Introduction
Pancreatic cancer (PC) is a major public health
problem and is the eleventh most common cancer in the world, with
458,918 new cases and 432,242 deaths in 2018 (1). Risk factors for PC include the gut
microbiota, chronic pancreatitis and obesity (2). The 5-year survival rate is correlated
with tumor Tumor-Node-Metastasis stage. At stage 3, the 5-year
survival rate can reach 32%; however, ~52% of patients at stage 4
have a 5-year survival rate of only 3% (3). Hence, identifying PC in the early
stages is important for an improved prognosis. Unfortunately, the
symptoms of surgically resectable PC are non-specific, such as
epigastric or back pain, nausea and satiety (4). As the aforementioned symptoms can be
indications of benign and alterative diseases, for example acute
and chronic pancreatitis and cholelithiasis, PC may be misdiagnosed
and is therefore difficult to diagnose in early stages. Most
patients are diagnosed in advanced stages, which makes surgery
challenging, and current chemotherapy does not yield satisfactory
outcomes (5–9), due to epithelial-to-mesenchymal
transition, increased cancer stem cells population, (hypovascular
tumor microenvironment and chemotherapy-resistance protein
(10). Therefore, understanding
oncogenesis, identifying a guaranteed early diagnostic marker and
investigating new treatments are important to improve the diagnosis
and treatment of PC.
Anillin (ANLN) was first identified by Field and
Albert in 1995 (11). ANLN encodes
an actin-binding protein and is an important cellular component for
cytokinesis. The localization of ANLN changes during the cell cycle
(12). During interphase, ANLN
located at nucleus, while during mitosis, ANLN located at cell
cortex (11). Furthermore, by
binding to other key proteins for mitosis, such as F-actin, myosin
II and septins, ANLN forms cleavage furrows (12–16).
Recently, the important roles of ANLN in the proliferation and
invasion of various cancer cells and tumor development have been
described. Downregulating the expression of ANLN arrests cancer
cells at the G2/M stage of mitosis, decreasing the
capacity for proliferation and invasion in small-cell lung cancer.
The expression of ANLN is associated with 5-year cancer-specific
survival (17). In addition,
correlations between ANLN and other tumors, including breast,
pancreatic, prostate, bladder and colorectal cancer, have been
reported (18–22). However, the expression of ANLN and
its association with the clinical features of PC are unclear.
Therefore, the expression of ANLN and its clinical characteristics
in PC were examined in the present study. The Cancer Genome Atlas
(TCGA), Oncomine, Gene Expression Ontology (GEO) and Gene
Expression Database of Normal and Tumor tissues (GENT) databases
were used to obtain samples and the clinical datasets were
analyzed. Afterwards, Gene Set Enrichment Analysis (GSEA) was
employed to analyze the pathways of ANLN and cancer
development.
Materials and methods
Gene expression data and survival data
resources
The Oncomine database was used to identify the
expression levels of ANLN between normal tissue and cancer tissue
in diverse cancer types (http://www.oncomine.org/).
Gene expression of ANLN in normal and cancerous
pancreatic tissues was downloaded from TCGA database (https://portal.gdc.cancer.gov/) and the Gene
Expression database of Normal and Tumor tissues 2 (GENT2)
(http://gent2.appex.kr/) (23). Bioinformatics and survival data were
downloaded from TCGA database. In total, 182 samples, including
four normal and 178 tumor samples, with clinical information were
download from TCGA database in February 2020. Gene expression of
ANLN in normal (n=106) and pancreatic cancer (n=324) of GENT2
database was obtained from the GEO public repository using the
U133Plus2 (GPL570) platform (23).
Cell lines and culture
Human pancreatic cancer cell lines, PANC-1 and
AsPC-1, were purchased from CHI Scientific, Inc. Human pancreatic
cancer cell line, MIA PaCa-2, and normal pancreatic cell line,
hTRET-HPNE, were purchased from Shanghai Zhongqiaoxinzhou Biotech
Company (https://www.zqxzbio.com/). The human
pancreatic cancer cell line, CFPAC-1, was purchased from The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
hTRET-HPNE, MIA PaCa-2 and PANC-1 cells were cultured in DMEM
(HyClone; Cyvita), supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and penicillin G (100 U/ml) (Beyotime Institute
of Biotechnology). AsPC-1 and CFPAC-1 cells were cultured in
RPMI-1640 and IMDM (Gibco; Thermo Fisher Scientific, Inc.),
respectively, with 10% FBS and penicillin G (100 U/ml). All cells
were maintained at 37°C with 5% CO2. Cells were
harvested at 80% confluence.
Reverse transcription-quantitative
(RT-q)PCR
All cells were collected, and total RNA was
extracted using RNAiso PLUS (Takara Bio, Inc.). According to the
instructions, reverse-transcription was performed using a
PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio, Inc.).
PCR was performed using Rotor-Gene Q (Qiagen GmbH) with TB
Green™Premix Ex Taq™ II (Tli RNase H Plus) (Takara Bio, Inc.). The
PCR cycling conditions were initial denaturation at 95°C for 30
sec, followed by 40 of cycles of denaturation at 95°C for 5 sec,
annealing at 55°C for 30 sec and elongation at 72°C, 30 sec and one
cycle of final extension at 72°C for 5 min. GAPDH was used as a
housekeeping gene. ANLN gene expression was analyzed using the
2−ΔΔCq formula (24). The
primer sequences are as follows: ANLN, Forward:
5′-CAAGATGTATCCAATGACT-3′ and reverse: 5′-TGACTGAAGAATGAATGTT-3′;
GAPDH, forward: 5′-CTCTCCACGGATCAGCTGTC-3′ and reverse:
5′-CAGGGAGGACACGAAGGAT-3′. All experiments were repeated three
times.
Genovariation of ANLN
cBioPortal (https://www.cbioportal.org/) was used to examine the
types and frequency of ANLN alterations and methylation of ANLN in
PC. The sequencing data for PC from cBioPortal was from GDAC
firehose in TCGA database.
GSEA
GSEA (https://www.gsea-msigdb.org/gsea/datasets.jsp) was
performed to conduct Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway biological process enrichment analysis to explore the gene
sets.
Statistical analysis
P<0.0001, fold-change >2 and top 10% gene rank
were used as the significance threshold values for the Oncomine
database. Data from TCGA and GEO databases were analyzed using R
(version 3.6.2) (25) and GENT2,
respectively. Spearman's correlation analysis was utilized to
determine the relationship between the levels of ANLN mRNA
expression and genovariation of ANLN. Gene expression levels over
the median were considered high expression and those below the
median were low expression (median value, 3.648). To examine the
association between 5-year survival rate and ANLN expression, K-M
survival analysis and log rank test was used. Comparison of
biological information and ANLN expression between two groups was
performed using Mann Whitney U tests, while comparison between
three groups or more was performed using Kruskal Wallis and Dunn's
post hoc tests. Logistic regression was used to analyze the
association between clinical variables and ANLN expression.
Univariate and multivariate Cox regression was used to determine
whether ANLN expression level was a prognostic factor. Other
differences between two groups was analyzed using unpaired or
paired Student's t-tests as appropriate, whereas differences
between three or more groups was performed by ANOVA followed by
Dunnett's post hoc test. P<0.05 was considered to indicate a
statistical significant difference.
Results
Gene expression of ANLN is increased
in PC
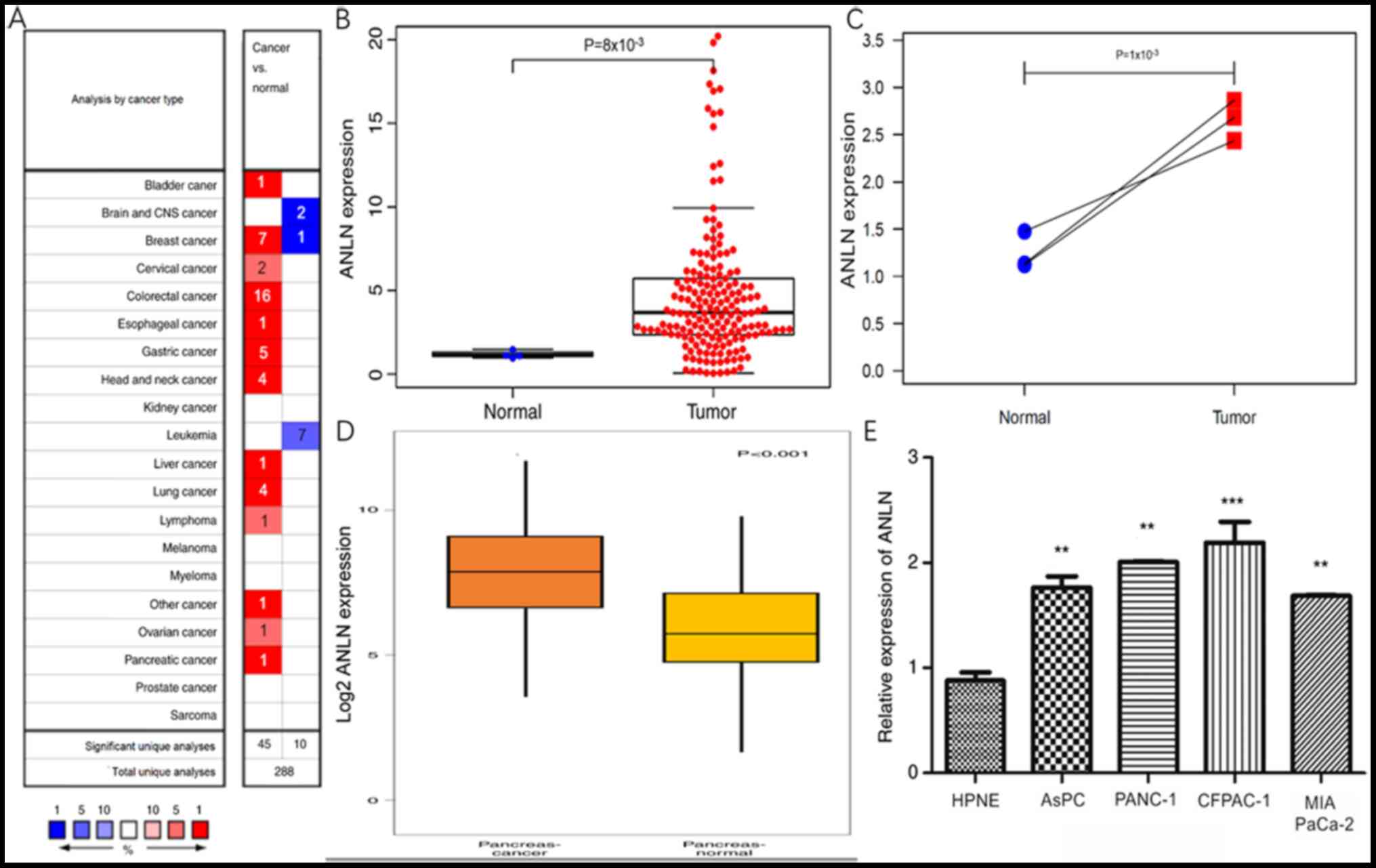
The Oncomine database revealed that the expression
of ANLN was increased in bladder, breast, cervical, colorectal,
esophageal, gastric, head and neck, liver, lung, lymphoma, ovarian
and pancreatic cancer (Fig. 1A).
TCGA database was also searched and obtained similar results for PC
(Fig. 1B). ANLN expression in normal
pancreas tissue was much lower than in pancreatic tumor tissue
(P=0.008). To rule out bias, each adjacent normal sample was paired
with its tumor tissue and the expression of ANLN was analyzed. As
expected, ANLN was more highly expressed in tumor tissues compared
with in normal tissues (P=0.001; Fig.
1C). As the quantity of normal tissue in TCGA database is
notably different from tumor tissue and there was no more data for
normal and tumor tissues in TCGA, the GEO database was used to
verify these results. The log2 fold-change was 1.849, ANLN was
significantly more highly expressed in tumor tissue compared with
in normal tissue (P<0.001; Fig.
1D). Moreover, the differences in ANLN mRNA expression between
normal pancreas cell line and each of PC cell lines were analyzed.
RT-PCR indicated that all cancer cell lines presented higher
expression levels of ANLN compared with the normal cell line
(P<0.01; Fig. 1E).
Mutations in ANLN are not common, and
mRNA expression of ANLN is related to methylation
In total, there were 186 samples from 185 patients
in cBioPortal. As shown in Table I,
only 9.73% of patients exhibited genetic alterations in ANLN.
Mutation, amplification and multiple alterations occurred in one
case each. mRNA levels were high in 15 cases, with a frequency of
8.11%.
 | Table I.Summary of genovariation of ANLN in
pancreatic cancer. |
Table I.
Summary of genovariation of ANLN in
pancreatic cancer.
| Alterations | Frequency, n
(%) |
|---|
| Mutation | 1 (0.54) |
| Amplification | 1 (0.54) |
| mRNA high | 15 (8.11) |
| Multiple
alteration | 1 (0.54) |
| Total | 18 (9.73) |
The patient with multiple alterations in the ANLN
gene had three missenses mutations dispersed at different locations
that caused changes in the protein (Fig.
2A). Most patients had no mutations. As illustrated in Fig. 2B, it was concluded that there was no
significant difference in the mRNA expression of ANLN among the
different genotypes. Additionally, the methylation and mRNA
expression of ANLN was analyzed. Spearman's correlation analysis
showed a weak but statistically significant negative correlation
between gene methylation and mRNA expression of ANLN, with the
coefficient and P-value equaling −0.22 and 0.002664, respectively
(Fig. 2C).
High expression of ANLN predicts poor
survival and grade, representing an independent survival index
As shown in Fig. 3A,
high expression levels of ANLN predicted poor survival (P=0.002).
ANLN expression was not associated with some clinical features,
such as stage (P=0.229; Fig. 3B),
lymph node metastasis (P=0.524; Fig.
3E), distant metastasis (P=0.726; Fig. 3F) and tumor size (P=0.127; Fig. 3D) but was associated with higher
grade (P<0.01; Fig. 3C). Logistic
regression analysis revealed the same conclusion; high ANLN
expression is significantly associated with worse grade [grade 3
vs. 1 odds ratio (OR): 5.662; 95% confidence interval (CI), 3.590,
35.527; P<0.001] while the other clinical information including
stage, lymph node metastasis and distant metastasis was not related
to ANLN expression with P=0.262, 0.612 and 0.218, respectively
(Table II).
 | Table II.Association between the clinical
characteristics and anillin expression using logistic
regression. |
Table II.
Association between the clinical
characteristics and anillin expression using logistic
regression.
| Clinical
characteristic | Total | Odds ratio (95%
CI) | P-value |
|---|
| Grade, 3 vs. 1 | 182 | 5.662
(3.590–35.527) |
4.85×10−5 |
| Stage, 4 vs. 1 | 182 | 1.370
(0.430–88.643) | 0.262 |
| Lymph node
metastasis, positive vs. negative | 180 | 1,187
(0.612–2.315) | 0.612 |
| Distant metastasis,
positive vs. negative | 90 | 3.237
(0.395–66.980) | 0.218 |
Univariate analysis demonstrated that higher
expression of ANLN indicated worse prognosis [hazard ratio (HR):
1.107, 95% CI (1.061, 1.156); P<0.001; Table III). Multivariate analysis also
confirmed that ANLN was an independent factor for prognosis (HR:
1.090, 95% CI (1.043, 1.139), P<0.001) The age and lymph node
metastasis are another prognostic factors with P<0.05.
Univariate and multivariate analysis confirmed other factors
including sex, grade, stage and tumor size were not related to
prognosis (Table III).
 | Table III.Relationship between clinical
characteristics and overall survival in pancreatic cancer using
univariate and multivariate Cox regression. |
Table III.
Relationship between clinical
characteristics and overall survival in pancreatic cancer using
univariate and multivariate Cox regression.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinical
characteristic | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 1.028
(1.005–1.051) | 0.016 | 1.024
(1.001–1.047) | 0.041 |
| Sex, male vs.
female | 0.781
(0.509–1.200) | 0.259 | 0.891
(0.570–1.395) | 0.615 |
| Grade | 1.331
(0.981–1.806) | 0.066 | 1.160
(0.855–1.574) | 0.341 |
| Stage | 1.294
(0.862–1.942) | 0.214 | 0.994
(0.521–1.895) | 0.984 |
| Tumor size | 1.624
(0.977–2.701) | 0.061 | 1.266
(0.638–2.513) | 0.500 |
| Lymph node
metastasis, positive vs. negative | 2.258
(1.308–3.898) | 0.003 | 1.820
(1.028–3.220) | 0.040 |
| ANLN | 1.107
(1.061–1.156) | 0.000 | 1.090
(1.054–1.139) | <0.001 |
GSEA identified ANLN-relevant
pathways
GSEA was used to identify KEGG signaling pathways
associated with ANLN in PC. GSEA demonstrated that high ANLN
expression was correlated with the ‘neuroactive ligand receptor
interaction pathway’ [Normalized Enrichment Score (NES): 1.826,
nominal P<0.001 Fig. S1], while
low expression of ANLN was correlated with various pathways
associated with cancer development, such as ‘p53 signaling’
(Fig. S2), the ‘cell cycle’
(Fig, S3), ‘DNA replication’
(Fig. S4), ‘mismatch repair’
(Fig. S5), ‘nucleotide excision
repair’ (Fig. S6) and ‘PC pathways’
(Fig. S7) pathway (Table IV).
 | Table IV.Gene sets enriched in different ANLN
phenotypes. |
Table IV.
Gene sets enriched in different ANLN
phenotypes.
| ANLN expression
phenotype | Gene set name | NES | Nom P-value | FDR q-value |
|---|
| High
expression |
neuroactive_ligand_receptor_interaction | 1.826 | <0.001 | 0.123 |
| Low expression | cell_cycle | −2.255 | <0.001 | <0.001 |
|
|
P53_signaling_pathway | −2.158 | <0.001 | <0.001 |
|
|
DNA_replication | −1.879 | <0.001 | 0.016 |
|
|
Mismatch_repair | −1.927 | <0.001 | 0.011 |
|
|
Nucleotide_excision_repair | −1.872 | <0.001 | 0.016 |
|
|
Pancreatic_cancer | −1.743 | 0.008 | 0.031 |
Discussion
Upregulated expression of ANLN and its function have
been reported in numerous cancer types, including PC (19,20,22,26–30).
Some studies have also demonstrated that PC progression is
attributed to ANLN by enhancing EZH2 expression and manipulating
the microRNA-218-5p/LIM and SH3 domain protein 1 signaling axis
(31). The present bioinformatics
analysis revealed that the expression of ANLN was elevated in PC,
and increased expression is associated with higher grade. Moreover,
ANLN represents was an independent predictor of prognosis. In
addition, GSEA reported that ANLN was associated with ‘p53’, ‘cell
cycle’, ‘DNA replication’, ‘mismatch repair’, ‘nucleotide excision
repair’ and ‘PC pathways’.
Previous studies have demonstrated that ANLN is
overexpressed in pancreatic ductal adenocarcinoma and that its
expression is correlated with differentiation in bladder cancer
(20,32). These are consistent with the present
results, in which ANLN expression was elevated in PC tissues
compared with normal pancreatic tissues. TCGA database showed that
expression of ANLN was increased in PC tissue. Due to the notable
difference in the number of tissues between cancer and normal
specimens, paired and GEO database analyses were performed and
reached the same conclusion. In addition to the database analysis,
RT-qPCR also confirmed that ANLN was more highly expressed in PC
cells.
Subsequently, databases were used to analyze whether
elevated ANLN expression affects the biological features of PC.
Enhanced expression of ANLN was correlated with advanced
differentiation. However, Wang et al (31) reported that the levels of ANLN
expression were correlated with tumor size, differentiation,
Tumor-Node-Metastasis stage, lymph node metastasis and distant
metastasis, which may be mediated by cell-cell adhesion-related
genes, for example the catenin β-1 gene promoting the proliferation
of hepatoblastoma (33). The results
of Wang et al (31)are
slightly different from the present data. The quantity of original
data downloaded from TCGA database might not be sufficient, which
could have contributed to this difference. Some clinical data are
still unknown in TCGA database, for instance nearly half of
patients' distant metastasis data was unclear.
According to the present univariate and multivariate
Cox regression results, it was concluded that ANLN is an
independent prognostic factor for PC. There is a possibility that
ANLN may impact prognosis by contributing to cancer cell
propagation, invasion, aggressiveness and chemotherapy sensitivity.
Previous studies also support this hypothesis, for example, when
treated with chemotherapy, patients of breast cancer with high
expression of ANLN had worse prognosis (21). Wang et al (31) reported that knockdown of ANLN
inhibits the invasion of pancreatic cancer cells and high
expression of ANLN is related to poor prognosis of PC. In addition,
another study demonstrated that high expression of ANLN increases
the migration of non-small cell lung cancer cells and is associated
with a worse prognosis of (17).
Chemotherapy sensitivity was not confirmed in the bioinformatics
analysis, because treatment for each patient was not mentioned in
TCGA database. Hence, the study (21) was reviewed and how ANLN affects
treatment was identified. Knockdown ANLN decreases the expression
of cyclin D1, stalls the cell cycle in the G2/M phase
and inhibits breast cancer cell proliferation. Therefore, the study
demonstrated that high expression of ANLN limits the efficacy of
chemotherapy in breast cancer; however, the reason for this is
still unclear (21). However, a
direct association between ANLN and chemotherapy in PC is yet to be
reported.
Mutation of ANLN is uncommon, with only a few
patients (9.73%) presenting with ANLN gene alterations in the
present study. Among all alterations, increased mRNA was most
common and a significant correlation was identified between
methylation and ANLN mRNA expression. Methylation at different
positions causes diverse results, including regulating gene
expression (34). However, the
present study did not identify where ANLN methylation occurs or
whether the high levels of ANLN mRNA are attributed to alterations
in methylation. Further investigation needed to resolve the exact
association between methylation and ANLN mRNA expression.
From GSEA, high expression of ANLN was correlated
with the ‘neuroactive ligand receptor interaction pathway’.
However, to the best of our knowledge no study has investigated
this relationship in PC. Low expression of ANLN was associated with
‘p53 signaling’, ‘cell cycle’, ‘DNA replication’, ‘mismatch
repair’, ‘nucleotide excision repair’ and ‘PC pathways’. Various
reports have demonstrated that ANLN serves an essential role as a
regulator in the cell cycle pathway during which eukaryotic cells
divide (12,35). Chromosome-derived Ran-GTP signals
decrease local ANLN expression in the cortex of proliferating
cells, which causes asymmetric membrane elongation during mitosis
(36). As reported, under
pathological circumstances, the development of multiple cancer
types can be caused by genetic mutations and dysregulated mitotic
proliferation (37). In other words,
abnormal expression of ANLN during the cell cycle may promote the
development of cancer. Further studies, including investigation of
the relative pathways and treatment associated with ANLN
expression, are needed in PC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was funded by The Cuiying Innovation
Project of Science and Technology of Lanzhou University (grant no.
CY2017-BJ04).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request. Additional datasets generated and/or analyzed during the
current study are available in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/), Oncomine
(http://www.oncomine.org/), Gene Expression
database of Normal and Tumor tissues 2 (GENT2) (http://gent2.appex.kr/), cBioPortal (https://www.cbioportal.org/) and GSEA (https://www.gsea-msigdb.org/gsea/datasets.jsp)
repositories.
Authors' contributions
CW conceived and designed the study. YN, ZZ, MC, FM,
YF, YK and BK obtained the bioinformation data and analyzed the
data. YN wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PC
|
pancreatic cancer
|
|
ANLN
|
anillin
|
|
GSEA
|
Gene Set Enrichment Analysis
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuigan A, Kelly P, Turkington RC, Jones
C, Coleman HG and McCain RS: Pancreatic cancer: A review of
clinical diagnosis, epidemiology, treatment and outcomes. World J
Gastroenterol. 24:4846–4861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
American Cancer Society, . Cancer Facts
& Figures 2020. American Cancer Society, Inc.; Atlanta, GA:
2020, simplehttps://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2020.htmlFebruary.
2020
|
|
4
|
Mizrahi JD, Surana R, Valle JW and Shroff
RT: Pancreatic cancer. Lancet. 395:2008–2020. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Avgerinos DV and Björnsson J: Malignant
neoplasms: Discordance between clinical diagnoses and autopsy
findings in 3,118 cases. Apmis. 109:774–780. 2010. View Article : Google Scholar
|
|
6
|
Bosetti C, Bertuccio P, Negri E, La
Vecchia CL, Zeegers MP and Boffetta P: Pancreatic cancer: Overview
of descriptive epidemiology. Mol Carcinog. 51:3–13. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lambe M, Eloranta S, Wigertz A and
Blomqvist P: Pancreatic cancer; reporting and long-term survival in
Sweden. Acta Oncol. 50:1220–1227. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oberstein PE and Olive KP: Pancreatic
cancer: Why is it so hard to treat? Therap Adv Gastroenterol.
6:321–337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sens MA, Zhou XD, Weiland T and Cooley AM:
Unexpected neoplasia in autopsies: Potential implications for
tissue and organ safety. Arch Pathol Lab Med. 133:1923–1931.
2009.PubMed/NCBI
|
|
10
|
Uzunparmak B and Sahin IH: Pancreatic
cancer microenvironment: A current dilemma. Clin Transl Med.
8:2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Field CM and Alberts BM: Anillin, a
contractile ring protein that cycles from the nucleus to the cell
cortex. J Cell Biol. 131:165–178. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oegema K, Savoian MS, Mitchison TJ and
Field CM: Functional analysis of a human homologue of the
drosophila actin binding protein anillin suggests a role in
cytokinesis. J Cell Biol. 150:539–552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gould GW: Animal cell cytokinesis: The
role of dynamic changes in the plasma membrane proteome and
lipidome. Semin Cell Dev Biol. 53:64–73. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goyal A, Takaine M, Simanis V and Nakano
K: Dividing the spoils of growth and the cell cycle: The fission
yeast as a model for the study of cytokinesis. Cytoskeleton
(Hoboken). 68:69–88. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
D'Avino PP: How to scaffold the
contractile ring for a safe cytokinesis-lessons from
Anillin-related proteins. J Cell Sci. 122:1071–1079. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song K, Russo G and Krauss M: Septins as
modulators of endo-lysosomal membrane traffic. Front Cell Dev Biol.
4:1242016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang G, Shen W, Cui L, Chen W, Hu X and Fu
J: Overexpression of Anillin (ANLN) is correlated with colorectal
cancer progression and poor prognosis. Cancer Biomark. 16:459–465.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng S, Yu X, Ma C, Song R, Zhang Z, Zi X,
Chen X, Wang Y, Yu Y, Zhao J, et al: Transcriptome sequencing
identifies ANLN as a promising prognostic biomarker in bladder
urothelial carcinoma. Sci Rep. 7:31512017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Olakowski M, Tyszkiewicz T, Jarzab M, Król
R, Oczko-Wojciechowska M, Kowalska M, Kowal M, Gala GM, Kajor M,
Lange D, et al: NBL1 and anillin (ANLN) genes over-expression in
pancreatic carcinoma. Folia Histochem Cytobiol. 47:249–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Chen J, Zhong MZ, Huang J, Hu YP,
Feng DY, Zhou ZJ, Luo X, Liu ZQ, Jiang WZ and Zhou WB:
Overexpression of ANLN contributed to poor prognosis of
anthracycline-based chemotherapy in breast cancer patients. Cancer
Chemother Pharmacol. 79:535–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tamura K, Furihata M, Tsunoda T, Ashida S,
Takata R, Obara W, Yoshioka H, Daigo Y, Nasu Y, Kumon H, et al:
Molecular features of hormone-refractory prostate cancer cells by
genome-wide gene expression profiles. Cancer Res. 67:5117–5125.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park SJ, Yoon BH, Kim SK and Kim SY:
GENT2: An updated gene expression database for normal and tumor
tissues. BMC Med Genomics. 12 (Suppl 5):1012019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Team R: RStudio: Integrated Development
for R, 2015. RStudio, Inc.; Boston, MA: 2015, simplehttp://www.rstudio.comPubMed/NCBI
|
|
26
|
Long X, Zhou W, Wang Y and Liu S:
Prognostic significance of ANLN in lung adenocarcinoma. Oncol Lett.
16:1835–1840. 2018.PubMed/NCBI
|
|
27
|
Sadi AM, Wang DY, Youngson BJ, Miller N,
Boerner S, Done SJ and Leong WL: Clinical relevance of DNA
microarray analyses using archival formalin-fixed paraffin-embedded
breast cancer specimens. Bmc Cancer. 11:2532011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Mo Y, Midorikawa K, Zhang Z, Huang
G, Ma N, Zhao W, Hiraku Y, Oikawa S and Murata M: The potent tumor
suppressor miR-497 inhibits cancer phenotypes in nasopharyngeal
carcinoma by targeting ANLN and HSPA4L. Oncotarget. 6:35893–35907.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weinberger P, Ponny SR, Xu H, Bai S,
Smallridge R, Copland J and Sharma A: Cell Cycle M-Phase genes are
highly upregulated in anaplastic thyroid carcinoma. Thyroid.
27:236–252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xia L, Su X, Shen J, Meng Q, Yan J, Zhang
C, Chen Y, Wang H and Xu M: ANLN functions as a key candidate gene
in cervical cancer as determined by integrated bioinformatic
analysis. Cancer Manag Res. 10:663–670. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang A, Dai H, Gong Y, Zhang C, Shu J, Luo
Y, Jiang Y, Liu W and Bie P: ANLN-induced EZH2 upregulation
promotes pancreatic cancer progression by mediating
miR-218-5p/LASP1 signaling axis. J Exp Clin Cancer Res. 38:3472019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zeng S: The mechanism of ANLN in the
carcinogenesis of bladder cancer. PhD dissertationChinese People's
Liberation Army Naval Medical University Shanghai 211 Engineering
College, Shanghai, China2017.
|
|
33
|
Liu L, Wang J, Sun G, Wu Q, Ma J, Zhang X,
Huang N, Bian Z, Gu S, Xu M, et al: m6A mRNA methylation
regulates CTNNB1 to promote the proliferation of hepatoblastoma.
Mol Cancer. 18:1882019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ghoshal K, Majumder S, Li Z, Dong X and
Jacob ST: Suppression of metallothionein gene expression in a rat
hepatoma because of promoter-specific DNA methylation. J Biol Chem.
275:539–547. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nigg EA: Mitotic kinases as regulators of
cell division and its checkpoints. 2:21–32. 2001.
|
|
36
|
Kiyomitsu T and Cheeseman IM: Cortical
dynein and asymmetric membrane elongation coordinately position the
spindle in anaphase. Cell. 154:391–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brinkley BR: Managing the centrosome
numbers game: From chaos to stability in cancer cell division.
Trends Cell Biol. 11:18–21. 2001. View Article : Google Scholar : PubMed/NCBI
|