Introduction
Glioma is the most common type of primary brain
tumor that originates from glial cells, with an overall incidence
of ~5/100,000 individuals per year worldwide (1,2).
According to the World Health Organization (WHO) grading criteria,
gliomas are categorized into low-grade gliomas (grade I and II) and
high-grade gliomas (grade III and IV) (1). Glioma exhibits several malignant
characteristics, such as strong invasiveness and a high recurrence
rate (3,4). Despite considerable advances in
surgery, radiotherapy and chemotherapy, the median survival time of
patients with glioma has only marginally improved (5). In particular, grade IV glioma, also
known as glioblastoma, is the most aggressive form of glioma with a
median survival of 12–15 months (6,7).
Therefore, the identification of novel cancer-associated biomarkers
may be beneficial for improving the prognosis of patients with
glioma.
microRNAs (miRNAs/miRs) are a class of highly
conserved small non-coding RNAs that post-transcriptionally
modulate the expression of mRNAs by binding to their
3′-untranslated region (UTR) (8).
Studies have shown that miRNAs play a crucial role in various
biological processes, including cell proliferation,
differentiation, apoptosis and migration (9,10).
Aberrant miRNA expression is involved in the progression of various
diseases, including cancer (11,12). To
date, several miRNAs have been identified to function as oncogenes
or suppressor genes in cancer development, such as miR-21 in
colorectal cancer (13) and miR-338
in glioma (14), which suggests that
they are potential therapeutic targets and prognostic biomarkers
(15). Abnormal miR-665 expression
has been reported in multiple human diseases, including cancer
(16–18). For example, a recent study identified
several glioma-associated miRNAs, including miR-665 (19). To the best of our knowledge, the
clinical significance and functional role of miR-665 in glioma have
not been investigated. High mobility group box 1 (HMGB1) is
upregulated in tumor tissues or cancer cell lines and contributes
to the development of a variety of cancer types, such as gastric
and colorectal cancer and hepatocellular carcinoma (20–22).
Previous studies have demonstrated an association between HMGB1 and
the Wnt/β-catenin pathway in esophageal squamous cell carcinoma
(23) and colorectal carcinoma
(24). Accordingly, it was
hypothesized the miR-665/HMGB1 axis may be involved in the
progression of glioma through regulating the Wnt/β-catenin
pathway.
The present study aimed to examine the expression of
miR-665 in glioma using reverse transcription quantitative PCR
(RT-q)PCR and explore its clinical significance using Kaplan-Meier
method. The functional role of miR-665 was also investigated using
cellular experiments in vitro, demonstrating the role of the
miR-665/HMGB1 axis in regulating the progression of glioma through
the Wnt/β-catenin pathway.
Materials and methods
Patients and tissue specimens
The present study was approved by The Ethics
Committee of Affiliated Hospital of Zunyi Medical University
(approval no. SYXK Qin 2018-0006), and written informed consent was
obtained from the patients with glioma. Paired fresh glioma tissue
specimens and adjacent non-cancerous tissue specimens (3-cm away
from glioma) were obtained from a total of 113 glioma patients who
underwent resection at Affiliated Hospital of Zunyi Medical
University between March 2010 and May 2014. Inclusion criteria were
as follows: i) patients had not received anticancer treatment
before surgical resection; ii) complete clinicopathological
characteristic information; and iii) patients had no other
diseases. All tissue specimens were made anonymous, confirmed by
experienced pathologists independently and immediately snap-frozen
and stored in liquid nitrogen until RNA extraction. Detailed
clinicopathological characteristics of all the patients, including
age, sex, tumor size, Karnofsky performance scale (KPS) and WHO
grade (25,26), were obtained. For prognostic
analyses, the 5-year overall survival information of patients was
collected after surgery and recorded by telephone every 3 months
during the first 2 years, every 6 months during the next 2 years
and one year thereafter until death events occurred or the cut-off
follow-up date. In the clinical analysis, glioma patients were
divided into low miR-665 expression group and high miR-665
expression group according to the mean miR-665 expression level
(0.2168) in glioma tissues.
Cell culture and transfection
The human glioma cell lines (U251MG, A172, LN18 and
T98G) were purchased from The Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences. All glioma cells were incubated
in DMEM (Gibco; Thermo Scientific, Inc.) containing 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.). Normal human astrocytes (NHAs;
cat. no. CC-2565) were purchased from Lonza Group, Ltd. and
cultured in astrocyte growth media (Lonza Group, Ltd.) supplemented
with 10% FBS (Thermo Fisher Scientific, Inc.). All cells were
incubated at 37°C in a humidified atmosphere containing 5%
CO2.
The miR-665 mimic (5′-ACCAGGAGGCUGAGGCCCCU-3′),
miR-665 inhibitor (5′-AGGGGCCUCAGCCUCCUGGU-3′), mimic non-targeting
negative control (NC; 5′-UUCUCCGAACGUGUCACGUTT-3′), inhibitor
non-targeting NC (5′-CAGUACUUUUGUGUAGUACAA-3′), pcDNA3.1 (negative
control for pcDNA3.1-HMGB1) and pcDNA3.1-HMGB1 were
synthesized by Shanghai GenePharma Co., Ltd. For cell transfection,
2×105 glioma cells were seeded into 6-well plates, and
miR-665 mimic, mimic NC, miR-665 inhibitor, inhibitor NC, pcDNA3.1
or pcDNA3.1-HMGB1 were transfected into A172 and T98G cells
using Lipofectamine® 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocols.
After transfection for 48 h, the transfection efficiency was
confirmed by RT-qPCR and transfected cells were collected for
further analysis.
RNA extraction and reverse
transcription quantitative PCR (RT-q)PCR
Total RNA was extracted from glioma tissues and
normal cells and glioma cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. Afterwards, the RNA was reverse
transcribed to cDNA using the PrimeScript™ RT Reagent kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
The miR-665 reverse transcription primer was
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGGGC-3′. Then, qPCR was
performed using the SYBR-Green PCR kit (Thermo Fisher Scientific,
Inc.) on an ABI 7900 thermocycler (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The thermocycling conditions for RT-qPCR
were as follows: Initial denaturation at 95°C for 3 min, followed
by 35 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30
sec. The primers used were as follows: miR-665, forward:
5′-GCCGAGACCAGGAGGCTGAG-3′ and reverse: 5′-CTCAACTGGTGTCGTGGA-3′;
U6, forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse:
5′-AACGCTTCACGAATTTGCGT-3′; HMGB1, forward:
5′-TCCAGCAGTTTGTTGGGGTT-3′ and reverse: 5′-AACAAGGGCCTAAGCGAGTC-3′;
β-actin, forward: 5′-AAGACGTACTCAGGCCATGTCC-3′ and reverse:
5′-GACCCAAATGTCGCAGTCAG-3′. Relative miR-665 and HMGB1 mRNA
expression levels were analyzed using the 2−ΔΔCq method
(27) and normalized to U6 and
β-actin, respectively.
Cell proliferation assay
The cell proliferation rate was analyzed using a
Cell Counting Kit (CCK)-8 (Beyotime Institute of Biotechnology). A
total of 2×103 transfected cells/well were seeded into
96-well plates and incubated for 0, 24, 48 and 72 h at 37°C before
the addition of 10 µl CCK-8 reagent. Cells were incubated for a
further 2 h, and optical density of each well was measured at a
wavelength of 450 nm using a microplate reader (Bio-Rad
Laboratories, Inc.).
Cell migration and invasion assay
The migration and invasion capabilities of glioma
cells were evaluated using Transwell inserts (8-µm pores; BD
Biosciences). In the invasion assays, the membranes were pre-coated
with Matrigel (BD Biosciences) at 37°C for 1 h, while a
non-Matrigel insert was used for the migration assays. After
transfection, 5×104 transfected cells were re-suspended
in FBS-free medium and placed into the upper chambers, while the
medium with 10% FBS was added into the lower chambers. After 24 h,
migratory and invasive cells in the lower membrane were fixed with
4% paraformaldehyde at room temperature for 20 min and stained with
0.1% crystal violet at 37°C for 15 min. Stained cells were counted
from five random fields under a light microscope (magnification,
×200; Olympus Corporation).
Bioinformatics analysis and
Dual-luciferase reporter assay
Bioinformatics analyses were performed using
TargetScan (www.targetscan.org) and miRWalk
(mirwalk.umm.uni-heidelberg.de/) to predict the
potential target genes of miR-665. The predicted wild-type (wt) or
mutant (mut) 3′-UTR of HMGB1 was amplified and cloned into a
pmirGLO plasmid (Promega Corporation), generating the plasmids
HMGB1 3′-UTR-wt and HMGB1 3′-UTR-mut, respectively.
Then, the generated vectors were cotransfected with miR-665 mimic,
mimic NC, miR-665 inhibitor or inhibitor NC into A172 cells using
Lipofectamine® 3000 reagent as aforementioned. After 48
h of transfection, the firefly luciferase activity was determined
using the Dual-Luciferase Reporter Assay system (Promega
Corporation) by normalizing to Renilla luciferase
activity.
Western blotting
Transfected A172 cells were cultured and prepared in
6-well plates overnight. The cells were then harvested and washed
twice with PBS and lysed with RIPA lysis buffer (Beyotime Institute
of Biotechnology) to extract total protein. The protein
concentration was determined using a BCA protein assay kit
(Beyotime Institute of Biotechnology). For western blot analysis,
proteins were separated via 10% SDS-PAGE and then blotted onto
nitrocellulose membranes. The membranes were blocked in 5% milk for
1 h at room temperature and were then incubated with primary
antibodies against HMGB1 (cat. no. ab18256; 1:1,000
dilution), β-catenin (cat. no. ab32572; 1:1,000 dilution), cyclin
D1 (cat. no. ab16663; 1:2,000 dilution), C-myc (cat. no. ab32072;
1:1,000 dilution) (all from Abcam) and β-actin (cat. no. sc-47778;
1:1,000 dilution; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
After that, the membranes were incubated with corresponding
secondary antibody goat anti-rabbit HRP (cat. no. ab205718; 1:5,000
dilution; Abcam) or goat anti-mouse HRP (cat. no. ab6789; 1:5,000
dilution; Abcam) for 1 h at room temperature. The signals were
visualized using an electrochemiluminescence kit (GE Healthcare)
and protein levels were quantified by densitometry using ImageJ
version (1.42 software; National Institutes of Health).
Statistical analysis
All experiments were repeated at least three times
and data are presented as the mean ± SD, unless otherwise shown.
The statistical analyses were performed using SPSS 20.0 (IMB Corp.)
and GraphPad Prism 5 software (GraphPad Software Inc.).
χ2 tests were used to analyze the association between
miR-665 expression and clinical characteristics of patients with
glioma. Paired Student's t-tests were used to compare differences
between tumor and adjacent non-tumor tissues and one-way ANOVA
followed by a Tukey's post hoc test was performed to analyze the
difference among three or more groups. Kaplan-Meier curves with
log-rank tests and Cox regression analysis were performed to
investigate the prognostic significance of miR-665. P<0.05 was
considered to indicate a statistically significant difference.
Results
Decreased miR-665 expression in glioma
tissues and cells
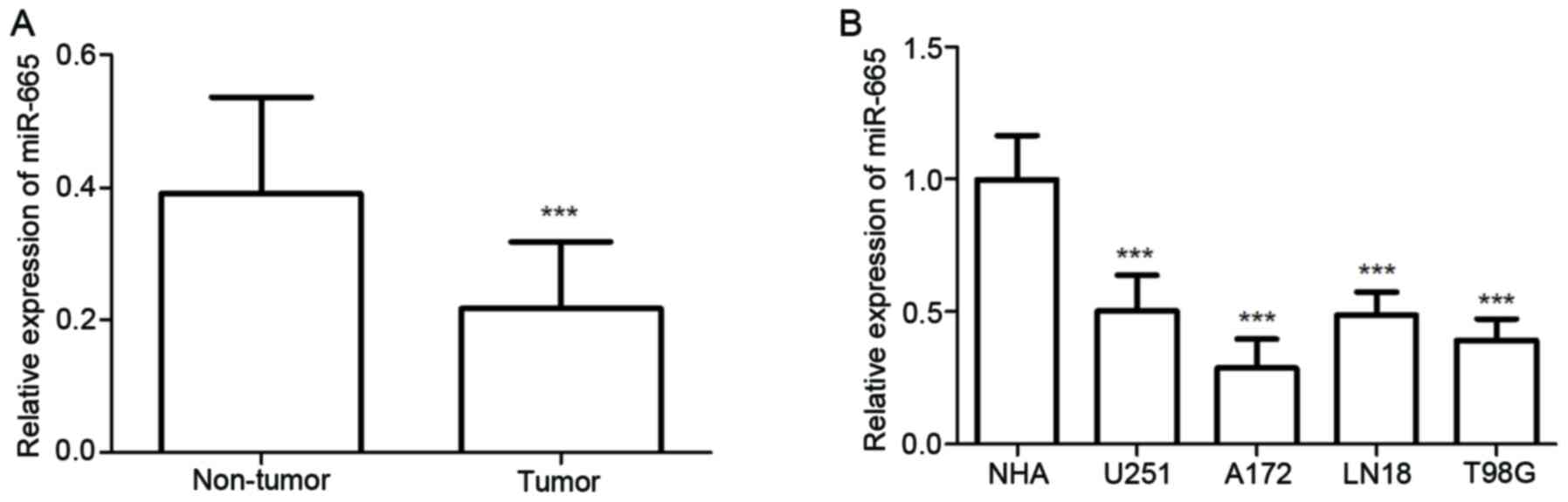
The expression of miR-665 in both glioma tissues and
cell lines was detected using RT-qPCR. The results showed glioma
tissues exhibited significantly decreased relative miR-665
expression compared with normal tissues (P<0.001; Fig. 1A). As demonstrated in Fig. 1B, miR-665 expression was
significantly decreased in glioma cells, including U251MG, A172,
LN18 and T98G cells, compared with expression in NHA cells (all
P<0.001). These results suggested that miR-665 may be a tumor
suppressor in glioma. Among these glioma cells, A172 and T98G
exhibited the lowest miR-665 expression levels, therefore these
cell lines were selected for further experiments.
Association between miR-665 expression
and clinicopathological characteristics and prognosis of patients
with glioma
Patients with glioma were classified into low (n=58)
and high (n=55) miR-665 expression groups based on the mean miR-665
expression level in glioma tissues. The present study demonstrated
that low miR-665 expression was significantly associated with KPS
(P=0.031) and WHO grade (P=0.006) in patients with glioma; however,
no significant association was observed between miR-665 expression
and other parameters, including age, sex and tumor size (Table I). These results suggested that
miR-665 expression may be involved in the development of
glioma.
 | Table I.Association of high (n=55) and low
(n=58) microRNA-665 with the clinicopathological features of 113
patients with glioma. |
Table I.
Association of high (n=55) and low
(n=58) microRNA-665 with the clinicopathological features of 113
patients with glioma.
|
|
| miR-665 expression,
n |
|
|---|
|
|
|
|
|
|---|
| Features | No. of cases | Low | High | P-value |
|---|
| Age, years |
|
|
| 0.383 |
|
≤50 | 52 | 29 | 23 |
|
|
>50 | 61 | 29 | 32 |
|
| Sex |
|
|
| 0.482 |
|
Female | 49 | 27 | 22 |
|
|
Male | 64 | 31 | 33 |
|
| Tumor size, cm |
|
|
| 0.636 |
|
≤5.0 | 57 | 28 | 29 |
|
|
>5.0 | 56 | 30 | 26 |
|
| KPS |
|
|
| 0.031 |
|
≤80 | 57 | 35 | 22 |
|
|
>80 | 56 | 23 | 33 |
|
| WHO grade |
|
|
| 0.006 |
|
I–II | 59 | 23 | 36 |
|
|
III–IV | 54 | 35 | 19 |
|
Furthermore, a Kaplan-Meier curve was constructed to
analyze 5-year follow-up data (the average follow-up time was 39.36
months), which suggested that patients with a low expression level
of miR-665 had a significantly shorter overall survival time
compared with the high expression group (P=0.008; Fig. 2). Furthermore, multivariate Cox
regression analysis revealed that the expression level of miR-665
and WHO grade were independent indicators for overall survival in
patients with glioma (P<0.05; Table
II). Overall, these results indicated that decreased miR-665
may be a prognostic biomarker in glioma.
 | Table II.Multivariate Cox regression analysis
for clinicopathological variables of patients with glioma. |
Table II.
Multivariate Cox regression analysis
for clinicopathological variables of patients with glioma.
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | HR | 95% CI | P-value |
|---|
| miR-665, low vs.
high expression | 1.878 | 1.058-3.333 | 0.031 |
| Age, ≤50 vs. >50
years | 0.801 | 0.452-1.418 | 0.447 |
| Sex, female vs.
male | 0.726 | 0.358-1.472 | 0.375 |
| Tumor size, >5.0
vs. ≤5.0 cm | 1.927 | 0.934-3.975 | 0.076 |
| KPS, >80 vs. ≤
80 | 0.564 | 0.317-1.006 | 0.052 |
| WHO grade, III–IV
vs. I–II | 1.784 | 1.013-3.144 | 0.045 |
miR-665 suppresses proliferation,
migration and invasion of glioma cells
To further investigate the functional relevance of
miR-665 in glioma, A172 and T98G cells were transfected with
miR-665 mimic, mimic NC, miR-665 inhibitor or inhibitor NC and were
analyzed using RT-qPCR. The relative expression of miR-665 was
significantly increased in glioma cells following transfection with
miR-665 mimic, and was significantly decreased following
transfection with the miR-665 inhibitor, compared with untreated
cells (P<0.001; Fig. 3A). The
mimic NC and inhibitor NC group were not significantly different
compared with the untreated group, therefore untreated cells were
used as control comparisons. Cell proliferation was examined using
CCK-8 assays, which revealed that overexpression of miR-665
significantly inhibited glioma cell proliferation, whereas
knockdown of miR-665 significantly promoted cell proliferation,
compared with untreated cells (P<0.05; Fig. 3B). Transwell migration and invasion
assays revealed that upregulation of miR-665 significantly
suppressed the migration and invasion of glioma cells, while
inhibition of miR-665 attenuated these abilities, compared with
untreated cells (P<0.01 or P<0.001; Fig. 4A-D). These results indicated that
miR-665 may exert an tumor suppressive role in glioma through
suppressing tumor cell proliferation, migration, and invasion.
miR-665 directly targets HMGB1 in A172
cells
According to the bioinformatics prediction, the
3′-UTR region of the HMGB1 gene was complementary with the seed
sequences of miR-665 (Fig. 5A).
Firstly, HMGB1 mRNA and protein expression levels were quantified
using RT-qPCR and western blotting, respectively. The results
showed that miR-665-overexpression inhibited both HMGB1 mRNA and
protein levels, while knockdown of miR-665 promoted both HMGB1 mRNA
and protein levels in A172 cells, compared with the untreated group
(P<0.001; Fig. 5B and C).
Subsequently, the luciferase reporter assay results revealed that
enhanced expression of miR-665 significantly inhibited the
luciferase activity of the HMGB1 3′-UTR wt, while downregulation of
miR-665 promoted the luciferase activity of the HMGB1 3′-UTR wt,
compared with the untreated group (P<0.05; Fig. 5D). Meanwhile, no significant changes
were observed in the HMGB1 3′-UTR mut. These results revealed that
miR-665 could directly target HMGB1 in A172 cells.
Effects of miR-665 and HMGB1 on the
activity of the Wnt/β-catenin signaling pathway
A previous study reported that miR-665 inhibits
tumorigenesis by directly targeting HMGB1 and inactivating
the Wnt/β-catenin pathway in retinoblastoma (28). Thus, the present study investigated
whether miR-665 and HMGB1 expression could affect the
Wnt/β-catenin signaling pathway in A172 cells. First, the
transfection efficiency was measured using RT-qPCR and the results
are shown in Fig. S1 (P<0.001
vs. untreated cells). The expression levels of the important
molecules in the Wnt/β-catenin signaling pathway, including
β-catenin, cyclin D1 and C-myc (29), were measured in A172 cells with the
β-actin as internal reference following the altered expression of
miR-665 and HMGB1. As presented in Fig.
6, overexpression of miR-665 significantly inhibited the
protein levels of β-catenin, cyclin D1 and C-myc, while recovery of
HMGB1 expression partially abrogated the inhibition of
β-catenin, cyclin D1 and C-myc protein levels caused by miR-665
upregulation, compared with untreated group (P<0.05). These
results suggested that miR-665 deactivated the Wnt/β-catenin
pathway in A172 cells through the negative mediation of
HMGB1.
Discussion
Glioma, a malignant brain tumor, is one of the most
common malignancies with high mortality rate in China, and there
were ~61,000 brain cancer-associated deaths in 2015 (30). To find more effective treatments for
glioma, there is a need to identify valuable cancer-related
biomarkers. In the present study, it was observed that miR-665
expression was significantly decreased in glioma tissues and cell
lines compared with normal tissues and NHAs, and miR-665 expression
was associated with KPS and WHO grade. Low expression levels of
miR-665 was associated with poor prognosis of patients with glioma.
Besides, overexpression of miR-665 significantly inhibited
proliferation, migration and invasion of glioma cells, while
knockdown of miR-665 promoted these cellular behaviors by targeting
HMGB1 through regulating the Wnt/β-catenin pathway.
In recent years, growing evidence has demonstrated
that miRNAs function as diagnostic and/or prognostic biomarkers in
certain patients with cancer, such as colon cancer and
hepatocellular carcinoma (31,32). In
glioma, studies have indicated aberrantly expressed miRNAs are
correlated with clinical progression and patient survival, such as
miR-3653 (33), miR-362-3p (34) and miR-526b-3p (35). The present study focused on the
clinical significance and function of miR-665.
In the present study, miR-665 was significantly
downregulated in glioma tissues compared with normal tissues, as
well as in glioma cells compared with NHAs. Besides, downregulation
of miR-665 was associated with KPS and WHO grade, which suggested
that miR-665 acts as a negative regulator in the progression of
glioma. Furthermore, the Kaplan-Meier survival analyses results
showed that patients with low expression levels of miR-665 had a
shorter overall survival time compared with those with high
expression of miR-665. Of note, the multivariate analysis confirmed
that miR-665 was an independent predictor of poor prognosis,
suggesting that miR-665 may be a novel prognostic biomarker for
patients with glioma.
Recent studies have shown that miRNA can exert an
oncogenic or suppressor role in human cancer (36,37). For
instance, miR-6852 expression is downregulated in glioma and
suppresses cell proliferation, migration and invasion by targeting
lymphoid enhancer-binding factor 1 (38). miR-454-3p exerts tumor-suppressive
functions in glioblastoma, inhibiting cell proliferation and
inducing apoptosis by targeting nuclear factor of activated
T-cells, cytoplasmic 2 (39).
Several studies have reported that miR-mediates disease progression
in various types of cancer. For instance, Zhao et al
(40) demonstrated that miR-665
expression is upregulated and associated with poor survival in
patients with breast cancer, and promotes the progression of breast
cancer by targeting nuclear receptor subfamily 4 group A member 3.
Hu et al (41) reported that
miR-665 expression is increased in hepatocellular carcinoma tissues
and cells, and promotes tumor proliferation, migration and
invasion, suggesting that miR-665 is a potential target for
hepatocellular carcinoma treatment. On the other hand, the
suppressor effects of miR-665 have also been observed in several
other types of cancer, such as retinoblastoma (28), cervical (42) and ovarian cancer (43). In ovarian cancer, miR-665 suppresses
the proliferation and migration of ovarian cancer cells by
targeting homeobox protein Hox-A10, which may act as a
tumor-suppressing gene (43). These
studies suggest that miR-665 may exert an oncogenic or suppressor
role in cancer, depending on the cancer type.
In the present study, overexpression of miR-665
inhibited cell proliferation, migration and invasion of glioma
cells, while the opposite effects were observed following knockdown
of miR-665. Taken together, the data suggested that miR-665
functioned as a tumor suppressor in glioma, which was consistent
with the effects of miR-665 in previous studies (28,42,43). In
retinoblastoma, miR-665 is markedly reduced and has as a
tumor-suppressive role by targeting HMGB1, suggesting that miR-665
is a candidate prognostic biomarker and therapeutic target for
patients with retinoblastoma (28).
The clinical role and oncogenic function role of HMGB1 have been
reported in non-small cell lung cancer (44) and gastric cancer (45,46). In
glioma, HMGB1 mediates tumor progression through promoting tumor
cell proliferation and the invasion of glioma cells (47,48). In
the present study, HMGB1 was predicted as a direct target of
miR-665. Furthermore, the present study also focused on the effects
of miR-665 and HMGB1 on the activity of the Wnt/β-catenin pathway,
which contributed to tumor progression. The results revealed that
miR-665 inactivated the Wnt/β-catenin signaling pathway by
targeting HMGB1 in A172 cells.
However, there are still some limitiations in the
present study. The design and fieldwork was conducted before
publication of the study, thus, the primary limitiation may be the
limited sample size. Besides, there are lack of some experimental
studies, such as in vivo studies. In the future studies,
more experimental verfication with large sample sizes are necessary
to understand the molecular mechanism of miR-665 in glioma.
Taken together, the present study demonstrated that
miR-665 was downregulated in glioma tissues and cell lines, and
predicted poor prognosis of patients with glioma. Besides, miR-665
inhibited the proliferation, migration and invasion of glioma cells
by targeting HMGB1 and thereby inactivating the Wnt/β-catenin
signaling pathway. The present study suggested that miR-665 may be
a novel prognostic biomarker and the miR-665/HMGB1 axis may provide
a potential therapeutic target for glioma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HS and MX conceived and designed the experiments and
contributed significantly to the analysis and manuscript
preparation. HS, LX, HT and CY performed the experiments. HS, LX
and HW performed the statistical analysis. HW and YZ the collected
data and interpreted the results. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Affiliated Hospital of Zunyi Medical University
(approval no. SYXK Qin 2018-0006), and written informed consent was
obtained from the patients with glioma.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hummel S, Kohlmann W, Kollmeyer TM,
Jenkins R, Sonnen J, Palmer CA, Colman H, Abbott D, Cannon-Albright
L and Cohen AL: The contribution of the rs55705857 G allele to
familial cancer risk as estimated in the Utah population database.
BMC Cancer. 19:1902019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iser IC, Pereira MB, Lenz G and Wink MR:
The epithelial-to-mesenchymal transition-like process in
glioblastoma: An updated systematic review and in silico
investigation. Med Res Rev. 37:271–313. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang
X, Jiang C, Kang C, Li X, Chen L, et al: CGCG clinical practice
guidelines for the management of adult diffuse gliomas. Cancer
Lett. 375:263–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Champ CE, Palmer JD, Volek JS,
Werner-Wasik M, Andrews DW, Evans JJ, Glass J, Kim L and Shi W:
Targeting metabolism with a ketogenic diet during the treatment of
glioblastoma multiforme. J Neurooncol. 117:125–131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Linz U: Commentary on Effects of
radiotherapy with concomitant and adjuvant temozolomide versus
radiotherapy alone on survival in glioblastoma in a randomised
phase III study: 5-year analysis of the EORTC-NCIC trial (Lancet
Oncol. 2009;10:459-466). Cancer. 116:1844–1846. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arvold ND and Reardon DA: Treatment
options and outcomes for glioblastoma in the elderly patient. Clin
Interv Aging. 9:357–367. 2014.PubMed/NCBI
|
|
8
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim DY and Sung JH: Regulatory role of
microRNAs in the proliferation and differentiation of
adipose-derived stem cells. Histol Histopathol. 32:1–10.
2017.PubMed/NCBI
|
|
10
|
Mahmoudian-Sani MR, Jalali A, Jamshidi M,
Moridi H, Alghasi A, Shojaeian A and Mobini GR: Long non-coding
RNAs in thyroid cancer: Implications for pathogenesis, diagnosis,
and therapy. Oncol Res Treat. 42:136–142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rivera-Barahona A, Pérez B, Richard E and
Desviat LR: Role of miRNAs in human disease and inborn errors of
metabolism. J Inherit Metab Dis. 40:471–480. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Paliouras AR, Monteverde T and Garofalo M:
Oncogene-induced regulation of microRNA expression: Implications
for cancer initiation, progression and therapy. Cancer Lett.
421:152–160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han
B, Bai Y, Li L, Zhang Y and Zhou L: MicroRNA-21 (Mir-21) Promotes
cell growth and invasion by repressing tumor suppressor PTEN in
colorectal cancer. Cell Physiol Biochem. 43:945–958. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu DZ, Zhao H, Zou QG and Ma QJ: MiR-338
suppresses cell proliferation and invasion by targeting CTBP2 in
glioma. Cancer Biomark. 20:289–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Möhnle P, Schütz SV, Schmidt M, Hinske C,
Hübner M, Heyn J, Beiras-Fernandez A and Kreth S: MicroRNA-665 is
involved in the regulation of the expression of the
cardioprotective cannabinoid receptor CB2 in patients with severe
heart failure. Biochem Biophys Res Commun. 451:516–521. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tan H, Zhao L, Song R, Liu Y and Wang L:
microRNA-665 promotes the proliferation and matrix degradation of
nucleus pulposus through targeting GDF5 in intervertebral disc
degeneration. J Cell Biochem. 119:7218–7225. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu NW, Huang X, Liu S and Lu Y: EXT1,
regulated by MiR-665, promotes cell apoptosis via ERK1/2 signaling
pathway in acute lymphoblastic leukemia. Med Sci Monit.
25:6491–6503. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ye X, Wei W, Zhang Z, He C, Yang R, Zhang
J, Wu Z, Huang Q and Jiang Q: Identification of microRNAs
associated with glioma diagnosis and prognosis. Oncotarget.
8:26394–26403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tesarova P, Kalousova M, Zima T and Tesar
V: HMGB1, S100 proteins and other RAGE ligands in cancer-markers,
mediators and putative therapeutic targets. Biomed Pap Med Fac Univ
Palacky Olomouc Czech Repub. 160:1–10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Jiang Z, Yan J and Ying S: HMGB1
as a potential biomarker and therapeutic target for malignant
mesothelioma. Dis Markers. 2019:41831572019.PubMed/NCBI
|
|
22
|
Wu T, Zhang W, Yang G, Li H, Chen Q, Song
R and Zhao L: HMGB1 overexpression as a prognostic factor for
survival in cancer: A meta-analysis and systematic review. Dis
Markers. 7:50417–50427. 2016.
|
|
23
|
Zhao Y, Yi J, Tao L, Huang G, Chu X, Song
H and Chen L: Wnt signaling induces radioresistance through
upregulating HMGB1 in esophageal squamous cell carcinoma. Cell
Death Dis. 9:4332018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reed KR, Song F, Young MA, Hassan N,
Antoine DJ, Gemici NB, Clarke AR and Jenkins JR: Secreted HMGB1
from Wnt activated intestinal cells is required to maintain a crypt
progenitor phenotype. Oncotarget. 7:51665–51673. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nagane M, Kobayashi K, Ohnishi A, Shimizu
S and Shiokawa Y: Prognostic significance of O6-methylguanine-DNA
methyltransferase protein expression in patients with recurrent
glioblastoma treated with temozolomide. Jpn J Clin Oncol.
37:897–906. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li C, Zou H, Wang Z, Tang X, Fan X, Zhang
K, Liu J and Li Z: REST, not REST4, is a risk factor associated
with radiotherapy plus chemotherapy efficacy in glioma. Drug Des
Devel Ther. 12:1363–1371. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Du S, Lv Y, Zhang F and Wang W:
MicroRNA-665 inhibits the oncogenicity of retinoblastoma by
directly targeting high-mobility group box 1 and inactivating the
Wnt/β-catenin pathway. Cancer Manag Res. 11:3111–3123. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang H, Li X, Meng W, Zhang L, Zhu X, Bai
Z, Yan J and Zhou W: Overexpression of p16ink4a regulates the
Wnt/beta-catenin signaling pathway in pancreatic cancer cells. Mol
Med Rep. 17:2614–2618. 2018.PubMed/NCBI
|
|
30
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dehghan F, Boozarpour S, Torabizadeh Z and
Alijanpour S: miR-21: A promising biomarker for the early detection
of colon cancer. Onco Targets Ther. 12:5601–5607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ning S, Liu H, Gao B, Wei W, Yang A, Li J
and Zhang L: miR-155, miR-96 and miR-99a as potential diagnostic
and prognostic tools for the clinical management of hepatocellular
carcinoma. Oncol Lett. 18:3381–3387. 2019.PubMed/NCBI
|
|
33
|
Chen Y, Li ZH, Liu X, Liu GX, Yang HM and
Wu PF: Reduced expression of miR-3653 in glioma and its
correlations with clinical progression and patient survival. Eur
Rev Med Pharmacol Sci. 23:6596–6601. 2019.PubMed/NCBI
|
|
34
|
Xu G, Fang P, Chen K, Xu Q, Song Z and
Ouyang Z: MicroRNA-362-3p Targets PAX3 to Inhibit the Development
of Glioma through Mediating Wnt/beta-Catenin Pathway.
Neuroimmunomodulation. 26:119–128. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu M, Li X, Liu Q, Xie Y, Yuan J and
Wanggou S: miR-526b-3p serves as a prognostic factor and regulates
the proliferation, invasion, and migration of glioma through
targeting WEE1. Cancer Manag Res. 11:3099–3110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun X, Cui S, Fu X, Liu C, Wang Z and Liu
Y: MicroRNA-146-5p promotes proliferation, migration and invasion
in lung cancer cells by targeting claudin-12. Cancer Biomark.
25:89–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen S, Shi F, Zhang W, Zhou Y and Huang
J: miR-744-5p inhibits non-small Cell lung cancer proliferation and
invasion by directly targeting PAX2. Technol Cancer Res Treat.
18:15330338198769132019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang J, Liu H, Zheng K, Zhang S and Dong
W: MicroRNA-6852 suppresses glioma A172 cell proliferation and
invasion by targeting LEF1. Exp Ther Med. 18:1877–1883.
2019.PubMed/NCBI
|
|
39
|
Zuo J, Yu H, Xie P, Liu W, Wang K and Ni
H: miR-454-3p exerts tumor-suppressive functions by down-regulation
of NFATc2 in glioblastoma. Gene. 710:233–239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao XG, Hu JY, Tang J, Yi W, Zhang MY,
Deng R, Mai SJ, Weng NQ, Wang RQ, Liu J, et al: miR-665 expression
predicts poor survival and promotes tumor metastasis by targeting
NR4A3 in breast cancer. Cell Death Dis. 10:4792019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu Y, Yang C, Yang S, Cheng F, Rao J and
Wang X: miR-665 promotes hepatocellular carcinoma cell migration,
invasion, and proliferation by decreasing Hippo signaling through
targeting PTPRB. Cell Death Dis. 9:9542018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao L, Jin H, Zheng Y, Mao Y, Fu Z, Li X
and Dong L: DANCR-mediated microRNA-665 regulates proliferation and
metastasis of cervical cancer through the ERK/SMAD pathway. Cancer
Sci. 110:913–925. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu J, Jiang Y, Wan Y, Zhou S, Thapa S and
Cheng W: MicroRNA665 suppresses the growth and migration of ovarian
cancer cells by targeting HOXA10. Mol Med Rep. 18:2661–2668.
2018.PubMed/NCBI
|
|
44
|
Ma Y, Kang S, Wu X, Han B, Jin Z and Guo
Z: Up-regulated HMGB1 in the pleural effusion of non-small cell
lung cancer (NSCLC) patients reduces the chemosensitivity of NSCLC
cells. Tumori. 104:338–343. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y and Qin C: MiR-1179 inhibits the
proliferation of gastric cancer cells by targeting HMGB1. Hum Cell.
32:352–359. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang J, Zhang R, Lu WW, Zhu JS, Xia LQ,
Lu YM and Chen NW: Clinical significance of hmgb1 expression in
human gastric cancer. Int J Immunopathol Pharmacol. 27:543–551.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Angelopoulou E, Piperi C, Adamopoulos C
and Papavassiliou AG: Pivotal role of high-mobility group box 1
(HMGB1) signaling pathways in glioma development and progression. J
Mol Med (Berl). 94:867–874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang J, Liu C and Hou R: Knockdown of
HMGB1 improves apoptosis and suppresses proliferation and invasion
of glioma cells. Chin J Cancer Res. 26:658–668. 2014.PubMed/NCBI
|