Introduction
Ovarian cancer (OC), a common gynecological tumor,
is the fifth leading cause of tumor-associated death in women, and
there were ~22,240 new cases and ~14,070 deaths associated with OC
every year worldwide (1). Most
patients with OC were diagnosed at the advanced stage with tumor
metastasis and recurrence (2).
Despite achieving great progress in the technologies for the
detection and therapy of OC, the overall survival rate of patients
with OC remains low. Therefore, it is urgent to find the effective
therapeutic targets for OC.
Long non-coding RNAs (lncRNAs), with over 200
nucleotides, are a group of conserved RNAs that play pivotal
function in cancer progression, such as cell proliferation,
mobility, apoptosis and autophagy (3–6). In
recent years, lncRNAs were reported to be involved in various human
cancer types, including OC (7,8). Small
nucleolar RNA host gene 20 (SNHG20), 8,275 bases in length, was
originally identified as an oncogene in hepatocellular cancer
(9). A previous study indicated that
SNHG20 knockdown suppressed the growth of OC cells through
modulating the levels of downstream genes (10). However, the functional mechanism of
SNHG20 is largely unknown in OC.
MicroRNAs (miRNAs/miRs), ~22 nucleotides, exert
crucial roles via inhibiting translation or promoting degradation
(11,12). Multiple studies demonstrated that
miRNAs participated in the progression of various cancer types,
including lung cancer (13),
osteosarcoma (14), thyroid cancer
(15) and OC (16). A recent study suggested that
miR-338-3p level was decreased in OC, and upregulation of
miR-338-3p repressed OC cell proliferation (17). Moreover, Liu et al (18) demonstrated that miR-338-3p acted as a
tumor suppressor to regulate cell proliferation, mobility and
apoptosis in OC. These data revealed that miR-338-3p played an
important role in OC. Thus, the present explored the underlying
mechanism of miR-338-3p in OC.
Myeloid cell leukemia 1 (MCL1) belongs to B-cell
lymphoma 2 (BCL-2) family, which is considered as a class of cell
apoptosis regulators in human cancer (19). It was reported that many factors,
including deubiquitinase and miRNAs, mediated the level of MCL1 to
affect cell growth in OC. For instance, USP13 regulated MCL
stabilization to modulate tumor growth (20). Su et al (21) demonstrated that miR-142-5p mediated
cisplatin-induced cell apoptosis via regulating MCL expression.
Thus, MCL pathway is associated with OC development. Therefore, it
is important to examine the function of MCL in OC cells.
Here, we determined the levels of SNHG20, miR-338-3p
and MCL1 in OC tissues and cells. Furthermore, the function of
SNHG20 was investigated in OC in vitro and in vivo.
Besides, the underlying mechanism of SNHG20 in OC progression was
explored.
Materials and methods
Tissues and cell culture
30 OC tissues and 30 adjacent normal tissues were
obtained from the patients at the hospital of The Affiliated Renhe
Hospital of China Three Gorges University from April 2016 to
January 2019. The average age of OC patients was (52.02 ± 10.88)
years, in which there were 18 patients ≥50 years and 12 patients
<50 years. These tissue samples were stored at −80°C until use.
This research was approved by the Ethics Review Committees of The
Affiliated Renhe Hospital of China Three Gorges University. All
participates provided written informed consent.
Normal human ovarian surface epithelial (HOSE) cell
line was provided by ScienCell Research Laboratories, Inc. OC cell
lines (OVCAR3, SKOV3, A2780 and CAOV-3) were bought from the Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
These cells were cultured in RPMI-1640 medium (cat. no. 22400089;
Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (FBS; cat. no. SH30068.03; Hyclone; Cytvia) and 1%
penicillin/streptomycin (cat. no. 516106; EMD Millipore) in a
humidified incubator at 37°C with 5% CO2.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA was isolated from OC tissues or cells
using TRIzol reagents (Invitrogen; Thermo Fisher Scientific, Inc.),
and subsequently used to synthesize complementary DNA using the
SuperScript III® (cat. no. 18080-044; Invitrogen; Thermo
Fisher Scientific, Inc.) based on the recommended protocol (65°C
for 5 min, 50°C for 30–60 min and 70°C for 15 min). Subsequently,
an SYBR Premix ExTaq kit (cat. no. RR047A; Takara Bio, Inc.) was
applied to perform qPCR. The thermocycling conditions were as
follows: 94°C for 5 min, followed by 40 cycles of 94°C for 30 sec,
60°C for 1 min, and 72°C for 30 sec. The data were normalized by U6
or GAPDH level, and analyzed using the 2−ΔΔCq method
(22). The primers used in the study
are listed in Table I.
 | Table I.Reverse transcription-quantitative
PCR primer sequences. |
Table I.
Reverse transcription-quantitative
PCR primer sequences.
| Gene | Sequence |
|---|
| SNHG20 | F:
5′-ATGGCTATAAATAGATACACGC-3′ |
|
| R:
5′-GGTACAAACAGGGAGGGA-3′ |
| miR-338-3p | F:
5′-AACCGGTCCAGCATCAGTGATT-3′ |
|
| R:
5′-CAGTGCAGGGTCCGAGGT-3′ |
| MCL1 | F:
5′-GGGCAGGATTGTGACTCTCATT-3′ |
|
| R:
5′-GATGCAGCTTTCTTGGTTTATGG-3′ |
| U6 | F:
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ |
|
| R:
5′-CCAGTGCAGGGTCCGAGGT-3′ |
| GAPDH | F:
5′-ATCACTGCCACCCAGAAGAC-3′ |
|
| R:
5′-TTTCTAGACGGCAGGTCAGG-3′ |
Cell transfection
Small interfering RNA against SNHG20 (si-SNHG20,
final concentration, 40 nM), small hairpin RNA against SNHG20
(sh-SNHG20, final concentration, 1 µg/ml), miR-338-3p mimic
(miR-338-3p, final concentration, 20 nM), miR-338-3p inhibitor
(anti-miR-338-3p, final concentration, 25 nM), and the relative
negative controls (si-NC, final concentration, 40 nM; sh-NC, final
concentration, 1 µg/ml; miR-NC, final concentration, 20 nM; and
anti-miR-NC, final concentration, 25 nM) were purchased from
Shanghai GenePharma Co. Ltd. (cat. no. B03001). For overexpression
of SNHG20 or MCL1 (pcDNA-SNHG20, final concentration, 0.8 µg/ml;
pcDNA-MCL1, final concentration, 1 µg/ml), its sequence was
inserted into the pcDNA3.1 vector (cat. no. GM-1013P001-P10;
GenePharma). The sequences of the constructs are listed in Table II and Data S1. Cell transfection was carried out
using Lipofectamine® 3000 (cat. no. L3000-015;
Invitrogen; Thermo Fisher Scientific, Inc.) in serum free Opti-MEM
medium (cat. no. 31985062; Thermo Fisher Scientific, Inc.) and
incubated in a humidified incubator at 37°C with 5% CO2
for 6 h, followed by medium being changed to RPMI-1640 medium
containing 10% fetal bovine serum and 1% penicillin/streptomycin.
Subsequent experimentation was performed 24 or 48 h
post-transfection.
 | Table II.Sequences of constructs used for
transfection. |
Table II.
Sequences of constructs used for
transfection.
| Gene | Sequence
(5′-3′) |
|---|
| si-SNHG20 |
GCCACUCACAAGAGUGUAUTT |
| si-NC |
GGATACGGAGTACTATAGC |
| sh-SNHG20 |
CACCGGCCCAGATTGGTACATTTC |
|
|
GAAAAATGTACCAATCTGG |
| miR-338-3p |
UUUGAGCAGCACUCAUUUUUGC |
| miR-NC |
CAGUACUUUUAGUGUGUACAA |
|
anti-miR-338-3p |
CAACAAAAUCACUGAUGCUGGA |
| anti-miR-NC |
UUGUAAGUUGCGACAGCCACUCA |
Cell proliferation assay
The MTT kit (M5655; Sigma-Aldrich; Merck KGaA) was
used to examine cell proliferation according to the manufacturers
manual. Briefly, SKOV3 and A2780 cells were transfected for 48 h,
and then cultured for 24, 48 or 72 h. Next, 10 µl MTT solution was
added to treat the cells at 37°C for 4 h, and then the
precipitation was incubated with dimethyl sulfoxide. Finally, the
absorbance was determined using the microplate reader (Bio-Rad
Laboratories, Inc.) at 490 nm.
Cell apoptosis assay
Cell apoptosis was measured using Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis
Detection kit (cat. no. A211-01/02; Vazyme Biotech Co., Ltd.)
according to the manufacturers manual. Briefly, SKOV3 and A2780
cells were cultured for 48 h. Then, the cells were collected,
washed and stained with 5 µl Annexin V-FITC and PI. Finally, cell
apoptosis was examined using the flow cytometer (BD Biosciences,
Inc.) and FlowJo software version v.10.0.6 (Becton, Dickinson and
Company).
Western blot assay
Tissue or cell lysates were obtained using lysis
buffer (cat. no. P0013B; Beyotime Institute of Biotechnology). A
BCA Protein Assay kit was employed to determine the protein
concentration. The samples (30 µg/lane) were separated by 10%
dodecyl sulfate, sodium salt-polyacrylamide gel electrophoresis
(SDS-PAGE), transferred to polyvinyl difluoride membrane (PVDF),
and blocked by 5% fat-free milk at room temperature. Next, the
membranes were incubated at 4°C overnight with the primary antibody
against MCL1 (1:1,000; cat. no. ab243136; Abcam), B-cell lymphoma 2
(Bcl-2; 1:1,000; cat. no. ab32124; Abcam), BCL2 associated X (Bax;
1:1,000; cat. no. ab32503; Abcam), cleaved caspase-3 (c-caspase-3;
1:1,000; ab32042; Abcam), procaspase-3 (1:1,000; cat. no. ab32499;
Abcam), vimentin (1:1,000; cat. no. ab227639; Abcam), E-cadherin
(1:1,000; cat. no. ab238099; Abcam), N-cadherin (1:1,000; ab76057;
Abcam) or GAPDH (1:1,000; cat. no. ab9485; Abcam). Membranes were
then incubated with the corresponding secondary antibody goat
anti-rabbit IgG H&L (HRP) (1:2,000; cat. no. ab214880; Abcam)
for 1 h at room temperature. Finally, an ECL detection system (EMD
Millipore) was applied to detect the signal, and quantified using
ImageJ software V1.8.0 (National Institutes of Health).
Cell migration and invasion assay
Transwell chamber (cat. no. 3450; Corning Life
Sciences) was applied to examine cell mobility. For transwell
migration assay, transfected SKOV3 and A2780 cells in 100 µl
RPMI-1640 medium without FBS were seeded into the top chamber. The
bottom chamber was filled with 500 µl RPMI-1640 medium with 10%
FBS. After 24 h incubation, the migratory cells on the bottom
chamber were counted using an IX91 inverted light microscope
(Olympus, Tokyo, Japan) (magnification, ×20). For transwell
invasion assay, the upper chamber was pre-coated with Matrigel
(cat. no. 356234; BD Biosciences), all other following steps were
the same as that for the cell migration assay.
Dual luciferase reporter assay
Potential binding sites between miR-338-3p and
SNHG20 or MCL1 were predicted using an online bioinformatics
database (starBase v.2.0; http://starbase.sysu.edu.cn/). The wide-type or
mutant-type of SNHG20 or MCL1 3′untranslated region (UTR)
(WT/MUT-SNHG20 or MCL1 3′UTR-WT/MUT) was cloned into the pGL3
vector (cat. no. E1761; Promega Corporations), and co-transfected
into SKOV3 and A2780 cells with miR-338-3p or miR-NC. After 48 h
incubation, the luciferase density was measured using the dual
luciferase assay system (cat. no. E1910; Promega Corporations).
Firefly luciferase activity was normalized to Renilla
luciferase activity and the average of triplicate samples was
calculated.
Mouse xenografts
Six BALB/c nude mice were randomly divided into the
following two groups (n=3 per group): i) sh-NC group; and ii)
sh-SNHG20. The mice (age, 6 weeks) were subcutaneously injected
with A2780 cells (1×106) transfected with sh-SNHG20 or
sh-NC into the left hind back of nude mice. After 7 days, tumor
volume (length × width2/2) was calculated every 4 days.
After 27 days, the mice were sacrificed by cervical dislocation,
following deep anesthesia with 2% isoflurane, and tumor tissues
were collected. Tumor weight was analyzed. This experiment was
carried out in line with the guidance of the National Animal Care
and Ethics Institution and was authorized by the Animal Research
Committee of the Affiliated Renhe Hospital of China Three Gorges
University.
Statistical analysis
Each experiment was conducted at least three times
independently. Experimental data were presented by mean ± standard
deviation (SD). Two independent groups were analyzed using
Student's-t test. ANOVA followed by Tukey's test was used to assess
the difference among more than two groups. Spearman's correlation
coefficient was calculated to analyze the association between the
levels of two genes. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of SNHG20 is higher in OC
tissues and cells
Firstly, RT-qPCR assay was used to detect the
expression of SNHG20 in OC tissues and the adjacent normal tissues.
As shown in Fig. 1A, SNHG20
expression was significantly higher in OC tissues. Moreover, SNHG20
level was higher in OC cells (OVCAR3, SKOV3, A2780 and CAOV-3)
compared with that in HOSE cells (Fig.
1B). These results indicate that SNHG20 level might be
associated with the development of OC.
SNHG20 knockdown represses the
proliferation, migration, invasion and EMT, whilst inducing
apoptosis in OC cells
To investigate the function of SNHG20 in OC cells,
si-SNHG20 was transfected into SKOV3 and A2780 cells to deplete
SNHG20 expression. Knockdown efficiency was confirmed by RT-qPCR
assay (Fig. 2A). As indicated in
Fig. 2B and C, SNHG20 knockdown
suppressed cell proliferation notably. Furthermore, flow cytometry
analysis demonstrated that cell apoptosis was significantly induced
by the downregulation of SNHG20 in SKOV3 and A2780 cells (Fig. 2D and E). Meanwhile, the levels of
three apoptosis-associated proteins (Bcl-2, Bax, and c-caspase-3)
were detected and found that Bcl-2 level was downregulated, while
Bax and c-caspase3 levels were upregulated by SNHG20 knockdown
(Fig. 2F and G). Next, the effect of
SNHG20 on cell mobility was explored. The data suggested that
SNHG20 knockdown suppressed cell migration and invasion (Fig. 3A and B). Furthermore, the decreased
levels of vimentin and N-cadherin, as well as the increased
E-cadherin level, were observed in SNHG20-depleted SKOV3 and A2780
cells (Fig. 3C). Taken together,
SNHG20 knockdown inhibited OC development.
miR-338-3p is a target of SNHG20
Bioinformatics tool starBase v2.0 predicted that
miR-338-3p was a potential target gene of SNHG20 (Fig. 4A). Subsequently, dual-luciferase
reporter assay was performed to verify the association between
miR-338-3p and SNHG20. The results suggested that miR-338-3p
decreased the luciferase activity of WT-SNHG20, whilst having no
effect on the luciferase activity of MUT-SNHG20 (Fig. 4B and C), revealing that SNHG20 could
bind to miR-338-3p. Successful overexpression efficiency of SNHG20
is demonstrated by Fig. 4D.
Moreover, miR-338-3p expression was significantly upregulated by
SNHG20 depletion and downregulated by SNHG20 overexpression
(Fig. 4E). The present study data
showed that miR-338-3p level was decreased in OC tissues and cells
(Fig. 4F and G). Furthermore,
miR-338-3p level was negatively correlated with SNHG20 level in OC
tissues (Fig. 4H). These data
suggest that SNHG20 targeted miR-338-3p and downregulated
miR-338-3p expression.
 | Figure 4.SNHG20 targets miR-338-3p. (A) The
interaction between SNHG20 and miR-338-3p was predicted by starBase
v2.0. Mutated sites were indicated by the red color. (B and C)
Dual-luciferase reporter assay was used to detect the luciferase
activity of SKOV3 and A2780 cells co-transfected with miR-NC or
miR-338-3p and WT-SNHG20 or MUT-SNHG20. (D) SNHG20 level in SKOV3
and A2780 cells transfected with pcDNA or SNHG20 was detected by
RT-qPCR. (E) RT-qPCR was used to measure miR-338-3p expression in
SKOV3 and A2780 cells transfected with si-NC, si-SNHG20, pcDNA, or
SNHG20, respectively. (F and G) miR-338-3p level was detected by
RT-qPCR in OC tissues and the adjacent normal tissues (F) as well
as OC cells and human ovarian surface epithelial cells (G). (H) The
correlation between SNHG20 and miR-338-3p was analyzed using linear
regression. *P<0.05. SNHG20, small nucleolar RNA host gene 20;
WT, wild type; MUT, mutant type; miR-NC, miR-negative control
mimic; pcDNA, pcDNA3.1 vector; si-NC, small interfering RNA against
negative control; si-SNHG20, small interfering RNA against SNHG20;
RT-qPCR, reverse transcription-quantitative PCR; OC, ovarian
cancer. |
SNHG20 depletion suppresses OC
progression by targeting miR-338-3p
To investigate whether SNHG20 affected OC cell
progression via suppressing miR-338-3p expression in OC cells,
SKOV3 and A2780 cells were transfected with si-NC, si-SNHG20,
si-SNHG20 + anti-miR-NC, or si-SNHG20 + anti-miR-338-3p,
respectively. Firstly, the data showed that transfection of
anti-miR-338-3p dramatically downregulated the expression of
miR-338-3p, while it was upregulated by miR-338-3p mimic
transfection (Fig. 5A). MTT assay
suggested that SNHG20 knockdown suppressed cell proliferation,
which was reversed by downregulation of miR-338-3p (Fig. 5B and C). In addition, it was found
that cell apoptosis induced by SNHG20 knockdown was rescued by
miR-338-3p inhibitor (Fig. 5D). As
shown in Fig. 5E and F, miR-338-3p
inhibitor weakened the effect of SNHG20 knockdown on Bcl-2, Bax and
c-caspase-3 protein levels. On the other hand, the inhibition
effects of SNHG20 knockdown on cell migratory and invasive
abilities were blocked by miR-338-3p inhibitor (Fig. 5G and H). Besides, the regulatory
effects of SNHG20 knockdown on the levels of EMT markers were
weakened by miR-338-3p inhibitor (Fig.
5I and J). Taken together, SNHG20 regulates OC cell progression
through suppressing miR-338-3p expression.
 | Figure 5.SNHG20 regulates OC progression
through sponging miR-338-3p. (A) miR-338-3p level was investigated
using reverse transcription-quantitative PCR in SKOV3 and A2780
cells transfected with miR-338-3p or anti-miR-338-3p. (B-J) SKOV3
and A2780 cells were transfected with si-NC, si-SNHG20, si-SNHG20 +
anti-miR-NC, or si-SNHG20 + anti-miR-338-3p, respectively. (B and
C) MTT assay was employed to measure cell proliferation ability.
(D) Cell apoptosis rate was analyzed using flow cytometry. (E and
F) The levels of cell apoptosis-associated proteins were determined
by western blot assay. (G and H) Cell migration and invasive
abilities were assessed by transwell assay. (I and J) Western blot
assay was used to detect the relative protein levels of EMT
markers. *P<0.05. SNHG20, small nucleolar RNA host gene 20; OC,
ovarian cancer; anti-miR-NC, miR-negative control inhibitor; si-NC,
small interfering RNA against negative control; si-SNHG20, small
interfering RNA against SNHG20; anti-miR-338-3p, miR-338-3p
inhibitor; EMT, epithelial-mesenchymal transition. |
SNHG20 upregulates MCL1 level via
inhibiting miR-338-3p expression
Using bioinformatics tool starBase v2.0, it was
found that MCL1 possessed a complementary sequence with miR-338-3p
(Fig. 6A). Dual-luciferase reporter
assay showed that the luciferase activity of MCL1 3′UTR-WT, but not
MCL1 3′UTR-MUT, was reduced by miR-338-3p, confirming that
miR-338-3p interacted with MCL1 (Fig. 6B
and C). Moreover, the level of MCL1 was significantly
downregulated by miR-338-3p overexpression, which was reversed by
SNHG20 upregulation (Fig. 6D and E).
These data indicate that SNHG20 inhibits miR-338-3p expression to
increase MCL1 level in OC cells.
 | Figure 6.SNHG20 regulates miR-338-3p
expression to modulate MCL1 level. (A) The interaction between
miR-338-3p and MCL1 was predicted by starBase v2.0. Mutated sites
were indicated by the red color. (B and C) The luciferase activity
of SKOV3 and A2780 cells co-transfected with MCL1 3′UTR-WT or MCL1
3′UTR-MUT and miR-NC or miR-338-3p was examined by dual-luciferase
reporter assay. (D and E) MCL1 expression was measured by western
blot analysis in SKOV3 and A2780 cells transfected with miR-NC,
miR-338-3p, miR-338-3p + pcDNA or miR-338-3p + SNHG20,
respectively. (F and G) Relative mRNA level and protein level of
MCL1 were detected by RT-qPCR and western blotting in OC tissues
and the adjacent normal tissues. (H) The association between MCL1
level and SNHG20 level was analyzed using linear regression. (I and
J) Relative mRNA level and protein level of MCL1 were determined by
RT-qPCR and western blotting in OC cells and human ovarian surface
epithelial cells. *P<0.05. SNHG20, small nucleolar RNA host gene
20; MCL1, myeloid cell leukemia 1; WT, wild type; MUT, mutant type;
miR-NC, miR-negative control mimic; RT-qPCR, reverse
transcription-quantitative PCR; OC, ovarian cancer. |
Subsequently, the level of MCL1 in OC tissues was
detected. The results demonstrated that MCL1 mRNA and protein level
were dramatically higher in OC tissues (Fig. 6F and G). Moreover, MCL1 level was
positively correlated with SNHG20 level in OC tissues (Fig. 6H). Besides, increased MCL1 mRNA and
protein expression were observed in OC cells (Fig. 6I and J). Thus, MCL1 might act as an
oncogene in OC development.
SNHG20 mediates MCL1 expression to
regulate OC cell progression
To explore whether SNHG20 exerted its function via
regulating MCL1 expression in OC cells, SKOV3 and A2780 cells were
transfected with si-NC, si-SNHG20, si-SNHG20 + pcDNA, or si-SNHG20
+ MCL1, respectively. RT-qPCR assay confirmed that transfection
with MCL1 significantly upregulated MCL1 expression (Fig. 7A and B). As demonstrated in Fig. 7C and D, cell proliferation was
suppressed by SNHG20 knockdown, which was rescued by MCL1
overexpression. SNHG20 depletion-induced cell apoptosis was
repressed by MCL1 upregulation in OC cells (Fig. 7E and F). Similarly, the effect of
SNHG20 knockdown on the levels of cell apoptosis-associated protein
was weakened by MCL1 upregulation (Fig.
7G and H). Furthermore, it was found that SNHG20 depletion
inhibited cell migration and invasion, while these effects were
reversed by MCL1 overexpression (Fig. 7I
and J). Besides, MCL1 overexpression weakened the effect of
SNHG20 depletion on the levels of EMT markers in OC cells (Fig. 7K and L). These results revealed that
SNHG20 promoted OC progression through modulating MCL1
expression.
 | Figure 7.SNHG20 depletion impeded OC
progression by regulating MCL1 level. (A and B) Relative mRNA level
and protein level of MCL1 were detected by reverse
transcription-quantitative PCR and western blotting in SKOV3 and
A2780 cells transfected with pcDNA or MCL1. (C-L) SKOV3 and A2780
cells were transfected with si-NC, si-SNHG20, si-SNHG20 + pcDNA or
si-SNHG20 + pcDNA-MCL1, respectively. (C and D) MTT assay was used
to measure cell proliferation. (E and F) Cell apoptosis rate was
detected using flow cytometry. (G and H) Relative levels of cell
apoptosis-associated proteins were measured by western blot assay.
(I and J) Transwell assay was used to determine cell migratory and
invasive abilities. (K and L) Western blot assay was carried out to
determine the protein levels of EMT markers. *P<0.05. SNHG20,
small nucleolar RNA host gene 20; OC, ovarian cancer; MCL1, myeloid
cell leukemia 1; pcDNA, pcDNA3.1 vector; si-NC, small interfering
RNA against negative control; si-SNHG20, small interfering RNA
against SNHG20; EMT, epithelial-mesenchymal transition. |
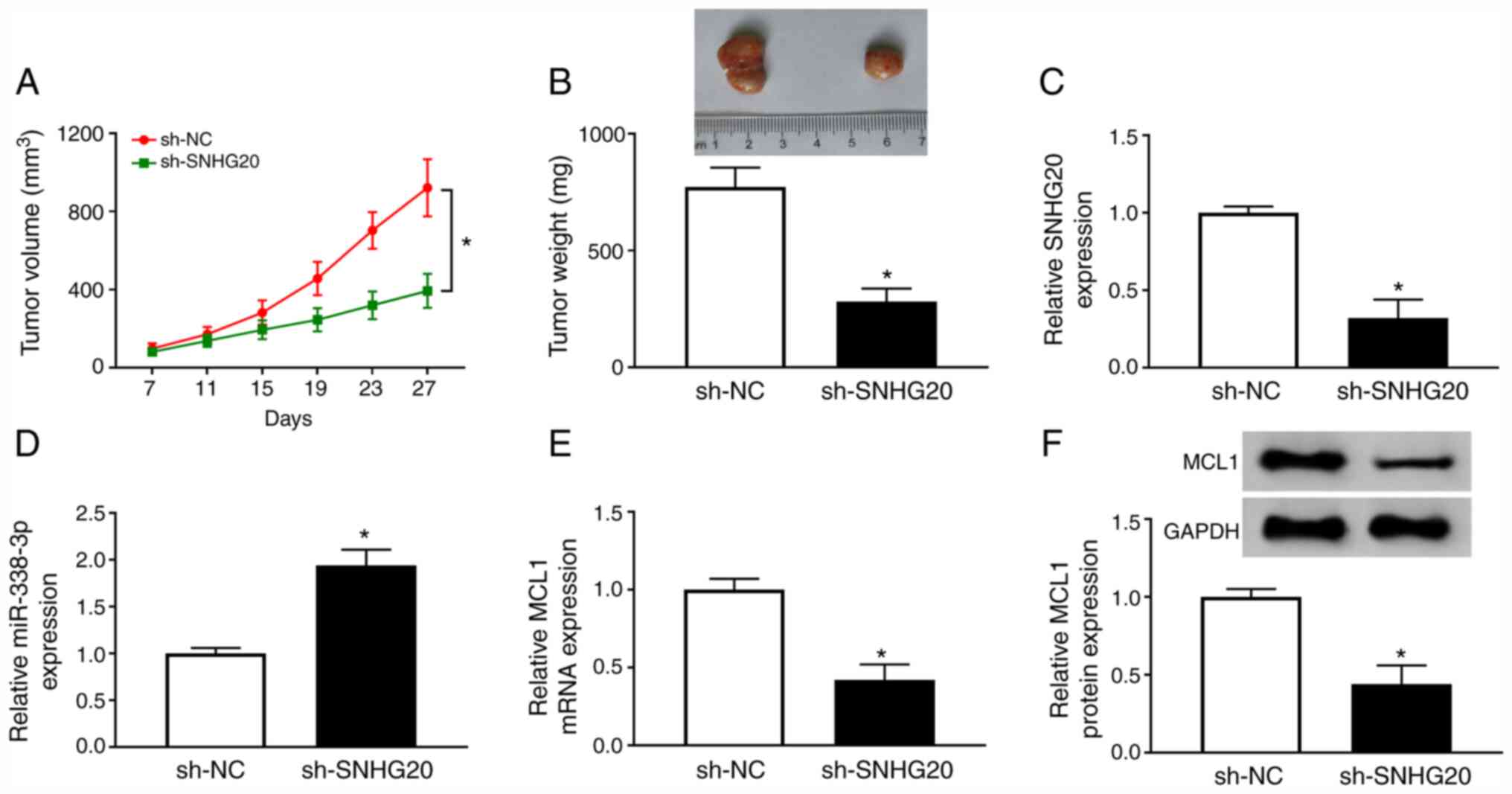
SNHG20 depletion attenuates tumor
growth in vivo
Finally, the effect of SNHG20 on tumor growth was
explored in vivo. A2780 cells transfected with sh-SNHG20 was
inoculated into the nude mice. The expression levels of SNHG20,
miR-338-3p and MCL1 in the A2780 cells transfected sh-SNHG20 are
shown in Fig. S1. The data
indicated that SNHG20 depletion remarkably repressed tumor volume
and weight (Fig. 8A and B). Next,
the levels of SNHG20, miR-338-3p, and MCL1 were determined in
sh-SNHG20 group and sh-NC group. As expected, SNHG20 level
(Fig. 8C) and MCL1 level (Fig. 8E and F) were downregulated, while
miR-338-3p level (Fig. 8D) was
upregulated in sh-SNHG20 group. Therefore, SNHG20 depletion
inhibited tumor growth in vivo.
Discussion
In recent years, increasing studies indicated that
lncRNAs were involved in OC metastasis. For example, lncRNA-MALAT1
knockdown repressed cell proliferation and elevated cell apoptosis
of OC cells via modulating miR-503-5p level (23). Wang et al (24) demonstrated that CDKN2B-AS1 depletion
inhibited OC cell proliferation and mobility through targeting
miR-411-3p. You et al (25)
demonstrated that DLX6-AS1 upregulation enhanced cell mobility by
binding to miR-613 in OC. These data show that lncRNAs play
essential roles in OC progression. Therefore, it is essential to
explore the role and mechanism of lncRNAs in OC.
SNHG20 was reported as a lncRNA to promote cell
development in various human cancer types, including prostate
cancer (26), glioma (27), glioblastoma (28), and nasopharyngeal carcinoma (29). In the present study, it was found
that SNHG20 level was remarkably elevated in OC tissues and cells.
This result corroborated with the previous data (30). Previous results indicated that SNHG20
positively regulated OC cell growth (10,31).
SNHG20 promoted cell proliferation and migration via silencing the
expression of proliferation regulator p21 in non-small cell lung
cancer (32). Another study
confirmed that SNHG20 promoted gastric cancer progression by
inhibiting p21 expression (33). In
glioma cells, downregulation of SNHG20 increased cell apoptosis but
inhibited cell proliferation by regulating PTEN/PI3K/AKT signaling
pathway (34). Moreover, SNHG20
could function as an oncogenic lncRNA by regulating
miR-140-5p/ADAM10 axis and MEK/ERK signaling pathway in cervical
cancer (35). Furthermore, knockdown
of SNHG20 remarkably inhibited cell proliferation, migration and
invasion via dysregulating the expression of p21, cyclin D1,
E-cadherin and vimentin in epithelial OC (31). The aforementioned results
demonstrated that SNHG20 was closely associated with cancer cell
progression. In the present study, it was observed that SNHG20
depletion repressed OC cell proliferation, mobility and promoted
apoptosis, suggesting that SNHG20 served as a positive regulator in
OC progression.
Present evidence suggested that lncRNA exerted its
function through binding to target genes and modulating the levels
of targets in cancer (36). For
instance, SNHG6 elevated chemoresistance in colorectal cancer cell
by targeting miR-26a-5p (37).
Bioinformatics tool showed that miR-338-3p is likely to interact
with SNHG20. Moreover, SNHG20 negatively regulated the expression
of miR-338-3p in OC. Besides, miR-338-3p level was reduced in OC
tissues and cells, this result was consistent with the previous
data (17). In human cancer, many
lncRNAs regulate cancer progression via regulating miR-338-3p
expression. For instance, LINC00689 downregulated miR-338-3p level
to accelerate glioma development and metastasis (38). LncRNA-SNHG15 repressed miR-338-3p
expression to promote colorectal cancer development (39). Furthermore, miR-338-3p was reported
to inhibit cell growth in OC (27,40). In
the present study, the results suggest that SNHG20 knockdown
suppressed OC progression by inhibiting miR-338-3p expression.
In the present study, the data demonstrated that
MCL1 was a potential target of miR-338-3p. Furthermore, miR-338-3p
was found to reduce MCL1 level in OC cells. Besides, MCL1 was
remarkably downregulated in OC tissues and cells. This result was
in agreement with the previous reporter (21). MCL1, as an oncogene, positively
regulated cell development in many human cancer types, such as
gastric cancer (41), lung cancer
(42), cervical cancer (43) and osteosarcoma (44). A recent study indicated that MCL1
depletion inhibited cell proliferation and invasion in OC cells
(21). The present data indicate
that SNHG20 repressed miR-338-3p expression to increase MCL1 level,
and MCL1 overexpression weakened the effect of SNHG20 depletion on
OC progression. These results suggest that SNHG20 depletion
suppressed OC progression via modulating the miR-338-3p/MCL1
axis.
Previous results demonstrated that SNHG20 depletion
remarkably repressed tumor growth of OC in vivo (10). The effect of SNHG20 on tumor growth
was also investigated in the present study. As expected, tumor
volume and weight were significantly suppressed by SNHG20
depletion. Therefore, SNHG20 knockdown attenuated OC tumor growth
in vivo.
In conclusion, the present results demonstrate that
SNHG20 knockdown represses the development of OC through modulating
the miR-338-3p/MCL1 axis, providing a potential target for the
therapy of patients with OC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DW, ZL, HL, JL and QQ made substantial contribution
to the concept and design, data analysis, and interpretation of the
data; DW, ZL and HL performed the experiments and interpreted the
data; DW and QQ drafted the manuscript; DW and JL revised the
manuscript critically for important intellectual content. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethical review
committee of the Affiliated Renhe Hospital of China Three Gorges
University. Written informed consent was obtained from all enrolled
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar
|
|
2
|
Doubeni CA, Doubeni AR and Myers AE:
Diagnosis and management of ovarian cancer. Am Fam Physician.
93:937–944. 2016.
|
|
3
|
Xue Y, Ma G, Gu D, Zhu L, Hua Q, Du M, Chu
H, Tong N, Chen J, Zhang Z and Wang M: Genome-wide analysis of long
noncoding RNA signature in human colorectal cancer. Gene.
556:227–234. 2015. View Article : Google Scholar
|
|
4
|
Xiao J, Lai H, Wei SH, Ye ZS, Gong FS and
Chen LC: lncRNA HOTAIR promotes gastric cancer proliferation and
metastasis via targeting miR-126 to active CXCR4 and RhoA signaling
pathway. Cancer Med. 8:6768–6779. 2019. View Article : Google Scholar
|
|
5
|
Liang H, Su X, Wu Q, Shan H, Lv L, Yu T,
Zhao X, Sun J, Yang R, Zhang L, et al: LncRNA promotes ischemic
myocardial injury by regulating autophagy through targeting.
Autophagy undefined. 1–15. 2019.
|
|
6
|
Li Q, Zhang J, Su DM, Guan LN, Mu WH, Yu
M, Ma X and Yang RJ: lncRNA TUG1 modulates proliferation,
apoptosis, invasion, and angiogenesis via targeting miR-29b in
trophoblast cells. Hum Genomics. 13:502019. View Article : Google Scholar
|
|
7
|
Terashima M, Ishimura A, Wanna-Udom S and
Suzuki T: MEG8 long noncoding RNA contributes to epigenetic
progression of the epithelial-mesenchymal transition of lung and
pancreatic cancer cells. J Biol Chem. 293:18016–18030. 2018.
View Article : Google Scholar
|
|
8
|
Lu YM, Wang Y, Liu SQ, Zhou MY and Guo YR:
Profile and validation of dysregulated long noncoding RNAs and
mRNAs in ovarian cancer. Oncol Rep. 40:2964–2976. 2018.
|
|
9
|
Zhang D, Cao C, Liu L and Wu D:
Up-regulation of LncRNA SNHG20 predicts poor prognosis in
hepatocellular carcinoma. J Cancer. 7:608–617. 2016. View Article : Google Scholar
|
|
10
|
He S, Zhao Y, Wang X, Deng Y, Wan Z, Yao S
and Shen H: Up-regulation of long non-coding RNA SNHG20 promotes
ovarian cancer progression via Wnt/β-catenin signaling. Biosci Rep.
38:BSR201706812018. View Article : Google Scholar
|
|
11
|
Trionfini P and Benigni A: MicroRNAs as
master regulators of glomerular function in health and disease. J
Am Soc Nephrol. 28:1686–1696. 2017. View Article : Google Scholar
|
|
12
|
Wang Y, Kim S and Kim IM: Regulation of
metastasis by microRNAs in ovarian cancer. Front Oncol. 4:1432014.
View Article : Google Scholar
|
|
13
|
Sun J, Qiao Y, Song T and Wang H: MiR-495
suppresses cell proliferation by directly targeting HMGA2 in lung
cancer. Mol Med Rep. 19:1463–1470. 2019.
|
|
14
|
Liu C, Cai L and Li H: miR185 regulates
the growth of osteosarcoma cells via targeting Hexokinase 2. Mol
Med Rep. 20:2774–2782. 2019.
|
|
15
|
Luo Y, Hao T, Zhang J, Zhang M, Sun P and
Wu L: MicroRNA-592 suppresses the malignant phenotypes of thyroid
cancer by regulating lncRNA NEAT1 and downregulating NOVA1. Int J
Mol Med. 44:1172–1182. 2019.
|
|
16
|
Wang C, Zhang W, Xing S, Wang Z, Wang J
and Qu J: MiR-342-3p inhibits cell migration and invasion through
suppressing forkhead box protein Q1 in ovarian carcinoma.
Anticancer Drugs. 30:917–924. 2019. View Article : Google Scholar
|
|
17
|
Zhang Y, Shi B, Chen J, Hu L and Zhao C:
MiR-338-3p targets pyruvate kinase M2 and affects cell
proliferation and metabolism of ovarian cancer. Am J Transl Res.
8:3266–3273. 2016.
|
|
18
|
Liu X, Wen J, Wang H and Wang Y: Long
non-coding RNA LINC00460 promotes epithelial ovarian cancer
progression by regulating microRNA-338-3p. Biomed Pharmacother.
108:1022–1028. 2018. View Article : Google Scholar
|
|
19
|
Yip KW and Reed JC: Bcl-2 family proteins
and cancer. Oncogene. 27:6398–6406. 2008. View Article : Google Scholar
|
|
20
|
Zhang S, Zhang M, Jing Y, Yin X, Ma P,
Zhang Z, Wang X, Di W and Zhuang G: Deubiquitinase USP13 dictates
MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat
Commun. 9:2152018. View Article : Google Scholar
|
|
21
|
Su J, Ruan S, Dai S, Mi J, Chen W and
Jiang S: NF1 regulates apoptosis in ovarian cancer cells by
targeting MCL1 via miR-142-5p. Pharmacogenomics. 20:155–165. 2019.
View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Sun Q, Li Q and Xie F: LncRNA-MALAT1
regulates proliferation and apoptosis of ovarian cancer cells by
targeting miR-503-5p. Onco Targets Ther. 12:6297–6307. 2019.
View Article : Google Scholar
|
|
24
|
Wang Y, Huang Y, Liu H, Su D, Luo F and
Zhou F: Long noncoding RNA CDKN2B-AS1 interacts with miR-411-3p to
regulate ovarian cancer in vitro and in vivo through
HIF-1a/VEGF/P38 pathway. Biochem Biophys Res Commun. 514:44–50.
2019. View Article : Google Scholar
|
|
25
|
You Q, Shi HY, Gong CF, Tian XY and Li S:
Long non-coding RNA DLX6-AS1 acts as an oncogene by targeting
miR-613 in ovarian cancer. Eur Rev Med Pharmacol Sci. 23:6429–6435.
2019.
|
|
26
|
Wu X, Xiao Y, Zhou Y, Zhou Z and Yan W:
lncRNA SNHG20 promotes prostate cancer migration and invasion via
targeting the miR-6516-5p/SCGB2A1 axis. Am J Transl Res.
11:5162–5169. 2019.
|
|
27
|
Liu J, Cheng LG and Li HG: LncRNA SNHG20
promoted the proliferation of glioma cells via sponging miR-4486 to
regulate the MDM2-p53 pathway. Eur Rev Med Pharmacol Sci.
23:5323–5331. 2019.
|
|
28
|
Gao XF, He HQ, Zhu XB, Xie SL and Cao Y:
LncRNA SNHG20 promotes tumorigenesis and cancer stemness in
glioblastoma via activating PI3K/Akt/mTOR signaling pathway.
Neoplasma. 66:532–542. 2019. View Article : Google Scholar
|
|
29
|
Sun C, Sun Y and Zhang E: Long non-coding
RNA SNHG20 promotes nasopharyngeal carcinoma cell migration and
invasion by upregulating TGF-β1. Exp Ther Med. 16:4967–4974.
2018.
|
|
30
|
Zhao W, Ma X, Liu L, Chen Q, Liu Z, Zhang
Z, Ma S, Wang Z, Li H, Wang Z and Wu J: SNHG20: A vital lncRNA in
multiple human cancers. J Cell Physiol. Jan 15–2019.(Epub ahead of
print).
|
|
31
|
Wang D, Dai J, Hou S and Qian Y: LncRNA
SNHG20 predicts a poor prognosis and promotes cell progression in
epithelial ovarian cancer. Biosci Rep. 39:BSR201821862019.
View Article : Google Scholar
|
|
32
|
Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B,
Zhang T, Zhou Y, Chen Q, Wei C, et al: Long non-coding RNA SNHG20
promotes non-small cell lung cancer cell proliferation and
migration by epigenetically silencing of P21 expression. Cell Death
Dis. 8:e30922017. View Article : Google Scholar
|
|
33
|
Liu J, Liu L, Wan JX and Song Y: Long
noncoding RNA SNHG20 promotes gastric cancer progression by
inhibiting p21 expression and regulating the GSK-3β/β-catenin
signaling pathway. Oncotarget. 8:80700–80708. 2017. View Article : Google Scholar
|
|
34
|
Guo LP, Zhang ZJ, Li RT, Li HY and Cui YQ:
Influences of LncRNA SNHG20 on proliferation and apoptosis of
glioma cells through regulating the PTEN/PI3K/AKT signaling
pathway. Eur Rev Med Pharmacol Sci. 23:253–261. 2019.
|
|
35
|
Guo H, Yang S, Li S, Yan M, Li L and Zhang
H: LncRNA SNHG20 promotes cell proliferation and invasion via
miR-140-5p-ADAM10 axis in cervical cancer. Biomed Pharmacother.
102:749–757. 2018. View Article : Google Scholar
|
|
36
|
Sun F, Liang W, Tang K, Hong M and Qian J:
Profiling the lncRNA-miRNA-mRNA ceRNA network to reveal potential
crosstalk between inflammatory bowel disease and colorectal cancer.
PeerJ. 7:e74512019. View Article : Google Scholar
|
|
37
|
Wang X, Lan Z, He J, Lai Q, Yao X, Li Q,
Liu Y, Lai H, Gu C, Yan Q, et al: LncRNA SNHG6 promotes
chemoresistance through ULK1-induced autophagy by sponging
miR-26a-5p in colorectal cancer cells. Cancer Cell Int. 19:2342019.
View Article : Google Scholar
|
|
38
|
Liu X, Zhu Q, Guo Y, Xiao Z, Hu L and Xu
Q: LncRNA LINC00689 promotes the growth, metastasis and glycolysis
of glioma cells by targeting miR-338-3p/PKM2 axis. Biomed
Pharmacother. 117:1090692019. View Article : Google Scholar
|
|
39
|
Li M, Bian Z, Jin G, Zhang J, Yao S, Feng
Y, Wang X, Yin Y, Fei B, You Q and Huang Z: LncRNA-SNHG15 enhances
cell proliferation in colorectal cancer by inhibiting miR-338-3p.
Cancer Med. 8:2404–2413. 2019. View Article : Google Scholar
|
|
40
|
Zhang R, Shi H, Ren F, Liu Z, Ji P, Zhang
W and Wang W: Down-regulation of miR-338-3p and Up-regulation of
MACC1 indicated poor prognosis of epithelial ovarian cancer
patients. J Cancer. 10:1385–1392. 2019. View Article : Google Scholar
|
|
41
|
Huang Y, Luo H, Li F, Yang Y, Ou G, Ye X
and Li N: LINC00152 down-regulated miR-193a-3p to enhance MCL1
expression and promote gastric cancer cells proliferation. Biosci
Rep. 38:BSR201716072018. View Article : Google Scholar
|
|
42
|
Suzuki J, Nakajima W, Suzuki H, Asano Y
and Tanaka N: Chaperone-mediated autophagy promotes lung cancer
cell survival through selective stabilization of the pro-survival
protein, MCL1. Biochem Biophys Res Commun. 482:1334–1340. 2017.
View Article : Google Scholar
|
|
43
|
Zhou C, Li G, Zhou J, Han N, Liu Z and Yin
J: miR-107 activates ATR/Chk1 pathway and suppress cervical cancer
invasion by targeting MCL1. PLoS One. 9:e1118602014. View Article : Google Scholar
|
|
44
|
Xu W, Li Z, Zhu X, Xu R and Xu Y: miR-29
family inhibits resistance to methotrexate and promotes cell
apoptosis by targeting COL3A1 and MCL1 in osteosarcoma. Med Sci
Monit. 24:8812–8821. 2018. View Article : Google Scholar
|