tumors to
PD-1 blockade. Science. 357:409–413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Overman MJ, McDermott R, Leach JL, Lonardi
S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al:
Nivolumab in patients with metastatic DNA mismatch repair-deficient
or microsatellite instability-high colorectal cancer (CheckMate
142): An open-label, multicentre, phase 2 study. Lancet Oncol.
18:1182–1191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagorsen D and Thiel E: Clinical and
immunologic responses to active specific cancer vaccines in human
colorectal cancer. Clin Cancer Res. 12:3064–3069. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Duan Q, Zhang H, Zheng J and Zhang L:
Turning cold into hot: Firing up the tumor microenvironment. Trends
Cancer. 6:605–618. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hazama S, Nakamura Y, Takenouchi H, Suzuki
N, Tsunedomi R, Inoue Y, Tokuhisa Y, Iizuka N, Yoshino S, Takeda K,
et al: A phase I study of combination vaccine treatment of five
therapeutic epitope-peptides for metastatic colorectal cancer;
safety, immunological response, and clinical outcome. J Transl Med.
12:632014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hazama S, Nakamura Y, Tanaka H, Hirakawa
K, Tahara K, Shimizu R, Ozasa H, Etoh R, Sugiura F, Okuno K, et al:
A phase II study of five peptides combination with
oxaliplatin-based chemotherapy as a first-line therapy for advanced
colorectal cancer (FXV study). J Transl Med. 12:1082014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hazama S, Takenouchi H, Tsunedomi R, Iida
M, Suzuki N, Iizuka N, Inoue Y, Sakamoto K, Nakao M, Shindo Y, et
al: Predictive biomarkers for the outcome of vaccination of five
therapeutic epitope peptides for colorectal cancer. Anticancer Res.
34:4201–4205. 2014.PubMed/NCBI
|
|
15
|
Shindo Y, Hazama S, Nakamura Y, Inoue Y,
Kanekiyo S, Suzuki N, Takenouchi H, Tsunedomi R, Nakajima M, Ueno
T, et al: miR-196b, miR-378a and miR-486 are predictive biomarkers
for the efficacy of vaccine treatment in colorectal cancer. Oncol
Lett. 14:1355–1362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kijima T, Hazama S, Tsunedomi R, Tanaka H,
Takenouchi H, Kanekiyo S, Inoue Y, Nakashima M, Iida M, Sakamoto K,
et al: MicroRNA-6826 and −6875 in plasma are valuable non invasive
biomarkers that predict the efficacy of vaccine treatment against
metastatic colorectal cancer. Oncol Rep. 37:23–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanaka H, Hazama S, Iida M, Tsunedomi R,
Takenouchi H, Nakajima M, Tokumitsu Y, Kanekiyo S, Shindo Y,
Tomochika S, et al: miR-125b-1 and miR-378a are predictive
biomarkers for the efficacy of vaccine treatment against colorectal
cancer. Cancer Sci. 108:2229–2238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kitahara M, Hazama S, Tsunedomi R,
Takenouchi H, Kanekiyo S, Inoue Y, Nakajima M, Tomochika S,
Tokuhisa Y, Iida M, et al: Prediction of the efficacy of
immunotherapy by measuring the integrity of cell-free DNA in plasma
in colorectal cancer. Cancer Sci. 107:1825–1829. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Crocker PR, Paulson JC and Varki A:
Siglecs and their roles in the immune system. Nat Rev Immunol.
7:255–266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fraschilla I and Pillai S: Viewing Siglecs
through the lens of tumor immunology. Immunol Rev. 276:178–191.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Matsumoto T, Takahashi N, Kojima T,
Yoshioka Y, Ishikawa J, Furukawa K, Ono K, Sawada M, Ishiguro N and
Yamamoto A: Soluble Siglec-9 suppresses arthritis in a
collagen-induced arthritis mouse model and inhibits M1 activation
of RAW264.7 macrophages. Arthritis Res Ther. 18:1332016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nicoll G, Ni J, Liu D, Klenerman P, Munday
J, Dubock S, Mattei MG and Crocker PR: Identification and
characterization of a novel siglec, siglec-7, expressed by human
natural killer cells and monocytes. J Biol Chem. 274:34089–34095.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Daly J, Carlsten M and O'Dwyer M: Sugar
free: Novel immunotherapeutic approaches targeting siglecs and
sialic acids to enhance natural killer cell cytotoxicity against
cancer. Front Immunol. 10:10472019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Suda T, Tsunoda T, Daigo Y, Nakamura Y and
Tahara H: Identification of human leukocyte antigen-A24-restricted
epitope peptides derived from gene products upregulated in lung and
esophageal cancers as novel targets for immunotherapy. Cancer Sci.
98:1803–1808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shimokawa T, Matsushima S, Tsunoda T,
Tahara H, Nakamura Y and Furukawa Y: Identification of TOMM34,
which shows elevated expression in the majority of human colon
cancers, as a novel drug target. Int J Oncol. 29:381–386.
2006.PubMed/NCBI
|
|
26
|
Uchida N, Tsunoda T, Wada S, Furukawa Y,
Nakamura Y and Tahara H: Ring finger protein 43 as a new target for
cancer immunotherapy. Clin Cancer Res. 10:8577–8586. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ishizaki H, Tsunoda T, Wada S, Yamauchi M,
Shibuya M and Tahara H: Inhibition of tumor growth with
antiangiogenic cancer vaccine using epitope peptides derived from
human vascular endothelial growth factor receptor 1. Clin Cancer
Res. 12:5841–5849. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wada S, Tsunoda T, Baba T, Primus FJ,
Kuwano H, Shibuya M and Tahara H: Rationale for antiangiogenic
cancer therapy with vaccination using epitope peptides derived from
human vascular endothelial growth factor receptor 2. Cancer Res.
65:4939–4946. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakashima-Nakasuga C, Hazama S, Suzuki N,
Nakagami Y, Xu M, Yoshida S, Tomochika S, Fujiwara N, Matsukuma S,
Matsui H, et al: Serum LOX-1 is a novel prognostic biomarker of
colorectal cancer. Int J Clin Oncol. 25:1308–1317. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fujiwara N, Usui T, Ohama T and Sato K:
Regulation of beclin 1 protein phosphorylation and autophagy by
protein phosphatase 2A (PP2A) and death-associated protein kinase 3
(DAPK3). J Biol Chem. 291:10858–10866. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Enjoji S, Yabe R, Tsuji S, Yoshimura K,
Kawasaki H, Sakurai M, Sakai Y, Takenouchi H, Yoshino S, Hazama S,
et al: Stemness is enhanced in gastric cancer by a SET/PP2A/E2F1
axis. Mol Cancer Res. 16:554–563. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yabe R, Tsuji S, Mochida S, Ikehara T,
Usui T, Ohama T and Sato K: A stable association with PME-1 may be
dispensable for PP2A demethylation - implications for the detection
of PP2A methylation and immunoprecipitation. FEBS Open Bio.
8:1486–1496. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Galon J, Costes A, Sanchez-Cabo F,
Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M,
Berger A, Wind P, et al: Type, density, and location of immune
cells within human colorectal tumors predict clinical outcome.
Science. 313:1960–1964. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuwahara T, Hazama S, Suzuki N, Yoshida S,
Tomochika S, Nakagami Y, Matsui H, Shindo Y, Kanekiyo S, Tokumitsu
Y, et al: Intratumoural-infiltrating CD4+ and
FOXP3+ T cells as strong positive predictive markers for
the prognosis of resectable colorectal cancer. Br J Cancer.
121:659–665. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Iizuka N, Oka M, Yamada-Okabe H, Nishida
M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, et al:
Oligonucleotide microarray for prediction of early intrahepatic
recurrence of hepatocellular carcinoma after curative resection.
Lancet. 361:923–929. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zimmerman DW: Some properties of
preliminary tests of equality of variances in the two-sample
location problem. J Gen Psychol. 123:217–231. 1996. View Article : Google Scholar
|
|
38
|
Kamarudin AN, Cox T and Kolamunnage-Dona
R: Time-dependent ROC curve analysis in medical research: Current
methods and applications. BMC Med Res Methodol. 17:532017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshikawa K, Shimada M, Miyamoto H,
Higashijima J, Miyatani T, Nishioka M, Kurita N, Iwata T and Uehara
H: Sonic hedgehog relates to colorectal carcinogenesis. J
Gastroenterol. 44:1113–1117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu M, Li X, Liu T, Leng A and Zhang G:
Prognostic value of hedgehog signaling pathway in patients with
colon cancer. Med Oncol. 29:1010–1016. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maréchal R, Bachet JB, Calomme A, Demetter
P, Delpero JR, Svrcek M, Cros J, Bardier-Dupas A, Puleo F, Monges
G, et al: Sonic hedgehog and Gli1 expression predict outcome in
resected pancreatic adenocarcinoma. Clin Cancer Res. 21:1215–1224.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pankov R and Yamada KM: Fibronectin at a
glance. J Cell Sci. 115:3861–3863. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yi W, Xiao E, Ding R, Luo P and Yang Y:
High expression of fibronectin is associated with poor prognosis,
cell proliferation and malignancy via the NF-κB/p53-apoptosis
signaling pathway in colorectal cancer. Oncol Rep. 36:3145–3153.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Inufusa H, Nakamura M, Adachi T, Nakatani
Y, Shindo K, Yasutomi M and Matsuura H: Localization of oncofetal
and normal fibronectin in colorectal cancer. Correlation with
histologic grade, liver metastasis, and prognosis. Cancer.
75:2802–2808. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miyazaki K, Sakuma K, Kawamura YI, Izawa
M, Ohmori K, Mitsuki M, Yamaji T, Hashimoto Y, Suzuki A, Saito Y,
et al: Colonic epithelial cells express specific ligands for
mucosal macrophage immunosuppressive receptors siglec-7 and −9. J
Immunol. 188:4690–4700. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dharmadhikari G, Stolz K, Hauke M, Morgan
NG, Varki A, de Koning E, Kelm S and Maedler K: Siglec-7 restores
β-cell function and survival and reduces inflammation in pancreatic
islets from patients with diabetes. Sci Rep. 7:453192017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ando M, Tu W, Nishijima K and Iijima S:
Siglec-9 enhances IL-10 production in macrophages via
tyrosine-based motifs. Biochem Biophys Res Commun. 369:878–883.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen DS and Mellman I: Oncology meets
immunology: The cancer-immunity cycle. Immunity. 39:1–10. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dong H, Strome SE, Salomao DR, Tamura H,
Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al:
Tumor-associated B7-H1 promotes T-cell apoptosis: A potential
mechanism of immune evasion. Nat Med. 8:793–800. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vermaelen K: Vaccine strategies to improve
anti-cancer cellular immune responses. Front Immunol. 10:82019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mlecnik B, Tosolini M, Kirilovsky A,
Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman
WH, Pagès F, et al: Histopathologic-based prognostic factors of
colorectal cancers are associated with the state of the local
immune reaction. J Clin Oncol. 29:610–618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Galon J, Mlecnik B, Bindea G, Angell HK,
Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C, et
al: Towards the introduction of the ‘Immunoscore’ in the
classification of malignant tumours. J Pathol. 232:199–209. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pagès F, Mlecnik B, Marliot F, Bindea G,
Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, et al:
International validation of the consensus Immunoscore for the
classification of colon cancer: A prognostic and accuracy study.
Lancet. 391:2128–2139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
O'Reilly MK and Paulson JC: Siglecs as
targets for therapy in immune-cell-mediated disease. Trends
Pharmacol Sci. 30:240–248. 2009. View Article : Google Scholar : PubMed/NCBI
|