Introduction
Colorectal cancer (CRC) is the third most common
malignant tumor, and it has high rates of global morbidity and
mortality. There were over 1.8 million newly diagnosed CRC cases
worldwide in 2018, and there were estimated to be over 880,000
mortalities (1). Affected by the
western diet and lifestyle, the incidence and mortality rates of
CRC have continued to increase in China, rising to the 3rd and 5th
highest, respectively (2). The
prognosis of CRC is closely associated with early diagnosis
(3,4). As reported in 2012, the 5-year survival
rate of early cancer was reported as 90%, and for the late stage,
it was <10% (5). Thus, it remains
crucial to identify accurate and non-invasive molecular biomarkers
for CRC diagnosis and treatment.
Circular RNAs (circRNAs), a class of newly
discovered endogenous non-coding RNAs that are formed by reverse
splicing (6–9), are characterized by high expression
levels (10,11). Their expression is often specific to
tissues or developmental stages (8,12). Thus,
circRNAs are considered effective biomarkers for the diagnosis and
prognosis of different types of cancer due to their abundance and
stability (13–17). Increasing evidence suggests that
circRNAs have a large number of miRNA binding sites and serve as
miRNA sponges to indirectly regulate gene expression (6,18,19).
CircRNAs play an important role in human diseases (20–22),
particularly in tumors, such as gastric cancer (23), papillary thyroid cancer (24) and lung cancer (25). However, research on circRNAs in human
CRC is limited.
In the present study, circRNA databases (http://circbase.org) andcirc2Traits (http://gyanxet-beta.com/circdb) were used to
screen two human circRNAs (hsa_circ_0001696 and hsa_circ_0001695)
that are associated with CRC. The results demonstrated that
hsa_circ_0001696 expression was downregulated in CRC tissues
compared with adjacent normal tissues, while no significant
differences were observed in hsa_circ_0001695 expression between
the two groups. Based on these results, human hsa_circ_0001696 was
identified as a candidate circRNA. Hsa_circ_0001696 is 347
nucleotides in spliced sequence length, its gene is located at
chr7:35707043-35712888 and the gene symbol is HERPUD2. Taken
together, the results of the present study suggest that suppressing
hsa_circ_0001696 expression may affect the proliferation and
migration of CRC cells.
Materials and methods
Patients and clinical specimens
A total of 18 paired CRC tissues and matched
adjacent normal tissues (5 cm from the edge of CRC) were collected
from resected surgical specimens at the Ningbo First Hospital,
Ningbo Hospital of Zhe Jiang University (Zhejiang, China), between
August 2017 and February 2018. The study included 7 men and 11
women (mean average age, 63 years; age range, 47–81). All tissue
samples were immediately frozen in RNA-fixer reagent (Bioteke
Corporation) following collection and stored at −80°C until RNA
extraction.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from CRC tissues and cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. RNA
integrity was assessed using a SmartSpec Plus spectrophotometer
(Bio-Rad Laboratories, Inc.), and RNA concentrations were
determined using a NanoDrop ND2000 (Thermo Fisher Scientific,
Inc.). Total RNA (1 µg) was reverse transcribed into cDNA using the
GoScript RT System kit, with random primers (Promega Corporation;
cat. no. A5001). The following thermocycling conditions were used:
25°C for 5 min, 42°C for 60 min and 70°C for 15 min. qPCR was
subsequently performed using the SYBR-Green master mix (Promega
Corporation; cat. no. A6001) and Mx3005P real-time PCR System
(Stratagene; Agilent), according to the manufacturer's protocols.
The following primer sequences were used for qPCR:hsa_circ_0001696
forward, 5′-GGAAGCAGTCTGCCCGAATA-3′ and reverse,
5′-CCAAGCACAGAGTCACCAGT-3′; and GAPDH forward,
5′-TCGACAGTCAGCCGCATCTTCTTT-3′ and reverse,
5′-ACCAAATCCGTTGACTCCGACCTT-3′. Relative expression levels were
calculated using the 2−ΔΔCt method (26–28) and
normalized to the internal reference gene GAPDH. All experiments
were performed in triplicate.
Cell culture and transient
transfection
The human CRC cell lines, HCT116 and SW620, were
purchased from the Shanghai Institutes for Biological Sciences,
Chinese Academy of Sciences. Both cell lines were maintained in
DMEM supplemented with 10% fetal bovine serum (both purchased from
Gibco; Thermo Fisher Scientific, Inc.), at 37°C in a humidified
atmosphere with 5% CO2. The small interfering (si)RNA
and stable negative control (NC) sequences (Table I) were synthesized by Shanghai
GenePharma Co., Ltd. For siRNA and NC transfection with the 95%
purity and 20 uM initial concentration, cells were seeded into
6-well plates at 60% density and subsequently transfected at room
temperature for about 60 min with 80, 100 or 120 nM siRNA after 24
h of cultivation, using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Subsequent experiments were performed 24 h
post-transfection.
 | Table I.Primer sequences used for cell
transfection. |
Table I.
Primer sequences used for cell
transfection.
| Name | Primer sequence
(5′-3′) |
|---|
| siRNA-1 | Forward:
CCCGAAUACACCAAUCUCUTT |
|
(hsa_circ_0001696) | Reverse:
AGAGAUUGGUGUAUUCGGGTT |
| siRNA-2 | Forward:
CCUCAAUCCUUCUGAGGAATT |
|
(hsa_circ_0001696) | Reverse:
UUCCUCAGAAGGAUUGAGGTT |
| siRNA-1 (CDK4) | Forward:
GCAUGUAGACCAGGACCUATT |
|
| Reverse:
UAGGUCCUGGUCUACAUGCTT |
| siRNA-2 (CDK4) | Forward:
GCAGCACUCUUAUCUACAUTT |
|
| Reverse:
AUGUAGAUAAGAGUGCUGCTT |
| siRNA-1 (MMP9) | Forward:
CCACCACAACAUCACCUAUTT |
|
| Reverse:
AUAGGUGAUGUUGUGGUGGTT |
| siRNA-2 (MMP9) | Forward:
GCGCUGGGCUUAGAUCAUUTT |
|
| Reverse:
AAUGAUCUAAGCCCAGCGCTT |
| NC | Forward:
UUCUCCGAACGUGUCACGUTT |
|
| Reverse:
ACGUGACACGUUCGGAGAATT |
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) assay was performed
to assess cell proliferation (TransGen Biotech, Co., Ltd.),
according to the manufacturer's protocol. Transfected CRC cells
were seeded into 96-well plates at a density of 2,000 cells/well
and maintained in DMEM supplemented with 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.), and incubated for 24, 48,
72 and 96 h at 37°C in a humidified atmosphere with 5%
CO2. Subsequently, cells were incubated with CCK-8
solution (10 µl) for 2 h at 37°C and cell proliferation was
analyzed at a wavelength of 450 nm, using a microplate reader
(Applied Biosystems; Thermo Fisher Scientific, Inc.).
Colony formation assay
Transfected CRC cells were seeded into 6-well plates
at a density of 100 cells/well for 2 weeks to form colonies, and
DMEM was changed every 3 days. After 14 days, the supernatant was
discarded and cells were washed twice with phosphate-buffered
saline (PBS). Cells were fixed with 2 ml methanol (Geneslscherical
industry, Jinan, Shangdong, China, http://www.chembk.com/cn/chem/67-56-1) at room
temperature for 20 min and subsequently stained with 0.5% crystal
violet at room temperature for 15 min. Cells were re-washed with
PBS and the number of cell colonies were observed using an inverted
fluorescent light microscope (Olympus Corporation, CX33;
magnifications, 40×10).
Wound healing assay
Once the HCT116 and SW620 cells reached 95%
confluence, the monolayers were scratched (recorded at 0 h) and the
transfected CRC cells were cultured with serum-free medium for 24
h. Cells were observed under an inverted fluorescent light
microscope at 0 and 24 h (magnifications, 40×10). Image pro plus
6.0 software (Media Cybernetics, Inc.) was used to measure the
distance at 0 and 24 h. GraphPad Prismv8.3.0.538 (Graph Pad
Software, Inc.) was used to perform statistical analysis.
Western blotting
Total protein was extracted from CRC cells using
cell lysis buffer (Beijing Solarbio Science & Technology Co.,
Ltd.), and the BCA method was used to detect the protein. Total
protein (30 µg/lane) was separated by 12% SDS-PAGE and the
separated proteins were subsequently transferred onto
polyvinylidene fluoride (PVDF) membranes (EMD Millipore). The PVDF
membranes were blocked in blocking solution (Beyotime Institute of
Biotechnology) for 60 min in a shaker at room temperature and
incubated with primary antibodies against: Cyclin-dependent kinase
4 (CDK4; cat. no. ab137675), cyclin D (cat. no. ab62151), cyclin E
(cat. no. ab33911), matrix metalloproteinase 9 (MMP9; cat. no.
BA0573) and β-actin (cat. no. 4ab010745) overnight at 4°C on a
shaker, all of which used primary antibody dilution buffer and were
purchased from Beyotime Institute of Biotechnology. Following the
primary incubation, membranes were incubated with HRP-conjugated
affinipure rabbit anti-Goat IgG (H+L) (1:5,000; cat. no. SA00001-1;
ProteinTech Group, Inc) at room temperature for 60 min. Protein
bands were visualized using the bright ECL kit (Advansta, Inc.
K-12045-D20, http://advansta.com/products/western-blot-substrate-WesternBright-ECL)
and protein bands were detected using an Odyssey Infrared Imaging
system (LI-COR Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS 20.0
software (IBM Corp.). All experiments were performed in triplicate
and data are presented as the mean ± standard deviation.
Differences between paired groups were estimated by two-tailed
Student's t-test. One-way analysis of variance followed by Tukey's
post hoc test were used to compare differences between unpaired
multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Hsa_circ_0001696 is downregulated in
CRC tissues
RT-qPCR analysis was performed to detect
hsa_circ_0001696 expression in paired CRC tissues. The results
demonstrated that hsa_circ_0001696 expression was significantly
lower in CRC tissues compared with adjacent normal tissues
(P<0.01; Fig. 1A and B).
Furthermore, among the 18 paired CRC tissues, 14 samples (77.8%)
exhibited significantly downregulated hsa_circ_0001696 expression
in the cancerous tissues (Fig.
1C).
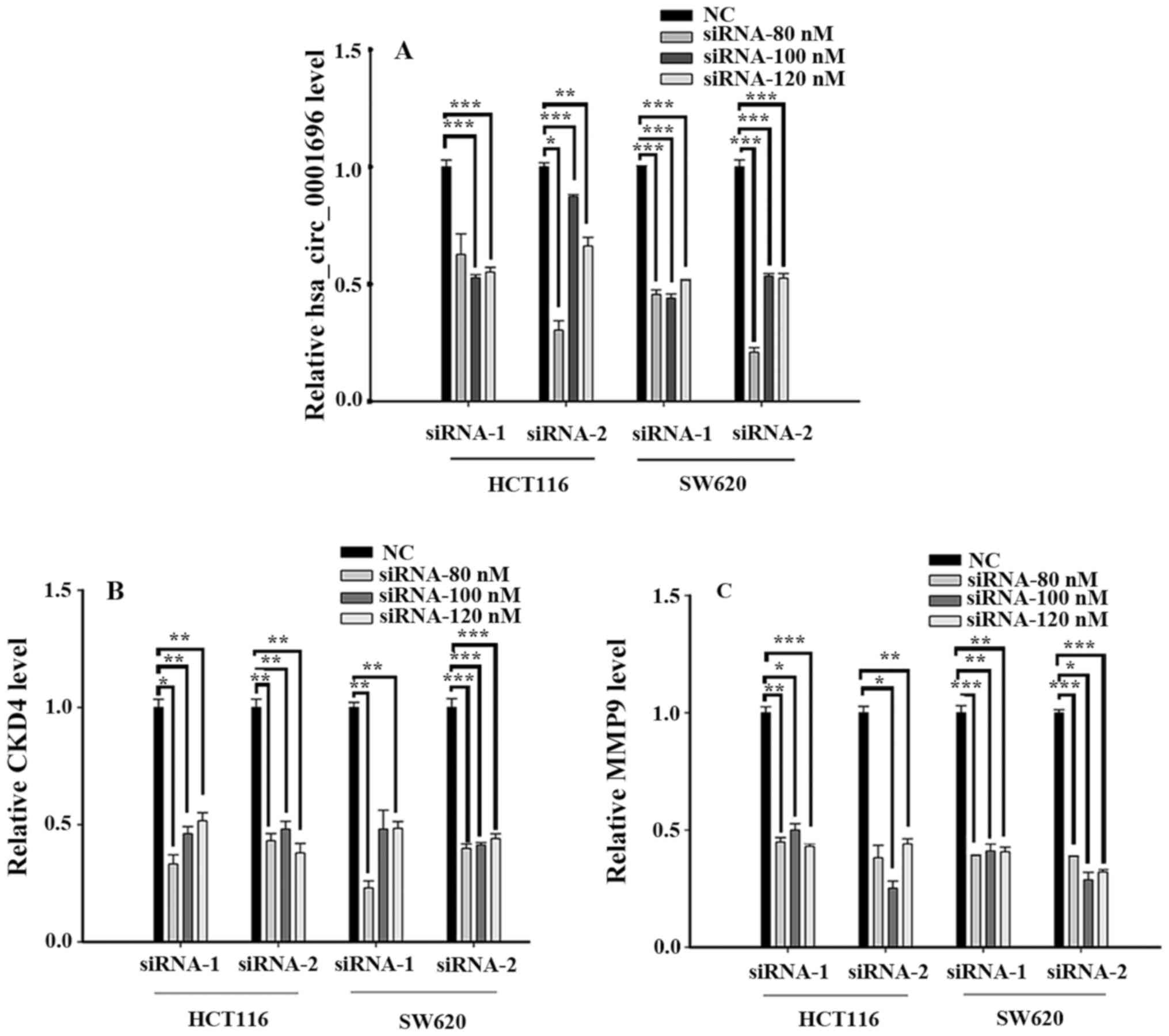
Transfection effect of siRNA
A total of two siRNAs were used to silence
hsa_circ_0001696, CDK4 and MMP9 expression in HCT116 and SW620
cells (Fig. 2). The results
demonstrated that the transfection efficiency of siRNA-2 was
higher, and siRNA-80 nM was used for subsequent experimentation, in
which hsa_circ_0001696 was transfected into HCT116 and SW620 cells.
With regards to CDK4, the transfection efficiency of siRNA-2 was
higher, and siRNA-80 nM was also used for subsequent
experimentation, whereas siRNA-100 nM of siRNA-1 was used for
MMP9.
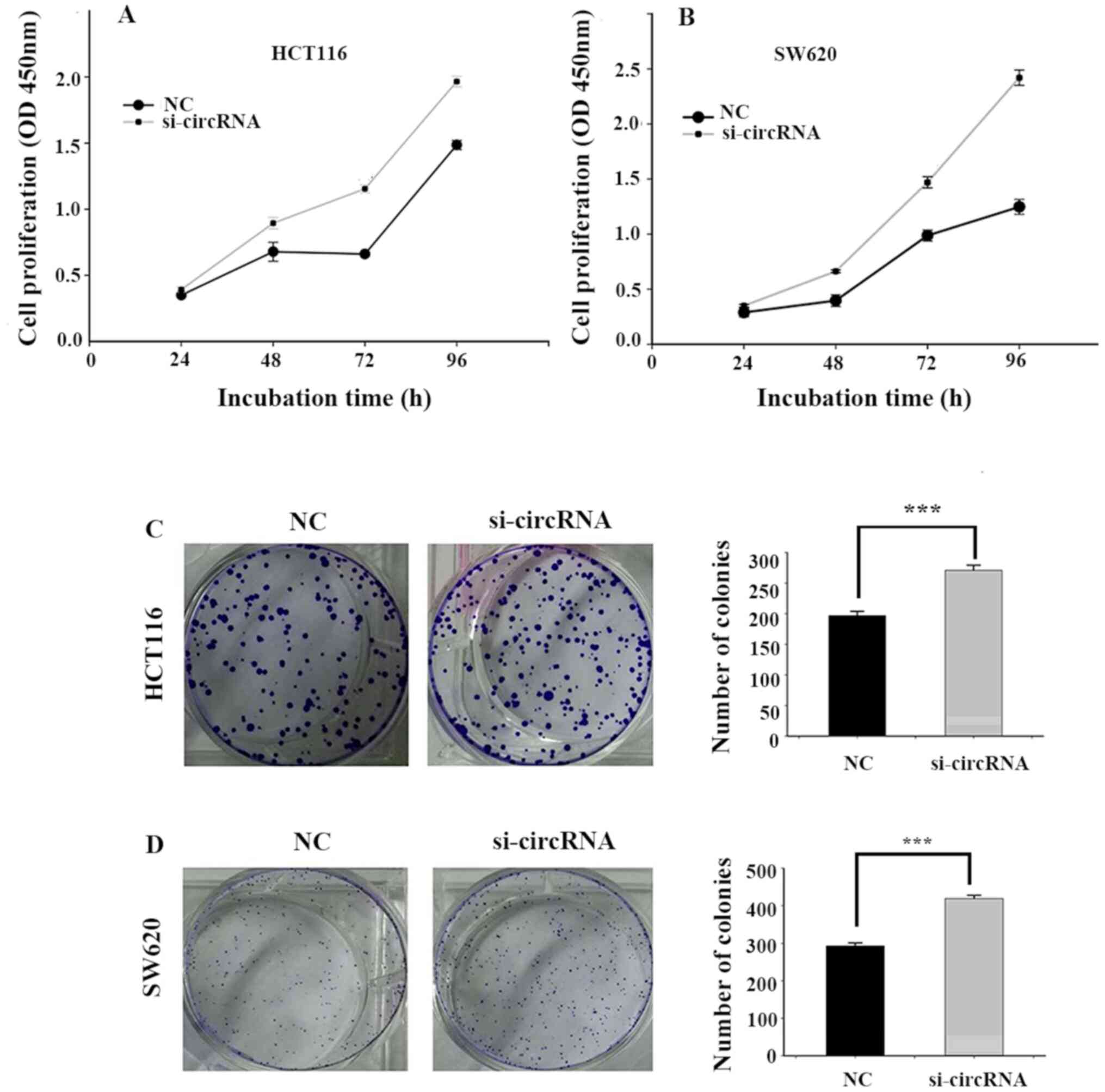
Silencing hsa_circ_0001696 promotes
CRC cell proliferation
To investigate the effect of hsa_circ_0001696 on the
biological behaviour of CRC cells, the CCK-8 and colony formation
assays were performed. The results of the CCK-8 assay demonstrated
that hsa_circ_0001696 knockdown significantly promoted HCT116 and
SW620 cell proliferation compared with the NC group (Fig. 3A and B). The results of the colony
formation assay demonstrated that hsa_circ_0001696 knockdown
significantly increased the number of CRC cell colonies (Fig. 3C and D).
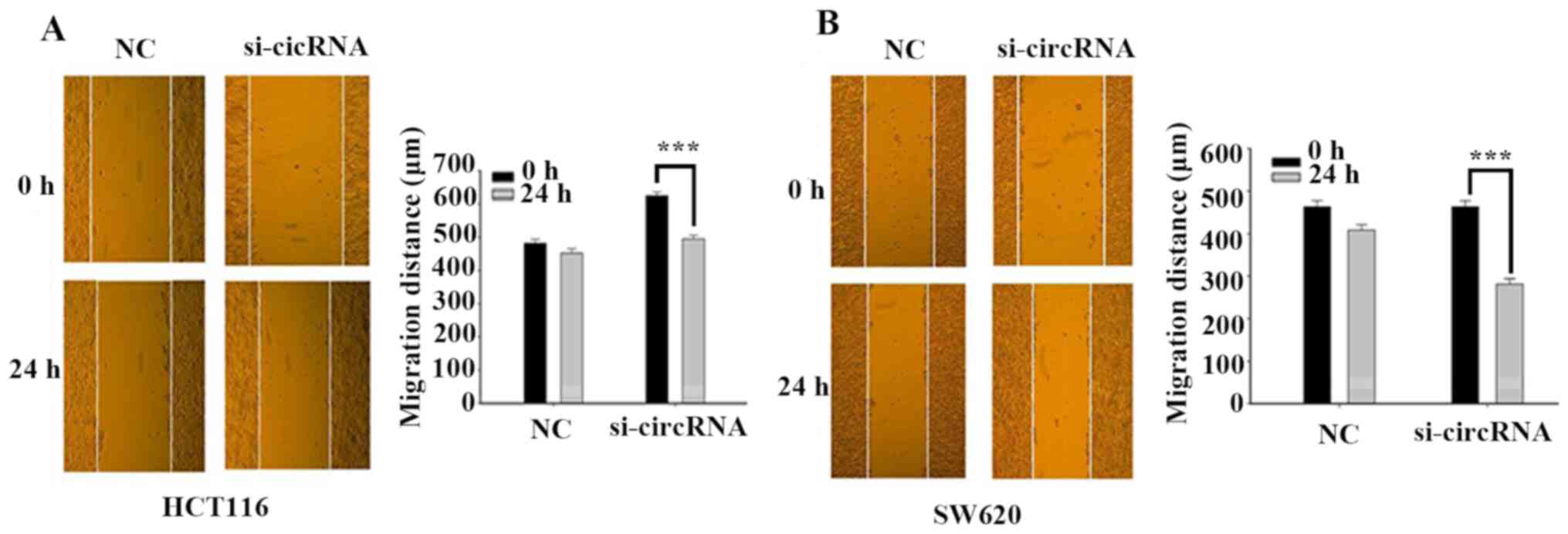
Silencing hsa_circ_0001696 promotes
CRC cell migration
The wound healing assay was performed to assess the
effect of hsa_circ_0001696 on CRC cell migration. The results
demonstrated that the migratory ability of HCT116 and SW620 cells
enhanced following transfection with si-circRNA, compared with the
control group (Fig. 4A and B). These
results suggest that hsa_circ_0001696 knockdown may promote the
migration of CRC cells.
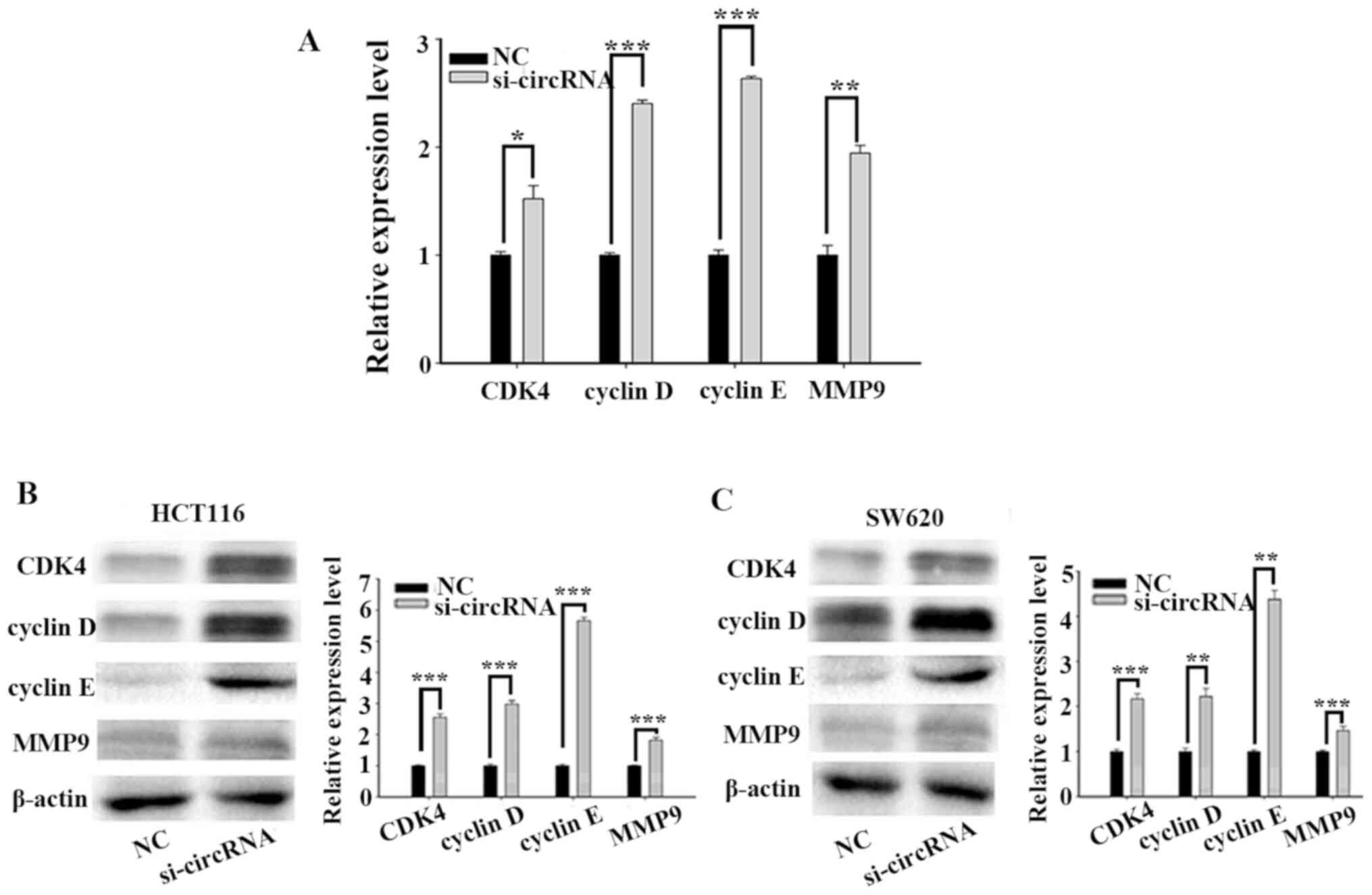
Mechanism of hsa_circ_0001696 in
CRC
To further investigate the molecular mechanism of
hsa_circ_0001696 on CRC cell proliferation and migration, the
expression levels of cell cycle-related proteins (CDK4, cyclin D
and cyclin E) and MMP9 were detected. RT-qPCR and western blot
analyses demonstrated that the expression levels of CDK4, cyclin D,
cyclin E and MMP9 significantly increased following transfection
with si-circRNA (Fig. 5).
Silencing CDK4 and MMP9 inhibits CRC
cell proliferation and migration, respectively
CDK4 and MMP9 expression levels were suppressed to
determine whether they attenuated the effect of hsa_circ_0001696 on
cell proliferation and migration. The results demonstrated that
silencing CDK4 expression inhibited CRC cell proliferation, while
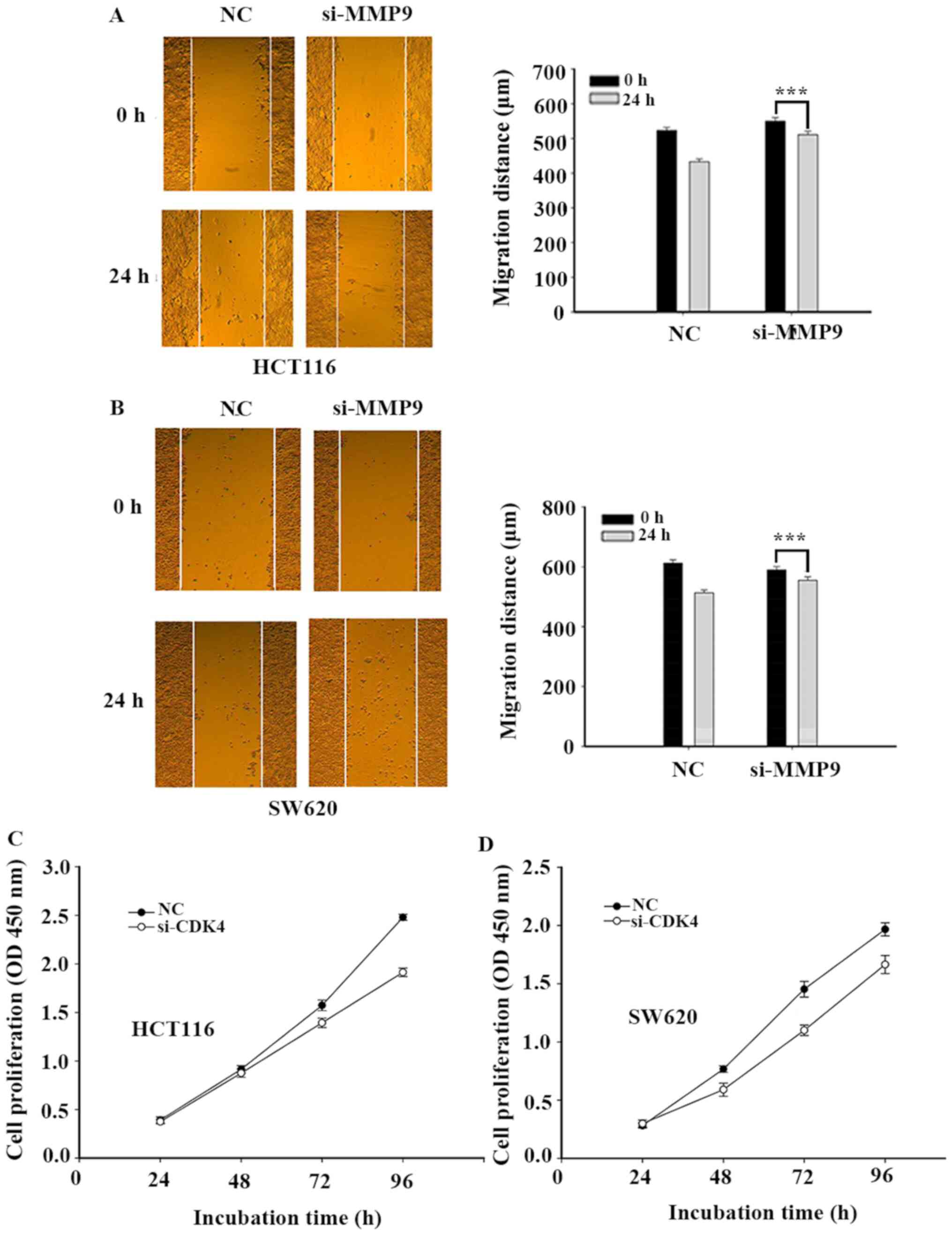
silencing MMP9 expression inhibited CRC cell migration (Fig. 6).
Discussion
CRC is one of the most common tumors of the
digestive system, and its mortality and morbidity rates rank third
worldwide (1). There were over 1.8
million newly diagnosed CRC cases worldwide in 2018, and there were
estimated to be over 880,000 mortalities (1). Although radical tumor resection and
adjuvant chemoradiotherapy have been extensively used in clinical
practice, the recurrence rate remains high at 25–40% (29), and 80% of recurrences occur within 3
years of radical surgery (30).
Thus, it remains critical to identify novel biomarkers for accurate
and non-invasive identification of CRC recurrence, diagnosis and
treatment.
CircRNAs are a class of novel non-coding RNAs
(6,7). Compared to other non-coding RNAs,
circRNAs have a unique ‘back-splicing’ structure, which is
generated from the joining of an upstream 3′ splice acceptor to a
downstream 5′ splice donor (31).
This unique structure can be resistant to exonucleases, such as
RNase, which makes circRNAs more stable in the human body compared
with linear RNA isoforms (7). Thus,
circRNAs are ideal molecular diagnostic markers that are more
advantageous than linear RNA (11,16).
Similar to long non-coding RNAs, there are subset of circRNAs
called ceRNAs (32–34) that act as miRNA sponge molecules to
regulate gene expression by adsorbing miRNAs (21,35,36).
Recently, a number of studies have demonstrated that circRNAs exert
a critical function in the progression of human CRC (35,37–39).
However, little is known about the role of hsa_circ_0001696 in the
development of CRC.
In the present study, circRNA databases and
bioinformatics analysis demonstrated that hsa_circ_0001696 (gene
symbol is HERPUD2) was significantly downregulated in CRC tissues
compared with adjacent normal tissues. These results suggest that
hsa_circ_0001696 may act as a suppressor gene in CRC
progression.
To further investigate the molecular mechanisms
involved in the regulation of hsa_circ_0001696, its expression was
assessed in two CRC cell lines, HCT116 and SW620. To detect the
effect of abnormally expressed hsa_circ_0001696 on CRC, cells were
transfected with siRNA to knockdown hsa_circ_0001696 expression.
The results demonstrated that hsa_circ_0001696 knockdown promoted
CRC cell proliferation and migration, and the formation of cell
colonies. In addition, CDK4, cyclin D, cyclin E and MMP9 mRNA and
protein expression levels increased in cells transfected with
siRNA. Furthermore, suppressing CDK4 and MMP9 expression inhibited
cell proliferation and migration, respectively.
The results of the present study identified a novel
association between hsa_circ_0001696 and CRC, whereby
hsa_circ_0001696 may act as a potential novel molecular diagnostic
marker. The results demonstrated that abnormal hsa_circ_0001696
expression affected CRC cell proliferation and migration, and the
formation of cell colonies by interfering with the protein
expression levels of CDK4, cyclin D, cyclin E and MMP9.
The present study is not without limitations. First,
the number of clinical samples was limited. The results indicated
that hsa_circ_0001696 was expressed at significantly lower levels
in 18 paired CRC tissues compared with adjacent normal tissues.
However, the number of clinical samples was not sufficient to
conclude that hsa_circ_0001696 can be used as a diagnostic marker.
Thus, prospective studies should focus on collecting samples from
multiple centres. Secondly, due to the structure of
hsa_circ_0001696, the present study only designed two siRNAs. The
interference effect may have been better if more siRNAs were
designed. CircRNAs serve as miRNA sponges to indirectly regulate
gene expression (14,21,22,40).
However, the present study failed to assess whether
hsa_circ_0001696 also has ceRNA functions or if it directly binds
proteins. In addition, the specific miRNAs that interact with
hsa_circ_0001696 to regulate the expression of miRNA target genes
remain unknown, and the protein binding partners have not yet been
investigated. Thus, these limitations will be the focus of
prospective studies.
In conclusion, the results of the present study
demonstrated that hsa_circ_0001696 expression was downregulated in
CRC tissues compared with adjacent normal tissues, thus it may act
as a molecular diagnostic marker. In addition, hsa_circ_0001696
knockdown promoted CRC cell proliferation and migration by
regulating the expression levels of CDK4, cyclin D, cyclin E and
MMP9.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Zhejiang
Provincial Natural Science Foundation of China (grant nos.
LQ18H160015 and LY19H030002), the Ningbo Natural Science Foundation
of China (grant no. 2018A610385) and the Medical Health Science and
Technology Project of Zhejiang Provincial Health Commission (grant
no. 2019KY571).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contribution
LX and XYD designed the present study. PFL drafted
the initial manuscript and performed statistical analysis. PFL, ZXZ
and XY performed the experiments. HJS helped perform statistical
analysis. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Human Research
Ethics Committee of Ningbo First Hospital (Ningbo, China; approval
no. 2019-R019) and performed in accordance with the World Medical
Association Declaration of Helsinki (41). Written informed consent was provided
by all patients prior to the study start.
Patient consent for publication
No applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
circRNA
|
circular RNA
|
|
CRC
|
colorectal cancer
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
siRNA
|
small interfering RNA
|
|
CCK-8
|
Cell Counting Kit-8
|
|
PBS
|
phosphate buffered saline
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
MMP9
|
matrix metalloproteinase 9
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Sun K, Zheng R, Zeng H, Zhang S,
Xia C, Yang Z, Li H, Zou X and He J: Cancer incidence and mortality
in China, 2014. Chin J Cancer Res. 30:1–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray C, Bell LN, Liang H, Collins D and
Yale SH: Colorectal Cancer Screening. WMJ. 116:27–33.
2017.PubMed/NCBI
|
|
4
|
Liang TJ, Wang HX, Zheng YY, Cao YQ, Wu X,
Zhou X and Dong SX: APC hypermethylation for early diagnosis of
colorectal cancer: A meta-analysis and literature review.
Oncotarget. 8:46468–46479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH and
Munschauer M: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ebbesen KK, Kjems J and Hansen TB:
Circular RNAs: Identification, biogenesis and function. Biochim
Biophys Acta. 1859:163–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang F, Nazarali AJ and Ji S: Circular
RNAs as potential biomarkers for cancer diagnosis and therapy. Am J
Cancer Res. 6:1167–1176. 2016.PubMed/NCBI
|
|
13
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weng W, Wei Q, Toden S, Yoshida K,
Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y and Goel A: Circular RNA
ciRS-7-A promising prognostic biomarker and a potential therapeutic
target in colorectal cancer. Clin Cancer Res. 23:3918–3928. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akhter R: Circular RNA and Alzheimer's
disease. Adv Exp Med Biol. 1087:239–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luan W, Shi Y, Zhou Z, Xia Y and Wang J:
circRNA_0084043 promote malignant melanoma progression via
miR-153-3p/Snail axis. Biochem Biophys Res Commun. 502:22–29. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou ZB, Huang GX, Fu Q, Han B, Lu JJ,
Chen AM and Zhu L: circRNA.33186 Contributes to the Pathogenesis of
Osteoarthritis by Sponging miR-127-5p. Mol Ther. 27:531–541. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bi W, Huang J, Nie C, Liu B, He G, Han J,
Pang R, Ding Z, Xu J and Zhang J: CircRNA circRNA_102171 promotes
papillary thyroid cancer progression through modulating
CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin
Cancer Res. 37:2752018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li
D and Zhang L: Increased expression of circRNA_102231 in lung
cancer and its clinical significance. Biomed Pharmacother.
102:639–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu
Z, Ye G, Zhang X, Xiao B and Guo J: Gastric juice long noncoding
RNA used as a tumor marker for screening gastric cancer. Cancer.
120:3320–3328. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen R, Wang Y, Wang CX, Yin M, Liu HL,
Chen JP, Han JQ and Wang WB: MiRNA-155 mediates TAM resistance by
modulating SOCS6-STAT3 signalling pathway in breast cancer. Am J
Transl Res. 7:2115–2126. 2015.PubMed/NCBI
|
|
28
|
Song H, Sun W, Ye G, Ding X, Liu Z, Zhang
S, Xia T, Xiao B, Xi Y and Guo J: Long non-coding RNA expression
profile in human gastric cancer and its clinical significances. J
Transl Med. 11:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Steele SR, Chang GJ, Hendren S, Weiser M,
Irani J, Buie WD, Rafferty JF; Clinical Practice Guidelines
Committee of the American Society of Colon, ; Rectal Surgeons:
Practice Guideline for the surveillance of patients after curative
treatment of colon and rectal cancer. Dis Colon Rectum. 58:713–725.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sargent D, Sobrero A, Grothey A, O'Connell
MJ, Buyse M, Andre T, Zheng Y, Green E, Labianca R, O'Callaghan C,
et al: Evidence for cure by adjuvant therapy in colon cancer:
Observations based on individual patient data from 20,898 patients
on 18 randomized trials. J Clin Oncol. 27:872–877. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun
S, Li J, Sun Y and Qin J: Hsa_circRNA_103809 regulated the cell
proliferation and migration in colorectal cancer via
miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 505:346–352.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL and
Yang Y: RNA sequencing reveals the expression profiles of circRNA
and indicates that circDDX17 acts as a tumor suppressor in
colorectal cancer. J Exp Clin Cancer Res. 37:3252018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yuan Y, Liu W, Zhang Y, Zhang Y and Sun S:
CircRNA circ_0026344 as a prognostic biomarker suppresses
colorectal cancer progression via microRNA-21 and microRNA-31.
Biochem Biophys Res Commun. 503:870–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: CircNT5E acts as a sponge of miR-422a to
promote glioblastoma tumorigenesis. Cancer Res. 78:4812–4825. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
World Medical Association: World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|