Introduction
Left ventricular dysfunction and heart failure (HF),
which are typically described as cardiotoxicity, are the most
concerning cardiovascular complications of anticancer
chemotherapies, which cause an increase in morbidity and mortality
(1). Based on previous studies of
anthracycline cardiotoxicity, increasing attention has focused on
tyrosine kinase inhibitors (TKIs), a type of pharmaceutical drug
inhibiting tyrosine kinases by competitively binding to and
inhibiting their ATP binding pocket (2–5). TKIs
are characterized by being multi-targeted anticancer agents that
lack sufficient specificity, which markedly increases the risk of
cardiotoxicity (5). In a prospective
study of patients with metastatic renal-cell carcinoma, 3–15% of
patients developed cardiac dysfunction induced by the TKIs
sunitinib, pazopanib and axitinib used after chemotherapy and 1–10%
of patients showed symptomatic HF (1). Nevertheless, there are no reliable
means to predict TKI-induced cardiotoxicity under development. As
the cancer patient population ages, these potential cardiac adverse
effects will become more prominent, and the need for improved
prediction and prognosis will become even more pressing (6). Single nucleotide polymorphisms (SNPs)
affecting disease risk can facilitate the identification of drug
targets (7). Similarly, if risk
alleles and drugs have equal functional implications, SNPs may
point to cardiac complications.
The present study aimed to integrate genes or gene
products interacting with TKIs in the Drug Gene Interaction
Database (DGIdb). In order to prioritize and identify potential
drug targets, the present study comprehensively investigated known
molecular targets of TKIs in HF-associated genome-wide association
studies (GWAS) databases and identified common SNPs of these genes
using multiple bioinformatics databases including PhenoScanner,
RegulomeDB, HaploReg v4.1, rSNPBase, the University of California
Santa Cruz (UCSC) Genome Browser and the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) database. The
present study successfully prioritized SNPs and annotated the
function of TKI-induced genes associated with HF, providing a
foundation for further understanding of TKI-induced pathogenesis
and the molecular mechanism of cardiotoxicity.
Materials and methods
DGIdb and GWAS datasets
Firstly, genes or gene products reported to interact
with TKIs were identified in DGIdb v2.22 (8–10). DGIdb
integrates existing resources to generate hypotheses on how mutant
genes may be targeted for therapy, which provides an interface for
a general search of genes for drug-gene interactions and
potentially drug-available genes (8). Subsequently, respective genes in
HF-associated GWAS datasets were screened from PhenoScanner
(11). Over the last decade, GWAS
datasets has facilitated our understanding of the potential role of
genetics in variable responses to drugs and provided significant
insight into genetic structure and the large number of SNPs
associated with complex diseases (12). A total of 549 HF-associated SNPs was
identified from previous GWAS with a P-value
<1×10−5.
Regulatory analysis using
RegulomeDB
SNPs interacting with TKIs for association with
cardiotoxicity were searched further in RegulomeDB (13). RegulomeDB is a database including
high-throughput experimental datasets from the Encyclopedia of DNA
Elements (ENCODE) project, Gene Expression Omnibus and other
sources, and is an important predictive tool to annotate and
prioritize potential regulatory SNPs in the human genome, as well
as to compute predictions and manual annotations to determine the
assumed regulatory potential and identify functional variations
(14). The known regulatory DNA
elements include DNAase hypersensitivity regions, transcription
factor binding sites and promoter regions, which have been
biochemically characterized as having the function of regulating
transcription (15).
Functional analysis using HaploReg
v4.1
The effects of these SNPs on chromatin structure and
allele-specific transcription factor binding were determined using
HaploReg v4.1 (16). HaploReg is
used to explore annotations of non-coding genomes in the results of
GWAS or novel variant sets (17).
Using Linkage disequilibrium information from the 1000 Genome
Project, linked SNPs and indels in nine cell types can be
visualized and their chromatin states and effects on regulatory
motifs can be predicted (17).
Functional analysis using
rSNPBase
rSNPBase is another tool for identifying potential
regulatory genes, focusing on regulatory SNPs involving multiple
regulatory types, including proximal, distal and
post-transcriptional regulation (18,19).
rSNPBase helps researchers to select candidate SNPs for further
genetic studies (especially for quantitative trait locus studies),
to identify SNPs with specific phenotypes and to explore in-depth
molecular mechanisms (20). Through
searching, SNPs are annotated by referring to experimentally
supported regulatory elements (ENCODE data), encompassing a wide
range of regulatory types (20).
Gene expression analysis of
platelet-derived growth factor receptor (PDGFR) α in
Genotype-Tissue Expression (GTEx) project
PDGFRα expression was assessed from the GTEx project
using RNA sequencing (RNA-Seq) datasets (21–23).
GTEx was designed based on data from 900 human donors from 53
sampling sites, including median gene expression levels in 51
tissues and two cell lines (24).
The present study was based on data from 8,555 tissue samples
obtained from 570 adult cadavers (25). Meanwhile, the UCSC Genome Browser was
used to visualize interactions and expression between genomic
regions (26).
Protein-protein interaction (PPI)
network analysis of PDGFRα using the STRING database
Functional interactions between proteins can provide
further insight on the molecular mechanisms of cellular processing.
The present study constructed a PPI network of PDGFRα using the
STRING database, which provides a key integration of PPIs,
including known and predicted interactions. The STRING database
aims to integrate and score all reported available sources of PPI
information, and to complement computational predictions (27). Interacting pairs with high confidence
(combined score >0.7) were selected to construct the PPI
network.
Results
Selection of SNPs
The main approach used in the present study is
illustrated in Fig. 1. A total of 90
genes that interacted with TKIs were identified in the DGIdb
(Table SI) and these genes were
studied in HF-associated GWAS datasets. A total of 549 common SNPs
of 60 selected genes were chosen to represent the association
between TKI molecular targets and cardiotoxicity
(P<1×10−5). Subsequently, all genome-wide association
analysis was performed on these SNPs on the basis of the
quantile-quantile plots. In total, 345 unique SNPs significantly
associated with cardiotoxicity below the genome-wide significant
threshold (P<5×10−8) are shown in the Manhattan plot
in Fig. 2.
 | Figure 1.Experimental strategy. 1, TKIs raise
the risk of HF. 2, All known TKI-targeting genes were extracted
from the DGIdb. 3, DGIdb revealed 90 genes interacting with TKIs.
4, GWAS was performed between all common SNPs in chromosomal
regions representing 90 genes and HF. 5, A total of 549 SNPs of 60
genes displayed significance for both TKI and HF risk. 6, These
genes were candidate risk genes for cardiotoxicity. 7, It can be
hypothesized that TKI-induced genes associated with cardiotoxicity
may be involved in adverse drug reactions. TKI, tyrosine kinase
inhibitor; DGIdb, Drug Gene Interaction Database; SNP, single
nucleotide polymorphism; HF, heart failure; GWAS, genome-wide
association study. |
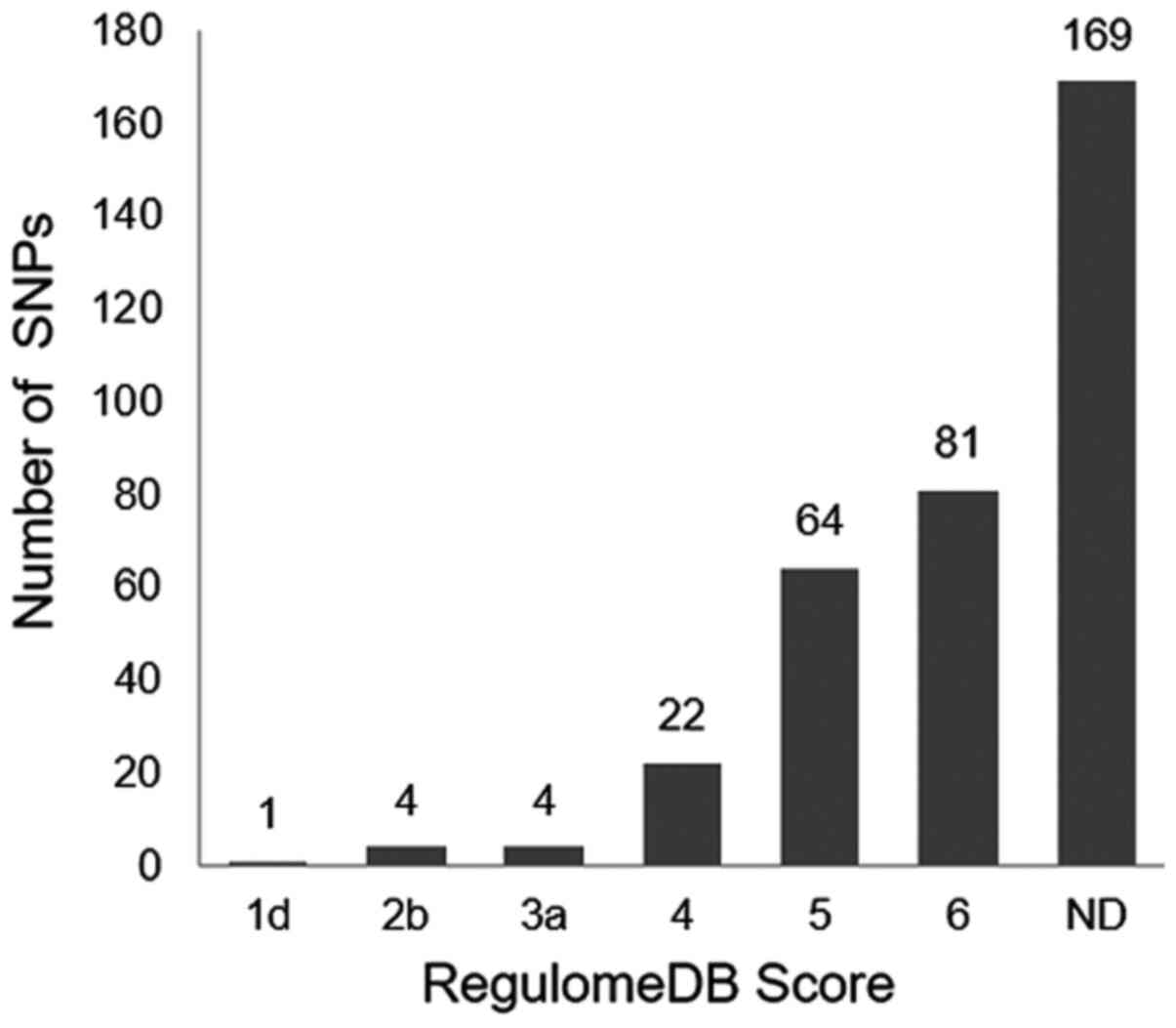
Regulatory analysis using RegulomeDB. In the
RegulomeDB scoring system, the 345 SNPs selected from DGIdb were
classified into six scores (scores 1–6), of which 176 SNPs had
annotated scores ranging between 1 and 6, and the remaining 169
SNPs had no annotated data (Fig. 3).
Notably, the lower the score, the more likely the variant was
located within the potential functional area (data not shown).
Among the 345 SNPs, 9 possessed strong regulatory potential (score
≤3; Table I). Among these 9 SNPs,
rs191188930 on PDGFRα was associated with numerous TKI drugs
(sunitinib, pazopanib, sorafenib, dasatinib and nilotinib; Table I), which will be the focus of the
present systematic analysis.
 | Table I.List of prioritized putative
regulatory single nucleotide polymorphisms. |
Table I.
List of prioritized putative
regulatory single nucleotide polymorphisms.
| Variant | Chr | Positiona |
LD(r2) | Ref | Alt | RegulomeDB
scoreb | Genes | Related tyrosine
kinase inhibitors |
|---|
| rs7115242 | 11 | 117037567 | 1 | G | A | 1d | SIK3 | Dasatinib |
| rs191188930 | 4 | 54256174 | 1 | G | T | 2b | PDGFRα | Sunitinib,
pazopanib, sorafenib, dasatinib and nilotinib |
| rs142136033 | 7 | 55100500 | 1 | G | A | 2b | EGFR | Sunitinib,
sorafenib, dasatinib and lapatinib |
| rs143160639 | 17 | 78214341 | 1 | G | T | 2b | BIRC5 | Lapatinib |
| rs113139165 | 2 | 212520566 | 1 | C | T | 2b | ERBB4 | Lapatinib |
| rs870064 | 1 | 36487326 | 1 | C | T | 3a | CSF3R | Dasatinib |
| rs117153772 | 8 | 127908194 | 1 | G | T | 3a | PVT1 | Imatinib
mesylate |
| rs141452045 | 6 | 152093883 | 1 | C | T | 3a | ESR1 | Lapatinib |
| rs111251321 | 2 | 212240009 | 1 | C | T | 3a | ERBB4 | Lapatinib |
Using histone modification analysis via RegulomeDB,
rs191188930 was predicted to localize in enhancer histone markers
(in adipose-derived mesenchymal stem cell cultured cells, IMR90
fetal lung fibroblasts cells, human endometrial stromal cells
(hESC)-derived CD56+ mesoderm cultured cells,
mesenchymal stem cell-derived adipocyte cultured cells, right
atrium, foreskin fibroblast primary cells skin01, placenta, ovary,
foreskin fibroblast primary cells skin02, fetal, muscle leg and
foreskin keratinocyte primary cells skin03; Table II). The histone modification
analysis of remaining SNPs is described in Table SII.
 | Table II.Key histone modification analysis of
rs191188930 using RegulomeDB. |
Table II.
Key histone modification analysis of
rs191188930 using RegulomeDB.
| Method | Location | Chromatin
function | Tissue/cells |
|---|
| ChromHMM |
chr4:55121000..55153200 | Genic
enhancers | Adipose-derived
mesenchymal stem cell cultured cells |
| ChromHMM |
chr4:55121000..55147600 | Strong
transcription | Human endometrial
stromal cell-derived CD56+ mesoderm cultured cells |
| ChromHMM |
chr4:55118600..55122400 | Genic
enhancers | IMR90 fetal lung
fibroblasts cells |
| ChromHMM |
chr4:55120800..55122400 | Enhancers | Mesenchymal stem
cell-derived adipocyte cultured cells |
| ChromHMM |
chr4:55121200..55125400 | Genic
enhancers | Foreskin fibroblast
primary cells skin01 |
| ChromHMM |
chr4:55120000..55124400 | Enhancers | Placenta |
| ChromHMM |
chr4:55122200..55124800 | Enhancers | Ovary |
| ChromHMM |
chr4:55121200..55122400 | Enhancers | Foreskin fibroblast
primary cells skin02 |
| ChromHMM |
chr4:55122000..55122400 | Enhancers | Foreskin
keratinocyte primary cells skin03 |
| ChromHMM |
chr4:55122200..55122400 | Enhancers | Fetal muscle
leg |
| ChromHMM |
chr4:55122200..55122400 | Enhancers | Right atrium |
Functional analysis using HaploReg
v4.1
Using HaploReg v4.1, rs191188930 was predicted to
localize in enhancer histone markers (IMR90 fetal lung fibroblasts
cells, foreskin fibroblast primary cells skin01, mesenchymal stem
cell derived adipocyte cultured cells, adipose derived mesenchymal
stem cells, foreskin fibroblast primary cells skin02, fetal muscle
leg, placenta, foreskin fibroblast primary cells skin03, ovary and
right atrium), DNase hypersensitivity (H1 derived mesenchymal stem
cells, IMR90 fetal lung fibroblasts cell line, psoas muscles,
foreskin fibroblast primary cells skin01, normal human epidermal
keratinocyte primary cells, primary cells skin02, placenta, normal
human adult dermal fibroblast primary cells and foreskin
fibroblast), and motifs changed (Dmbx1 and GCNF) as described in
Table III. The intronic annotation
of rs191188930 indicated that it affected bound proteins such as
CEBPB, but that there was no direct effect on promoter histone
markers. More detailed results are shown in Table SIII.
 | Table III.Functional analysis of related SNPs
using HaploReg. |
Table III.
Functional analysis of related SNPs
using HaploReg.
| Variant | Promoter histone
markers | Enhancer histone
markers | DNase
hyper-sensitivity | Bound proteins | Motifs changed | dbSNP functional
annotation |
|---|
| rs7115242 | BLD, GI, BONE | 20 tissues | 21 tissues | KAP1 | TBX5 | Intronic |
| rs191188930 |
| 7 tissues | 8 tissues | CEBPB | Dmbx1, GCNF | Intronic |
| rs142136033 | 4 tissues | 19 tissues | 14 tissues | POLR2A | Ets, FEV, HMG-IY,
IK-3, STAT | Intronic |
| rs143160639 | 23 tissues | 6 tissues | 19 tissues | 28 bound
proteins | BCL, BDP1, CTCFL,
EWSR1-FLI1, Ets, Hic1, INSM1, Myc, NERF1a, Rad21, STAT | Missense |
| rs113139165 |
| HRT | 13 tissues | CTCF, RAD21,
SMC3 | Arnt, BAF155, Mrg,
Mxi1, Myc, Pou2f2, Tgif1, Znf143 | Intronic |
| rs870064 | BLD, CRVX | 9 tissues | 11 tissues | AP2ALPHA, AP2GAMMA,
BAF155, HAE2F1 | PPAR, RXRA | Upstream |
| rs141452045 |
| ESDR, FAT,
SKIN | 6 tissues | SETDB1, TRIM28 | EBF | Intronic |
| rs117153772 | 5 tissues | 16 tissues | 23 tissues | CJUN |
| Intronic |
| rs111251321 |
|
|
|
| Gm397, Pou2f2,
Pou3f2, Pou3f3 | Intronic |
Functional analysis using
rSNPBase
Table IV integrates
the results of rSNPBase, in which regulatory characteristics of
SNPs retrieved for the four regulatory modes were assessed as ‘Yes’
or ‘No’. rs7115242, rs143160639 and rs870064 were involved in
proximal transcription regulation, which is associated with
regulatory elements associated with DNA accessibility. rs7115242,
rs143160639 and rs117153772 operated in distal regulation, which
can modify chromatin interactions. Additionally, most SNPs
interfered with the regulation of post-transcriptional RNA-binding
proteins.
 | Table IV.Regulatory features of retrieved SNPs
in four regulation manners using rSNPBase. |
Table IV.
Regulatory features of retrieved SNPs
in four regulation manners using rSNPBase.
| SNP_ID | rSNP | LD-proxy of rSNP
(r2>0.8) | Proximal
regulation | Distal
regulation | microRNA
regulation | RNA binding protein
mediated regulation | eQTL |
|---|
| rs7115242 | Yes | Yes | Yes | Yes | No | Yes | Yes |
| rs191188930 | Yes | No | No | No | No | Yes | No |
| rs142136033 | No | No | No | No | No | No | No |
| rs143160639 | Yes | No | Yes | Yes | No | Yes | No |
| rs113139165 | Yes | No | No | No | No | Yes | No |
| rs870064 | Yes | Yes | Yes | No | No | No | No |
| rs141452045 | Yes | No | No | No | No | Yes | No |
| rs117153772 | Yes | No | No | Yes | No | No | No |
| rs111251321 | Yes | No | No | No | No | Yes | No |
Functional analysis using PhenoScanner
in GWAS
A total of 465 significant associations (P<0.01)
were found using GWAS analysis in PhenoScanner. rs191188930 was
significantly associated with other diseases or phenotypes other
than cardiotoxicity, including acute pericarditis, aortic aneurysm
and dissection, other necrotizing vasculopathies, urticaria,
ascites and tubulo-interstitial nephritis (Table SIV).
Gene expression analysis of PDGFRα in
GTEx
The highest median PDGFRα expression was found using
GTEx datasets: Up to 26.96 reads per kilobase million (RPKM) in
‘Cells-Transformed fibroblasts’ (Ensembl gene ID,
ENSG00000134853.7; genomic position, hg19 chr4:55095264-55148145)
and the total median expression of all 53 tissues in the UCSC
genome browser was 197.39 RPKM (Fig.
4).
PPI network analysis and functional
pathway analysis of PDGFRα using the STRING database
Finally, the online STRING platform was used to
perform PPI network analysis and examine protein interaction
effects of PDGFRα. The PPI network included a total of 11 nodes and
39 edges (PPI enrichment P<3.56×10−7), as illustrated
in Fig. 5. Based on previous
research (4), three functional
pathway genes (PDGFRα, PDGFA and EGFR) affected by sunitinib were
integrated (Fig. 6), which manifest
downstream of the principle mechanism of sunitinib inhibition.
Potential associations were reported for EGFR with myocardial
infarction, congestive heart failure, hypertrophic cardiomyopathy
and myocarditis. PDGFA was strongly associated with cardiac
fibrosis and atrial fibrosis. PDGFRα was strongly associated with
cardiac fibrosis, Loeffler endocarditis, heart sarcoma and organic
heart disease.
Discussion
The present study comprehensively investigated the
possible functional relevance of the molecular targets of TKIs
associated with cardiotoxicity by performing integrative analyses
of publicly available datasets. The DGIdb listed 90 genes what
interacted with TKIs, a type of pharmaceutical drugs that inhibit
tyrosine kinases by competitively binding to and inhibiting their
ATP binding pocket (4). TKIs are
characterized by being multi-targeted anticancer agents that lack
sufficient specificity, which markedly increases the risk of
cardiotoxicity (5).
Over the last decade, GWAS has greatly advanced the
understanding of the genetic basis of HF. To identify genes
associated with TKIs, 549 HF-associated SNPs were identified from
GWAS. However, not all GWAS-recorded SNPs serve a role in the
pathogenesis of disease. Some SNPs are more likely to be functional
in TKI-induced HF with significantly high annotation scores.
Therefore, the present study performed comprehensive functional
analyses of these SNPs utilizing multiple bioinformatics databases,
including RegulomeDB, HaploReg v4.1, PhenoScanner v2, rSNPBase,
GTEx and STRING, to prioritize and identify potential drug
targets.
Using RegulomeDB, 9 SNPs possessed strong regulatory
potential with a score ≤3, and rs191188930 on PDGFRα (score 2b) was
associated with five TKI drugs (sunitinib, pazopanib, sorafenib,
dasatinib and nilotinib). Its annotation score (2b) suggested that
it is more likely to affect binding (transcription factor binding),
DNase footprint, DNase peak and any motif. Additionally,
rs191188930 was predicted to localize in enhancer histone markers
(adipose-derived mesenchymal stem cell cultured cells, IMR90 fetal
lung fibroblasts cells, hESC-derived CD56+ mesoderm
cultured cells, mesenchymal stem cell-derived adipocyte cultured
cells, right atrium, foreskin fibroblast primary cells skin01,
placenta, ovary, foreskin fibroblast primary cells skin02, fetal,
muscle leg and foreskin keratinocyte primary cells skin03). Using
HaploReg v4.1, rs191188930 was predicted to localize in enhancer
histone markers (IMR90 fetal lung fibroblasts cells, foreskin
fibroblast primary cells skin01, mesenchymal stem cell-derived
adipocyte cultured cells, adipose-derived mesenchymal stem cells,
foreskin fibroblast primary cells skin02, fetal muscle leg,
placenta, foreskin fibroblast primary cells skin03, ovary and right
atrium). Hence, the results obtained with HaploReg v4.1 were
similar to those with RegulomeDB. Additionally, functional
annotations by rSNPBase provided reliable evidence of the potential
of rs191188930 in the susceptibility of TKI-induced cardiotoxicity.
GWAS analysis in PhenoScanner revealed a total of 465 significant
associations (P<0.01). Additionally, rs191188930 was
significantly associated with other diseases or phenotypes other
than cardiotoxicity, including acute pericarditis, aortic aneurysm
and dissection, other necrotizing vasculopathies, urticaria,
ascites and tubulo-interstitial nephritis.
PDGFs and their tyrosine kinase receptors are
instrumental in adult organ diseases and embryonic organogenesis;
the biological effects of PDGFs are produced by the activation of
two tyrosine kinase receptors, PDGFRα and PDGFRβ (28,29).
PDGFRα plays significant roles in embryonic organogenesis,
differentiation, migration and function of specialized mesenchymal
cells (28,29). Particularly, PDGFRα is expressed by
multifunctional cardiovascular progenitor cells in mouse and human
embryonic stem cell systems (30).
Using 53 tissues from GTEx RNA-Seq data from 570
donors (8,555 samples), the highest median PDGFRα expression was
identified: Up to 26.96 RPKM in ‘Cells-Transformed fibroblasts’.
Concurrently, PPI analysis of PDGFRα using the STRING database
provided a direction for further research on the pathway of
cardiotoxicity caused by TKIs.
The present study successfully prioritized SNPs and
annotated the function of TKI-induced genes associated with HF,
providing a foundation for further understanding of TKI-induced
pathogenesis and the molecular mechanism of cardiotoxicity. The
present findings indicated that rs191188930 was significantly
associated with cardiotoxicity induced by TKIs. However, these
computational prediction findings should be pragmatically handled
and verified experimentally using appropriate systems before being
considered for genomic medicine. Polymorphism studies should be
followed up to assess the potential association of SNPs with more
complex, clinical disease-related endpoints. Further research is
required to elucidate the mechanisms of SNPs in their biological
function.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81770255 and
82000381) and the Heilongjiang Province Postdoctoral Science
Foundation (grant no. LBH-Z19188).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the following repositories: DGIdb
(http://dgidb.org/search_interactions), RegulomeDB
(https://regulomedb.org/regulome-search/), HaploReg
v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php),
rSNPBase, (http://rsnp.psych.ac.cn/),
PhenoScanner (http://www.phenoscanner.medschl.cam.ac.uk/?tdsourcetag=s_pctim_aiomsg),
GTEx (http://genome.ucsc.edu/) and STRING
(https://string-db.org/cgi/network.pl?taskId=xDOhbN0qKFY4&allnodes=1).
Authors' contributions
YL and YZ conceived and designed the study. YL, WW
and RG analyzed the data. YL and XX acquired the data. YL, RG and
XX contributed to the writing of the manuscript. All authors read
and approved the final manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zamorano JL, Lancellotti P, Rodriguez
Munoz D, Aboyans V, Asteggiano R, Galderisi M, Habib G, Lenihan DJ,
Lip GYH, Lyon AR, et al: 2016 ESC position paper on cancer
treatments and cardiovascular toxicity developed under the auspices
of the ESC committee for practice guidelines: The task force for
cancer treatments and cardiovascular toxicity of the European
society of cardiology (ESC). Eur Heart J. 37:2768–2801. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gyongyosi M, Lukovic D, Zlabinger K,
Spannbauer A, Gugerell A, Pavo N, Traxler D, Pils D, Maurer G,
Jakab A, et al: Liposomal doxorubicin attenuates cardiotoxicity via
induction of interferon-related DNA damage resistance. Cardiovasc
Res. 116:970–982. 2020.PubMed/NCBI
|
|
3
|
Gupta SK, Garg A, Bar C, Chatterjee S,
Foinquinos A, Milting H, Streckfuß-Bömeke K, Fiedler J and Thum T:
Quaking inhibits doxorubicin-mediated cardiotoxicity through
regulation of cardiac circular RNA expression. Circ Res.
122:246–254. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Force T, Krause DS and Van Etten RA:
Molecular mechanisms of cardiotoxicity of tyrosine kinase
inhibition. Nat Rev Cancer. 7:332–344. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Skubitz KM: Cardiotoxicity monitoring in
patients treated with tyrosine kinase inhibitors. Oncologist.
24:e600–e602. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Santoni M, Guerra F, Conti A, Lucarelli A,
Rinaldi S, Belvederesi L, Capucci A and Berardi R: Incidence and
risk of cardiotoxicity in cancer patients treated with targeted
therapies. Cancer Treat Rev. 59:123–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gill D, Georgakis MK, Koskeridis F, Jiang
L, Feng Q, Wei WQ, Theodoratou E, Elliott P, Denny JC, Malik R, et
al: Use of genetic variants related to antihypertensive drugs to
inform on efficacy and side effects. Circulation. 140:270–279.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Griffith M, Griffith OL, Coffman AC,
Weible JV, McMichael JF, Spies NC, Koval J, Das I, Callaway MB,
Eldred JM, et al: DGIdb: Mining the druggable genome. Nat Methods.
10:1209–1210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wagner AH, Coffman AC, Ainscough BJ, Spies
NC, Skidmore ZL, Campbell KM, Krysiak K, Pan D, McMichael JF,
Eldred JM, et al: DGIdb 2.0: Mining clinically relevant drug-gene
interactions. Nucleic Acids Res. 44:D1036–D1044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cotto KC, Wagner AH, Feng YY, Kiwala S,
Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL and Griffith
M: DGIdb 3.0: A redesign and expansion of the drug-gene interaction
database. Nucleic Acids Res. 46:D1068–D1073. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kamat MA, Blackshaw JA, Young R, Surendran
P, Burgess S, Danesh J, Butterworth AS and Staley JR: PhenoScanner
V2: An expanded tool for searching human genotype-phenotype
associations. Bioinformatics. 35:4851–4853. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liao X, Lan C, Liao D, Tian J and Huang X:
Exploration and detection of potential regulatory variants in
refractive error GWAS. Sci Rep. 6:330902016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boyle AP, Hong EL, Hariharan M, Cheng Y,
Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et
al: Annotation of functional variation in personal genomes using
RegulomeDB. Genome Res. 22:1790–1797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mesbah-Uddin M, Elango R, Banaganapalli B,
Shaik NA and Al-Abbasi FA: In-silico analysis of inflammatory bowel
disease (IBD) GWAS loci to novel connections. PLoS One.
10:e01194202015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang E, Zhao H, Zhao D, Li L and Du L:
Functional prediction of chronic kidney disease susceptibility gene
PRKAG2 by comprehensively bioinformatics analysis. Front Genet.
9:5732018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ward LD and Kellis M: HaploReg v4:
Systematic mining of putative causal variants, cell types,
regulators and target genes for human complex traits and disease.
Nucleic Acids Res. 44:D877–D881. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ward LD and Kellis M: HaploReg: A resource
for exploring chromatin states, conservation, and regulatory motif
alterations within sets of genetically linked variants. Nucleic
Acids Res. 40((Database Issue)): D930–D934. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo L, Du Y, Chang S, Zhang K and Wang J:
rSNPBase: A database for curated regulatory SNPs. Nucleic Acids
Res. 42((Database Issue)): D1033–D1039. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lou J, Gong J, Ke J, Tian J, Zhang Y, Li
J, Yang Y, Zhu Y, Gong Y, Li L, et al: A functional polymorphism
located at transcription factor binding sites, rs6695837 near LAMC1
gene, confers risk of colorectal cancer in Chinese populations.
Carcinogenesis. 38:177–183. 2017.PubMed/NCBI
|
|
20
|
Ganguly K, Saha T, Saha A, Dutta T,
Banerjee S, Sengupta D, Bhattacharya S, Ghosh S and Sengupta M:
Meta-analysis and prioritization of human skin
pigmentation-associated GWAS-SNPs using ENCODE data-based
web-tools. Arch Dermatol Res. 311:163–171. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Casper J, Zweig AS, Villarreal C, Tyner C,
Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et
al: The UCSC genome browser database: 2018 update. Nucleic Acids
Res. 46:D762–D769. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Haeussler M, Zweig AS, Tyner C, Speir ML,
Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN,
et al: The UCSC genome browser database: 2019 update. Nucleic Acids
Res. 47:D853–D858. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee CM, Barber GP, Casper J, Clawson H,
Diekhans M, Gonzalez JN, Hinrichs AS, Lee BT, Nassar LR, Powell CC,
et al: UCSC genome browser enters 20th year. Nucleic Acids Res.
48:D756–D761. 2020.PubMed/NCBI
|
|
24
|
McCall MN, Illei PB and Halushka MK:
Complex sources of variation in tissue expression data: Analysis of
the GTEx lung transcriptome. Am J Hum Genet. 99:624–635. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carithers LJ, Ardlie K, Barcus M, Branton
PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J,
Gelfand ET, et al: A novel approach to high-quality postmortem
tissue procurement: The GTEx project. Biopreserv Biobank.
13:311–319. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Karolchik D, Barber GP, Casper J, Clawson
H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L,
Haeussler M, et al: The UCSC genome browser database: 2014 update.
Nucleic Acids Res. 42:D764–D770. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tallquist MD and Soriano P: Cell
autonomous requirement for PDGFRalpha in populations of cranial and
cardiac neural crest cells. Development. 130:507–518. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hoch RV and Soriano P: Roles of PDGF in
animal development. Development. 130:4769–4784. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hong SP, Song S, Cho SW, Lee S, Koh BI,
Bae H, Kim KH, Park JS, Do HS, Im I, et al: Generation of
PDGFRα+ cardioblasts from pluripotent stem cells. Sci
Rep. 7:418402017. View Article : Google Scholar : PubMed/NCBI
|