Introduction
Multiple myeloma (MM) accounts for ~13% of
hematological cancers, with an estimated 24,280 to 30,330 new cases
and 12,650 deaths in 2016 (1), and
patients survive an average of 3 years (2). Although modern combination therapies
induce rapid, deep and sustainable responses, as well as prolonging
survival (3), MM remains an
incurable disease, and the majority of patients relapses and
requires further treatment (4).
Thus, novel targets for treating MM are urgently required.
Augmenter of liver regeneration (ALR), also known as
GFER, is a vital protein in various biological processes that was
first identified by Hagiya et al (5). ALR occurs in two isoforms: A long form
comprising 205 amino acid residues with a molecular weight of 23
kDa (6) that is present in the
intermembrane space of mitochondria (7), and a short form consisting of 125 amino
acids with a molecular weight of 15 kDa (8) that is secreted by hepatocytes and is
present in serum (9). Accumulating
evidence has revealed that ALR affects fundamental processes such
as energy transduction (10), cell
survival, cell regeneration (11),
metabolic homeostasis, iron metabolism (12) and stem cell maintenance (13). Different isoforms of ALR have been
associated with different subcellular locations and therefore
specific functions (14), but
despite considerable research on the overexpression and inhibition
of 23- and 15-kDa-ALR, their specific functions remain unclear.
A previous study has demonstrated that silencing ALR
can influence proliferation and apoptosis in human MM U266 cells
(15). However, little is known
about the role of 15-kDa-ALR in MM. Therefore, the present study
aimed to investigate the role and mechanism of extracellular
15-kDa-ALR in MM.
Materials and methods
Preparation of anti-ALR monoclonal
antibody
The 15-kDa-recombinant human ALR (rhALR) protein was
purchased from Abcam. A total of 5 female BALB/c mice (age, 6–10
weeks old; weight, 20–25 g) were obtained from the Experimental
Animal Center of Chongqing Medical University (Chongqing, China).
All mice received humane treatment according to the regulations of
the Institutional Animal Care and Use Committee of Chongqing
Medical University. All mice were housed in a specific
pathogen-free laboratory in an acclimatized room at standard room
conditions (25±2° and 55% humidity), with a 12-h light/dark cycle.
Mice were allowed free access to water and standard chow. The rhALR
protein was injected subcutaneously into five BALB/c mice for
immunization. Subsequently, anti-ALR monoclonal antibody (McAb) was
prepared using hybridoma technology as previously described
(16). Splenocytes were harvested
and fused with myeloma SP2/0 cells (obtained from the Institute of
Viral Hepatitis of Chongqing Medical University) to generate
hybridoma cells, which were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 20% FBS (HyClone;
Cytiva) and 100 U/ml penicillin and streptomycin at 37°C in a
humidified atmosphere with 5% CO2. Single
antibody-producing hybridoma cells were isolated using the limiting
dilution technique, and supernatants of growth-positive wells were
aspirated and screened for the presence of antibodies using ELISA.
The ratio of the absorbance of hybridoma cell culture supernatants
and myeloma cell SP2/0 culture supernatants was measured at 450 nm.
Absorbance >1.5 at optical density (OD)450 nm and ratio of the
absorbance of hybridoma cell culture supernatants to that of
negative controls >2.1 were considered positive. Single
McAb-producing hybridoma cells were isolated and were injected
intraperitoneally into ten BALB/c mice to produce ascites for
large-scale McAb production. All mice were anesthetized using 3%
isoflurane and sacrificed by cervical dislocation 10–14 days after
injection. Ascitic fluid containing McAb was harvested from the
intraperitoneal cavity using a 21-G needle and centrifuged at
10,000 × g at 4°C for 10 min. Isotypes of McAb were detected using
IsoQuick strips (Roche Molecular Diagnostics). Negative hybridoma
cells were also prepared as negative control ascites. HiTrap™ IgM
Purification HP (GE Healthcare Life Sciences) was used to purify
the monoclonal IgM according to the manufacturer's protocol.
ELISA
For ELISA, 96-well plates were coated with
15-kDa-rhALR (4 µg/ml) in 100 µl coating buffer overnight at 4°C
and incubated with blocking solution, consisting of 3% BSA (Beijing
Solarbio Science & Technology Co., Ltd.) in 0.05% Tween-20-PBS
(PBST) solution, at 4°C overnight. Ascites were diluted at the
ratio of 1:100, 1:1,000, 1:10,000, 1:100,000 and 1:1,000,000. After
washing with PBST, 100 µl ascites diluted at different ratios were
added and incubated at 4°C overnight. After incubation, the plates
were washed three times with PBST, and 100 µl 1:10,000-diluted
HRP-conjugated goat anti-mouse immunoglobulin (cat. no. 7076P2;
Cell Signaling Technology, Inc.) was added to each well and
incubated for 1 h at room temperature. After washing three times
with PBST, the enzymatic activity was determined by adding 100 µl
3,3′,5,5′-tetramethylbenzidine substrate solution and incubating in
the dark at 37°C for 30 min, followed by adding 5 µl 2M sulfuric
acid per well to terminate the reaction. The absorbance was
recorded at 450 nm using a microplate reader. For detecting ALR
protein in the medium, U266, RPMI8226 and MM1.S cells were cultured
in serum-free medium at 37°C for 48 h, and 96-well plates were
coated with cell medium and serial dilutions of rhALR protein
(2,000, 1,000, 500, 250, 125, 62.5, 31.25 and 0 ng/ml). The
quantity of protein was calculated by the standard curve.
Cell culture and treatment
The human MM U266, RPMI8226 and MM1.S cell lines
were obtained from the Institute of Viral Hepatitis of Chongqing
Medical University. U266 and MM1.S cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (HyClone; Cytiva) and 100 U/ml penicillin and streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.). RPMI8226 cells were
cultured in Iscove's modified Dulbecco's medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 20% FBS and 100 U/ml
penicillin and streptomycin. Cells were grown at 37°C in a
humidified atmosphere with 5% CO2. Ascites containing
anti-ALR McAb were added to the medium as the treatment group,
while ascites produced by negative hybridoma cells constituted the
negative control group, and PBS was used for the blank group.
Cell viability assay
Different dilutions of ascites containing anti-ALR
McAb (1:10, 1:50 and 1:100), negative ascites and PBS were
incubated with U266 cells for different time periods (24, 48 and 72
h). A Cell Counting Kit-8 (CCK-8; Beijing Solarbio Science &
Technology Co., Ltd.) assay was used to determine the cell survival
rate according to the manufacturer's protocol. Cells were plated in
96-well plates at a density of 1×104 cells/well. After
culturing at 37°C for 24, 48 or 72 h, the quantity of viable cells
was measured. A total of 10 µl CCK-8 reagent was added to each well
and incubated at 37°C for 2 h. The absorbance was recorded at 450
nm using a microplate reader, and the following formula was used to
calculate cell viability: Cell viability (%) = [A (ascites) - A
(Blank)] / [A (0 dosing) - A (blank)] × 100.
Apoptosis analysis via flow
cytometry
The apoptosis rates of human MM cells treated with
anti-ALR McAb, negative ascites and PBS were determined using an
Annexin V-FITC/PI staining kit (BD Biosciences). Cells were washed
twice with cold PBS and resuspended in 1X binding buffer at a cell
density of 1×106 cells/ml. Cells were incubated with 5
µl FITC and 10 µl PI for 15 min at room temperature in the dark. A
total of 1×104 cells were recorded by the FACSCanto II
flow cytometer (BD Biosciences) for each sample. Fluorescence data
were measured and analysed using FACSCanto II software (BD
Biosciences). Apoptosis detection in the presence of injury factors
was the same as aforementioned. Ascites containing anti-ALR McAb,
negative ascites and PBS were incubated with U266 cells at the
concentration of 1:10 for 48 h. Subsequently, 0.004 mg/ml
epirubicin (Pfizer, Inc.) was added to the medium for 24 h at 37°C.
Cells were harvested for apoptosis analysis. A cell cycle and
apoptosis analysis kit (Beijing Solarbio Science & Technology
Co., Ltd.) was used to determine the cell cycle according to the
manufacturer's protocol. U266 cells were treated with anti-ALR
McAb, negative ascites and PBS at a concentration of 1:10. After
culturing at 37°C for 72 h, cells were harvested and washed with
PBS, then fixed with 70% ethanol at 4°C for 2 h. A total of 100 µl
RNase A was added for 30 min at 37°C, then PI (0.5 ml) was added
and incubated for 10 min at 4°C in the dark for staining. Next,
cells were assessed using a FACSCanto II flow cytometer (BD
Biosciences). Cell cycle was analyzed using ModFit LT software
(version 3.2; Verity Software House, Inc.).
Western blotting
Proteins were extracted from cells using RIPA lysis
buffer (Beyotime Institute of Biotechnology) and quantified using a
BCA kit (Beyotime Institute of Biotechnology). Proteins (60
µg/lane) were separated via 15% SDS-PAGE, transferred to
polyvinylidene fluoride membranes, blocked with 5% skimmed milk in
TBS for 1 h at 37° and then incubated overnight at 4°C with primary
antibodies. The primary antibodies and dilutions used were as
follows: Anti-Bax (1:1,000; cat. no. 2774S), anti-Bcl-2 (1:1,000;
cat. no. 4223T), anti-caspase-3 (1:1,000; cat. no. 9662S),
anti-cleaved caspase-3 (1:1,000; cat. no. 9661T), anti-p44/42
(1:1,000; cat. no. 4695T), anti-phospho-p44/42 (1:1,000; cat. no.
4377T), anti-p38 (1:1,000; cat. no. 8690T), anti-phospho-p38
(1:1,000; cat. no. 4511T), anti-JNK (1:1,000; cat. no. 9252T),
anti-phospho-JNK (1:1,000; cat. no. 9251S), anti-STAT3 (1:1,000;
cat. no. 4904T), anti-phospho-STAT3 (1:1,000; cat. no. 9145T) and
anti-β-actin (1:1,000; cat. no. 4970T), all from Cell Signaling
Technology, Inc. Anti-β-tubulin (1:1,000; cat. no. 102053-T34) was
purchased from Sino Biological, Inc. Anti-CDK1 (1:1,000; cat. no.
ab201008) and anti-cyclin D1 (1:1,000; cat. no. ab40754) were
obtained from Abcam. After incubation for 1 h at 37°C with
HRP-conjugated secondary antibodies (1:10,000; cat. no. 7074P2;
Cell Signaling Technology, Inc.), the protein bands were visualized
using an enhanced chemiluminescence kit (Nanjing KeyGen Biotech
Co., Ltd.) to detect immunoreactive bands with a ChemiDoc Imaging
System (Bio-Rad Laboratories, Inc). Densitometry was analyzed using
Image lab™ V3.0 software (Bio-Rad Laboratories, Inc).
EdU assay
Cell proliferation was measured using an EdU assay
kit (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's
protocol. U266 cells were cultured at a density of 1×105
cells/well in 6-well plates and treated with 1:10 concentration of
anti-ALR McAb ascites, negative ascites or PBS at 37°C for 72 h. A
total of 50 µM EdU was added to the medium and incubated for 4 h at
37°C. Cells were harvested and fixed with 4% formaldehyde for 15
min at room temperature, centrifuged at 600 × g for 10 min at room
temperature, and washed with PBS. Subsequently, 2 mg/l glycine
solution was used to neutralise the response, and 0.5% Triton X-100
was added and incubated for 10 min at room temperature for
permeabilisation. The supernatant was discarded after
centrifugation (600 × g for 10 min at room temperature), and cells
were washed again with PBS and treated with 1X Apollo reaction
cocktail (100 µl/well; Guangzhou RiboBio Co., Ltd.) in the dark for
10 min at room temperature. Cells were washed three times with 0.5%
Triton X-100 and resuspended in 500 µl PBS. Next, 1×104
cells from each sample were assessed using a FACSCanto II flow
cytometer (BD Biosciences). Fluorescence data were measured and
analysed using FACSCanto II software (BD Biosciences).
RNA preparation, library construction
and RNA sequencing (RNA-seq)
Total RNA was extracted from U266 cells (n=3;
treatment and control groups) using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA quality was
checked using a Bioanalyzer 2200 instrument (Agilent Technologies,
Inc.), and cDNA libraries were constructed for each pooled RNA
sample using a NEBNext Ultra Directional RNA Library Prep Kit
(Illumina, Inc.) according to the manufacturer's protocol. Tagged
cDNA libraries were pooled in equal ratios and used for 150-bp
paired-end sequencing in a single lane of an Illumina HiSeqXTen
instrument (Illumina, Inc.).
Differentially expressed genes and
bioinformatics analysis
The DESeq algorithm (17) was applied to filter differentially
expressed genes. The results were filtered based on fold-change
(FC) and false discovery rate (FDR) using the following criteria:
log2FC >0.585 or <-0.585, and FDR <0.05. Volcano plots
were drawn using the R software package (ggplot2; version 3.2.1;
http://ggplot2.tidyverse.org/) based on
the analysis of differentially expressed genes, and colour was
determined by the filtering criteria. Gene Ontology (GO) analysis
was performed to predict the biological functions of differentially
expressed genes. GO annotations were downloaded from the National
Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), UniProt (http://www.uniprot.org/) and GO (http://www.geneontology.org/) databases. Fisher's
exact tests were used to identify significant GO categories, and
FDR was used to correct the P-values. FDR <0.05 was considered
to indicate a statistically significant difference. Pathway
analysis was used to identify significant pathways associated with
the differentially expressed genes according to the Kyoto
Encyclopedia of Genes and Genomes database (https://www.kegg.jp/). Fisher's exact tests were used
to select significant pathways, and the threshold of significance
was defined by the P-value and FDR <0.05. Protein-protein
interaction (PPI) network analysis of differentially expressed
genes was performed using the online Search Tool for the Retrieval
of Interacting Genes/Proteins (version 11.0; http://string-db.org/) (18). The top 10 hub genes were identified
using the cytoHubba plugin app of Cytoscape software (version
3.7.2) (19). Path-Act-Network
(https://www.kegg.jp/kegg/pathway.html) was employed to
select genes in enriched biological pathways, and Cytoscape was
used for the graphical representation of pathways.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from U266 cells using a
Total RNA Extraction kit (BioTeke Corporation) according to the
manufacturer's protocol. Total RNA was reverse transcribed to cDNA
using a PrimeScript II Reverse Transcriptase kit (Takara Bio, Inc.)
according to the manufacturer's protocol. RT-qPCR was performed
using SYBR Premix (Takara Bio, Inc.). For quantitative analysis,
hub genes, along with GAPDH as a positive control, were amplified
using SYBR Premix on a CFX Connect Real-Time System (Bio-Rad
Laboratories, Inc.). The qPCR thermocycling conditions were as
follows: One cycle at 94°C for 30 sec, followed by 40 cycles of 5
sec at 94°C for denaturation and 60°C for 30 sec for
annealing/extension using the two-step method. Relative gene
expression levels were calculated using CFX Manager software
(version 3.0; Bio-Rad Laboratories, Inc). The 2−ΔΔCq
method was used to evaluate the mRNA expression (20). Primer sequences are shown in Table I.
 | Table I.Primer sequences of hub genes. |
Table I.
Primer sequences of hub genes.
| Gene | Forward
sequence | Reverse
Sequence |
|---|
| CDK1 |
5′-GGAAACCAGGAAGCCTAGCATC-3′ |
5′-GGATGATTCAGTGCCATTTTGCC-3′ |
| SMC3 |
5′-ATGCGTGGAAGTCACTGCTGGA-3′ |
5′-GGCAGAAAAGTAACCTCTCCAGG-3′ |
| KIF11 |
5′-TACAGAAACCACTTAGTAGTGTCC-3′ |
5′-GAGTTCCTGTGAGAAGCCATCAG-3′ |
| SMC4 |
5′-GCCCAAGTAGCAATCAAGACTGC-3′ |
5′-GCTCTGCTGTTAGGTCATCCAC-3′ |
| NDC80 |
5′-CTGACACAAAGTTTGAAGAAGAGG-3′ |
5′-TAAGGCTGCCACAATGTGAGGC-3′ |
| NCAPG |
5′-GACGAACAGGAGGTGTCAGACT-3′ |
5′-TGCTGCGGTTTTGGCTCGTCTT-3′ |
| TTK |
5′-CCGAGATTTGGTTGTGCCTGGA-3′ |
5′-CATCTGACACCAGAGGTTCCTTG-3′ |
| CENPE |
5′-GGAGAAAGATGACCTACAGAGGC-3′ |
5′-AGTTCCTCTTCAGTTTCCAGGTG-3′ |
| NUF2 |
5′-TGGAGACTCAGTTGACTGCCTG-3′ |
5′-ATTTGGTCCTCCAAGTTCAGGCT-3′ |
| CENPU |
5′-CAGAAGGAATGAAAACCAGTGACA-3′ |
5′-ATGTGGCGATGGCTGCCTTACA-3′ |
| GAPDH |
5′-GGTGGTCTCCTCTGACTTCAACA-3′ |
5′-GTTGCTGTAGCCAAATTCGTTGT-3′ |
Statistical analysis
Each experiment was performed three times
separately. All quantitative data are expressed as the mean ±
standard deviation calculated using GraphPad Prism 5.0 software
(GraphPad Software, Inc.). One-way ANOVA followed by Tukey's post
hoc test was used to analyse differences among multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Anti-ALR McAb verification and ALR
expression in MM cell lines
One desirable and stable clone was screened via the
limiting dilution technique. Isotypes of McAb were IgM detected
using IsoQuick strips. ELISA was performed to test the ability of
anti-ALR McAb to bind 15-kDa-rhALR, with a control group comprising
ascites from the negative hybridoma. Absorbance >1.5 at OD450 nm
and ratio of the absorbance of anti-ALR ascites to that of negative
ascites >2.1 were considered to indicate specific binding of
antigen and antibody. As shown in Fig.
1A, the absorbance values in the anti-ALR(+) ascites and
negative ascites were 2.17±0.10 vs. 1.18±0.02, 2.30±0.16 vs.
0.88±0.14, 2.20±0.36 vs. 0.49±0.01, 1.90±0.52 vs. 0.28±0.09,
0.56±0.12 vs. 0.19±0.04 at dilutions ranging from 1:100 to
1:1,000,000, respectively. The results suggested that the antibody
titer in anti-ALR McAb ascites was 105. The absorbance
values of negative ascites was <1.5, and as the dilution of
negative ascites increased from 1:100 to 1:106, the
absorbance values decreased rapidly, which suggested that there was
a non-specific binding of negative ascites to rhALR protein.
However, after purification, the absorbance values in purified
antibody and negative control groups at OD450 nm at the dilutions
of 1:100 and 1:1,000 were 0.81±0.17 vs. 0.37+0.23 and 0.49±0.02 vs.
0.29±0.07, respectively (data not shown). The absorbance <1.5 at
OD450 nm suggested that the titer of the antibody was markedly
decreased, and therefore the purified antibody was not suitable for
subsequent experiments. ELISA was used to quantify 15-kDa-ALR in
the cell medium, and the concentration of 15-kDa-ALR in the medium
in MM1.S, RPMI8226 and U266 cells was 152.7±21.43, 200.7±31.56 and
179.5±68.92 ng/ml, respectively (data not shown), suggesting that
there were no differences in the quantity of ALR in the medium.
Anti-ALR McAb ascites were diluted to 1:10,000 as the primary
antibody for western blotting, and anti-ALR McAb was found to bind
specifically to 15-kDa-rhALR (Fig.
1B). The expression levels of 15-kDa-ALR in different MM cell
lines were investigated, and U266 cells displayed the highest
expression levels among the three MM cell lines (Fig. 1B); hence, this cell line was selected
for further experiments.
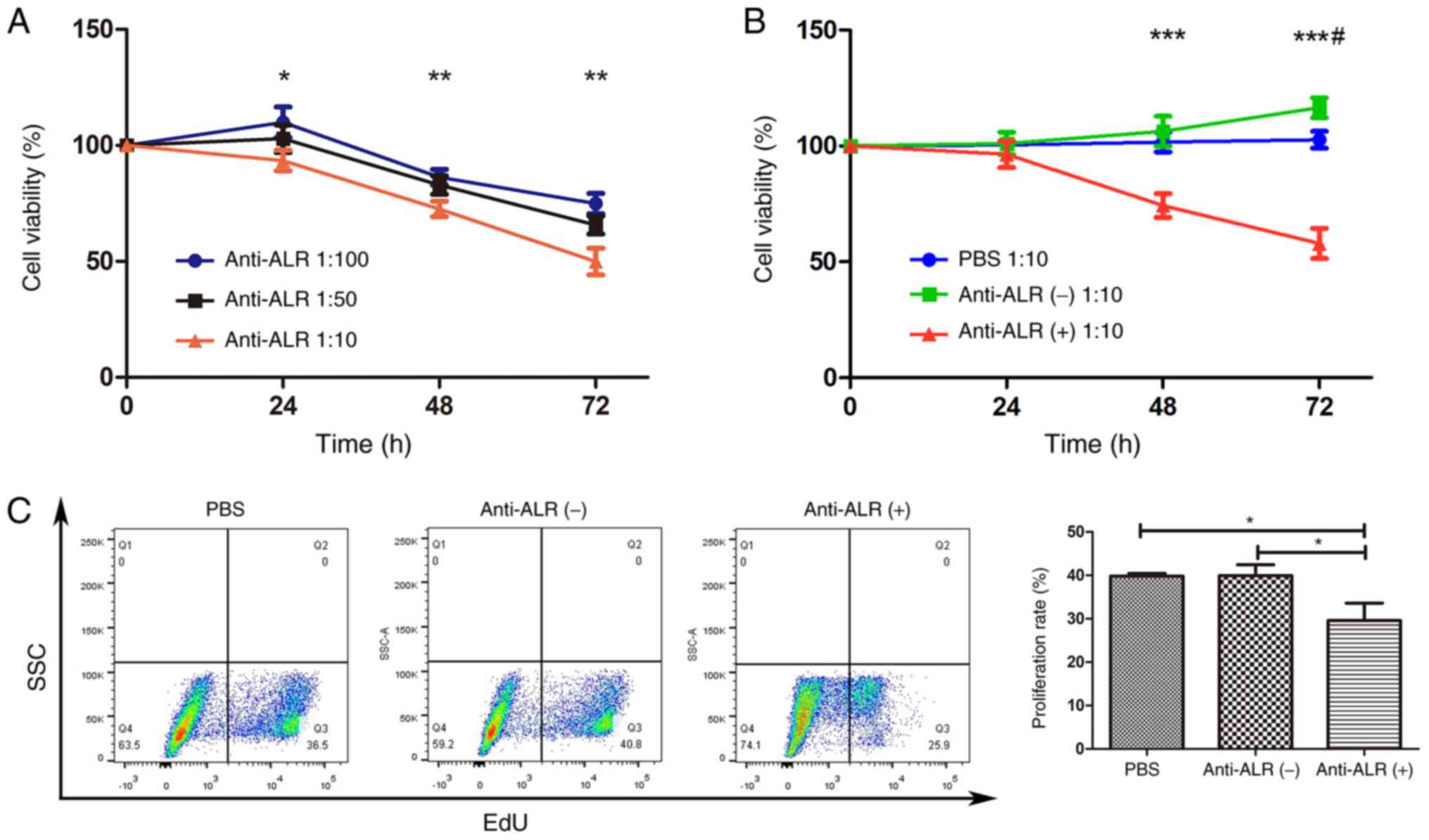
Effect of blocking 15-kDa-ALR on cell
survival and proliferation
To construct a cell model, U266 cells were treated
with different McAb concentrations (1:100, 1:50 and 1:10 ratio of
ascites to medium) for different durations (24, 48 and 72 h). The
CCK-8 assay results revealed that, when treated at 1:100 and 1:50
dilutions for 24 h, cell viability increased slightly, and then
decreased after 48 and 72 h. The higher the concentration, the
lower the cell viability when cells were treated for 48 and 72 h.
At a 1:10 concentration, cell viability decreased in a
time-dependent manner, and cell viability was significantly lower
than that at the other two concentrations at 24 h (96.43±5.78,
101.11±4.79 and 100.44±2.60% at 1:10, 1:50 and 1:100, respectively;
P<0.05), 48 h (72.62±3.35, 82.95±3.98 and 86.31±3.39% at 1:10,
1:50 and 1:100, respectively; P<0.01) and 72 h (49.97±3.31,
65.72±2.26 and 74.93±2.55% at 1:10, 1:50 and 1:100, respectively;
P<0.01) (Fig. 2A). Therefore, a
concentration of 1:10 was selected for subsequent experiments.
Next, cell viability was compared among the anti-ALR
treatment, negative ascites and PBS groups. The results revealed
that PBS had no influence on cell viability at a concentration of
1:10, regardless of treatment duration. Cell viability increased
slightly in the negative ascites group at a concentration of 1:10
after 72 h compared with 48 h, and there was a significant
difference between the negative ascites and PBS groups (P<0.05;
Fig. 2B). The effect of the anti-ALR
McAb on survival rate at 48 and 72 h was significantly lower than
that in the other two groups (P<0.001; Fig. 2B). Thus, an anti-ALR McAb
concentration of 1:10 for 72 h was selected for subsequent
experiments, with the same concentration of negative ascites as the
control and PBS blank groups.
Cell proliferation was measured using EdU assay, and
treatment with anti-ALR McAb, negative ascites and PBS yielded U266
cell proliferation rates of 23.77±1.39, 37.17±1.93 and 36.33±0.55,
respectively (P<0.01; Fig.
2C).
Effect of blocking 15-kDa-ALR on
apoptosis
Since the anti-apoptotic effect of exogenous ALR in
the liver has been demonstrated in various injury models (21,22),
apoptosis in U266 cells was investigated via flow cytometry and
western blotting (Fig. 3). When
treated with McAb for 72 h, the proportion of apoptotic cells (both
late and early apoptotic cells) detected by flow cytometry did not
differ between the three groups (7.70±0.29, 10.91±3.79 and
7.11±0.74% for the McAb, negative ascites and PBS groups,
respectively; Fig. 3A).
Additionally, the expression levels of the pro-apoptotic protein
Bax and the anti-apoptotic protein Bcl-2 were measured, and there
was no difference in the ratio of the two proteins between the
three groups (P>0.05; Fig. 3B).
Similarly, there were no differences in caspase-3 expression and in
the ratio of cleaved caspase-3/caspase-3 (P>0.05; Fig. 3B). However, following cell injury
induced by 0.004 mg/ml epirubicin treatment for 24 h, the
proportion of apoptotic cells was significantly increased after
blocking extracellular ALR with McAb (P<0.05; Fig. 3C).
 | Figure 3.Apoptosis of U266 cells treated with
anti-ALR McAb, negative ascites or PBS. (A) Flow cytometry analysis
of the apoptosis of U266 cells treated with anti-ALR McAb, negative
ascites or PBS at a concentration of 1:10 for 72 h, followed by
Annexin V-FITC/PI staining. (B) Analysis of apoptosis-associated
protein expression by western blotting. U266 cells were harvested
to examine the expression levels of Bax, Bcl-2, caspase-3 and
cleaved caspase-3 after treatment with anti-ALR McAb, negative
ascites or PBS at a concentration of 1:10 for 72 h. (C) U266 cells
treated with anti-ALR McAb, negative ascites or PBS at a
concentration of 1:10 for 48 h were co-treated with epirubicin at
0.004 mg/ml for 24 h, and apoptosis was examined by flow cytometry
after Annexin V-FITC/PI staining. *P<0.05 anti-ALR McAb group
vs. negative ascites and PBS groups. McAb, monoclonal antibody;
ALR, augmenter of liver regeneration. |
Differentially expressed genes
To clarify how extracellular ALR affected U266 MM
cells, RNA-Seq was performed to identify differentially expressed
RNAs between the anti-ALR McAb and negative ascites groups. A total
of 20,030 genes were subjected to differential gene analysis after
filtering low-abundance genes, and the results revealed 289 and 138
significantly upregulated and downregulated genes, respectively.
Hierarchical cluster analysis confirmed the quality of the
microarray data and the differences between the treatment and
control groups (Fig. 4A). The
distribution of RNAs is shown in a volcano plot (Fig. 4B).
 | Figure 4.Differentially expressed genes
between the anti-ALR McAb and negative control groups, as assessed
by RNA-sequencing. (A) Heat map analysis of differentially
expressed genes in U266 cells treated with anti-ALR McAb (trt) or
negative ascites (con). Red indicates genes with higher expression,
green indicates genes with lower expression and black indicates
genes not differing in terms of their expression levels between the
two groups. (B) Volcano plots showing the number, significance and
reliability of differentially expressed genes between the two
groups. Red dots indicate upregulation, while blue dots indicate
downregulation. McAb, monoclonal antibody; ALR, augmenter of liver
regeneration; FDR, false discovery rate; FC, fold-change; trt,
treatment group; con, control group. |
GO, pathway enrichment,
Path-Act-Network and PPI network analyses, and hub gene
validation
GO analysis revealed that differential genes in the
biological process category were primarily associated with ‘mitotic
nuclear division’, ‘cell cycle’ and ‘cell division’, while those in
the molecular function category were associated with ‘nucleic acid
binding’, ‘DNA binding’ and ‘RNA polymerase II core promoter
proximal region sequence-specific DNA binding’; the cellular
component category was primarily enriched in terms associated with
‘nucleus’, ‘centrosome’ and ‘intracellular’ functions (Fig. 5A). GO enrichment analysis revealed
that these differential genes were mainly associated with ‘cell
cycle’, ‘cell division’, ‘mitotic nuclear division’ and ‘mitotic
cell cycle’ (Fig. 5B), all of which
are associated with cell proliferation.
 | Figure 5.GO analysis of differentially
expressed genes. (A) GO divided into BP, MF and CC categories. The
top 15 -log10 values are shown. (B) GO enrichment analysis showing
the top 20 GO-term categories for differentially expressed genes.
The x-axis represents the rich factor, which refers to the ratio of
the number of differentially expressed genes enriched in the GO
category versus the total number of genes annotated in the GO
category. The larger the rich factor, the greater the degree of
enrichment. The colour of the dots indicates the P-value; the
smaller the P-value, the redder the colour, while larger P-values
are represented by greener colour. The size of the dots indicates
the number of differentially expressed genes contained in each GO
category; the larger the number of differentially expressed genes,
the larger the dots. GO, Gene Ontology; BP, biological process; MF,
molecular function; CC, cellular component. |
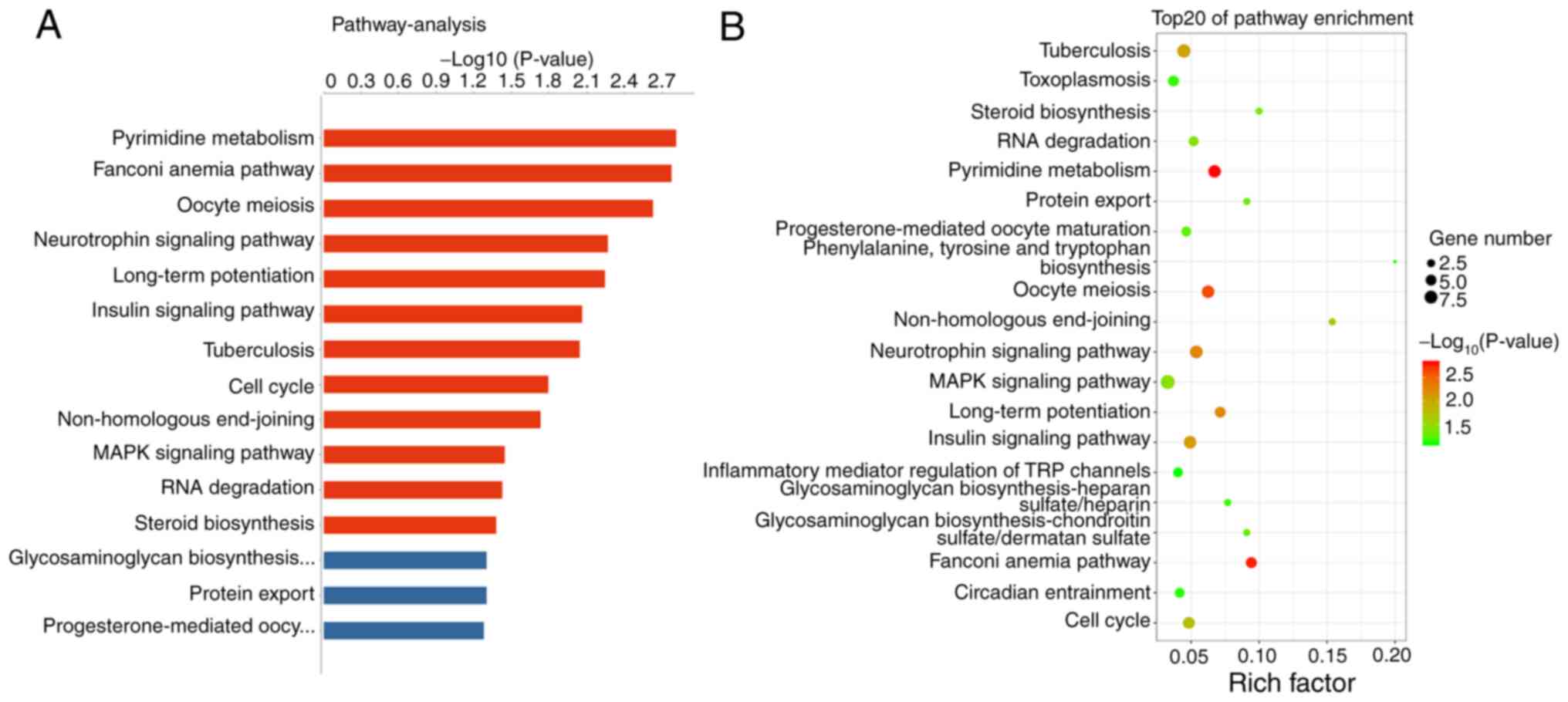
Pathway analysis based on gene annotation databases
can identify pathways in which differentially expressed genes are
significantly enriched, and pathways can directly reflect the
effects of genes on phenotypes (23). In the present study, 12 signaling
pathways were significantly different between groups (Fig. 6A), and the top three pathways were
‘pyrimidine metabolism’, ‘Fanconi anaemia pathway’ and ‘oocyte
meiosis’ (Fig. 6B).
The PPI network based on the identified
differentially expressed genes is shown in Fig. 7A. The top 10 genes were CDK1, SMC3,
KIF11, SMC4, NDC80, NCAPG, TTK, CENPE, NUF2 and CENPU, all of which
are potential hub genes according to the scores generated by the
cytoHubba plugin app (Fig. 7B).
Path-Act-Network analysis was used to probe signal transmission and
regulation processes among the various signaling pathways, and
MAPK, cell cycle and JAK-STAT were identified as the key signaling
pathways (Fig. 7C).
Hub gene and pathway validation
The top 10 hub genes were validated via RT-qPCR, and
the results are shown in Fig. 8. The
relative mRNA expression levels of CDK1 were downregulated, while
those of the other nine hub genes were upregulated in the anti-ALR
McAb group compared with in the negative ascites and PBS groups.
The changes in hub genes were consistent with the microarray
results. However, the changes in TTK and CENPU were not
statistically significant.
Signaling pathways were verified via western
blotting. The MAPK pathway includes the p44/42, p38 and JNK
cascades (24). The protein
expression levels of p44/42 were downregulated in the anti-ALR McAb
treatment group compared with in the negative ascites and PBS
groups, while p38 and phospho-p38 exhibited no significant
difference in expression. JNK expression was significantly
downregulated, while phospho-JNK expression was significantly
upregulated in the anti-ALR McAb group compared with in the
negative ascites and PBS groups (Fig.
9A). Additionally, STAT3 expression was significantly
downregulated in the treatment group compared with in the negative
ascites and PBS groups, (Fig. 9B),
as well as that of CDK1 and cyclin D1 (Fig. 9C). Cell cycle analysis revealed cell
cycle arrest at the S phase in the anti-ALR McAb group, but not in
the PBS or negative control groups (Fig.
9D).
 | Figure 9.Verification of changes in signaling
pathways after treatment with anti-ALR McAb, negative ascites or
PBS at a concentration of 1:10 for 72 h. (A) Expression levels of
important proteins in the MAPK signaling pathway, including p44/42,
p38 and JNK, as assessed by western blotting. (B) STAT3 protein
expression in the three groups. (C) Expression levels of cell
cycle-associated proteins, including CDK1 and cyclin D1, in the
three groups. (D) Flow cytometry analysis of the cell cycle after
PI staining. The x-axis represents the DNA content, while the
y-axis represents the number of cells. *P<0.05, **P<0.01,
***P<0.001 anti-ALR McAb vs. negative ascites and PBS groups.
McAb, monoclonal antibody; ALR, augmenter of liver regeneration; p,
phospho. |
Discussion
ALR mRNA expression has been previously detected in
several tissues, being relatively high in liver, kidney, testis and
brain (5,25). Additionally, ALR has been found in
the serum in both rat bacterial sepsis and mouse hemorrhagic shock
models (21). ALR expression is
upregulated in a number of solid tumours, such as hepatocellular
carcinoma (22,26), colon cancer (27) and T-cell leukaemia (28), and overexpression of ALR has a
protective effect on these tumour cells. Furthermore, a protective
effect of ALR was observed in neuroblastoma (29) and glioma (30). Our previous study revealed that ALR
expression was higher in U266 MM cells than in human peripheral
blood mononuclear cells (15). Our
research group has detected 15-kDa-ALR in patients with MM
(unpublished data). In small samples of bone marrow, the expression
levels of 15-kDa-ALR in myeloma cells were higher than those in
healthy volunteers (unpublished data). However, the sample size was
too small for statistical analysis, and it was difficult to obtain
bone marrow samples just for this experiment, so the data was
unpublished. In the present study, U266 cells exhibited the highest
expression levels of 15-kDa-ALR among the three cell lines
evaluated and the best response to the 15-kDa-ALR McAb.
A McAb was used to block 15-kDa-ALR, which is
secreted from cells, and can therefore avoid the influence of
23-kDa-ALR (9). In present study, an
IgM purification column was used to perform IgM purification.
However, after purification, the absorbance value of the purified
antibody was <1.5 at OD450 nm. The results suggested that the
purified antibody was not suitable for subsequent experiments. The
ascites containing McAb were selected for subsequent experiments.
Ascites produced by a negative hybridoma that cannot produce
anti-15-kDa-ALR McAb were used as a negative control. The
impurities in the ascites made it impossible to accurately estimate
the concentration of antibodies. Thus, the ascites were mixed for
the experiment to ensure the same quantity of antibody in the same
volume of ascites. The present CCK-8 results revealed that the
survival rate of U266 MM cells was negatively associated with the
concentration of McAb, which confirmed that low levels of
15-kDa-ALR confer a weaker protective effect. This information is
consistent with the results from a previous in vivo model of
acute liver injury (31) and a
previous in vitro model of acute injury (32). The cell viability was increased in
the negative ascites group compared with in the anti-ALR McAb group
at a concentration of 1:10 after 72 h, and the difference between
the negative ascites and PBS groups was statistically significant.
It was speculated that the reason for this may be that negative
ascites are rich in albumin and other factors, but not in anti-ALR
McAb, which may provide nutrition to cells without being inhibited
by anti-ALR McAb, which increased cell viability in the negative
ascites group. Previous research demonstrated that exogenous ALR
can induce proliferation in hepatocytes (33), and decrease apoptosis in liver cells
(32,34), kidney cells (35,36) and
lymphocytes (37). In order to
verify whether 15-kDa-ALR had the same effect in U266 MM cells, EdU
assays were used to assess cell proliferation. Treatment with the
15-kDa-ALR McAb decreased the cell proliferation rate compared with
that of the control group, which was consistent with the effect of
exogenous ALR on hepatocytes (38).
The current results of CCK-8 and EdU assays demonstrated that
15-kDa-ALR secreted by U266 cells induced cell proliferation and
that when autocrine ALR was blocked, cell proliferation
decreased.
The anti-apoptotic effect of exogenous ALR has been
demonstrated in different liver and kidney injury models (34,35).
Exogenous ALR decreases caspase-3 activity, which may be due to
decreased Bax expression (39).
Furthermore, a previous study revealed that exogenous ALR decreases
cell damage after partial hepatectomy by inducing anti-apoptotic
Bcl-2 expression, suggesting that exogenous ALR enhances liver
regeneration through apoptosis attenuation (34). Based on these findings, the present
study hypothesized that apoptosis would increase if extracellular
15-kDa-ALR was blocked using a McAb. However, when U266 cells were
treated with the McAb, apoptosis did not increase as expected. The
flow cytometry results revealed that there was no difference in
apoptosis, and western blotting indicated no differences in the
expression levels of Bax, Bcl-2 or caspase-3. Thus, the Bax/Bcl-2
ratio did not differ. The flow cytometry and western blotting
results on apoptosis were inconsistent with previous research
(32). This may be due to the fact
that, in the present study, there were no injury factors such as
TNFα, lipopolysaccharide or ischemia reperfusion, which may cause
apoptosis; hence, blocking extracellular 15-kDa-ALR with antibodies
alone may not be sufficient to stimulate apoptosis. However, a
previous study revealed that silencing ALR triggered apoptosis in
U266 cells without injury factors (15). Small hairpin RNA was used for
silencing ALR expression in a previous study (15), which influenced 23- and 15-kDa-ALR at
the same time. The 23-kDa-ALR is located in mitochondria and is
involved in the process of oxidative phosphorylation, which is
essential for life. When 23-kDa-ALR is silenced, apoptosis is
triggered (38). In the present
study, McAb blocked extracellular 15-kDa-ALR without influencing
23-kDa-ALR, which may be the reason for the inconsistent results
with previous studies. Therefore, epirubicin, a traditional
chemotherapy drug used to induce apoptosis (40), was employed in the present study.
When the injury factor was present, apoptosis was increased as
expected. These results confirmed that extracellular 15-kDa-ALR
promoted proliferation in the absence of external injury factors,
and the anti-apoptotic effect also occurred in the presence of
injury factors.
A previous study demonstrated that the mechanisms of
exogenous ALR affect the liver and that the MAPK signaling pathway
is important in cell proliferation (14). In the present study, 15-kDa-ALR in
the medium of U266 cells was detected by ELISA, which suggested
that U266 cells may secrete 15-kDa-ALR. In the early research of
our group, MTS assay was used to evaluate the influence of
exogenous ALR on the proliferation of U266 cells, and the results
suggested that after treating with exogenous ALR, the proliferation
of U266 cells was significantly increased (unpublished data);
however, the signal transduction mechanism in U266 cells was
unclear. The use of an antibody to block extracellular 15-kDa-ALR
in the present study was part of a research project on the signal
transduction mechanism of 15-kDa-ALR in U266 cells. GO and pathway
analysis of RNA-seq revealed that 15-kDa-ALR affected U266 cells
via the MAPK signaling pathway, similar to the findings in
hepatocytes (41). The MAPK
signaling pathway involves ERKs, JNKs and p38-MAPKs. In the present
study, western blotting indicated that the expression levels of
p44/42 were downregulated in the treatment group. p44/42 can be
activated by a variety of factors, such as inflammation (42) and heart failure (43), and this may be coincident with the
activation of other pathways that promote contradictory biological
responses (44). Combining the
Path-Act-Network analysis and western blotting results demonstrated
that p44/42 in the MAPK signaling pathway was downregulated by
blocking extracellular 15-kDa-ALR and decreasing the proliferation
of U266 cells. The expression levels of JNK and phospho-JNK were
also altered in the present study, demonstrating that the JNK
signaling pathway may be involved in the mechanism of blocking ALR
in U266 cells. A recent study revealed that xanthohumol exhibits
anti-myeloma activity via an ERK- and JNK-dependent mechanism in
RPMI8226 cells (45). Similar to the
aforementioned study, the present results indicated that the ERK1/2
and JNK signaling pathways may serve a role in anti-myeloma
effects, but changes in protein expression depended on different
stimuli. The expression levels of p38 and phospho-p38 were not
significantly altered among the three groups; hence, the p38
signaling pathway may not be involved in the mechanisms of blocking
15-kDa-ALR. In MM cells, when the activity of the STAT3 signaling
pathway was inhibited, cyclin D1 expression was downregulated,
which inhibited the proliferation of myeloma cells and induced
apoptosis (46). In the present
study, STAT3 and cyclin D1 expression was attenuated by blocking
extracellular 15-kDa-ALR, consistent with the results of a previous
study (47), suggesting that the
STAT3 signaling pathway may serve an important role in MM cells.
However, STAT3 is attenuated by exogenous ALR (47), which contradicts the current
findings, possibly due to the lack of injury factors in the present
study.
According to the PPI network, the top 10 hub genes
were associated with the cell cycle. In the current study, CDK1
expression was downregulated at both the mRNA and protein levels,
and cell cycle analysis indicated that blocking 15-kDa-ALR caused
cell cycle arrest in the S phase. A previous study on hematopoietic
stem cells demonstrated that binding of ALR to JAB1 blocks JAB1
interaction with p27kip1, which promotes cell cycle arrest
(48). Another study on leukemia
suggested that changes in the MAPK signaling pathway may lead to
S-phase cell cycle arrest (49). The
mechanism of the cell cycle arrest induced by blocking 15-kDa-ALR
may involve the combined effects of blocking the binding of ALR to
JAB1 and the regulation of the MAPK signaling pathway induced by
the anti-ALR McAb. The other hub genes identified in the present
study included members of the centromere protein family (CENPE and
CENPU), the nuclear division cycle 80 family (NDC80 and NUF2) and
the SMC family (SMC3, SMC4 and NCAPG), all of which are associated
with chromosome changes in the cell cycle. The expression levels of
these genes were upregulated following anti-ALR treatment, which
may be due to the fact that the cell cycle was arrested in S phase,
inducing the upregulation of these genes when blocking
15-kDa-ALR.
In conclusion, the current results provided evidence
that blocking extracellular 15-kDa-ALR inhibited the proliferation
of U266 MM cells through cell cycle arrest without increasing
apoptosis in the absence of injury factors. The ERK1/2 and JNK
branches of the MAPK signaling pathway, STAT3 signaling pathway and
the cell cycle appeared to be involved in the possible mechanism
(Fig. 10). Thus, 15-kDa-ALR may be
a novel target for myeloma treatment.
Acknowledgements
The authors would like to thank Mrs. Wanyan Deng and
Mr. Fang Jia (The Second Affiliated Hospital of Chongqing Medical
University, Chongqing, China) for their assistance in analysing the
data and formatting the manuscript.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81871608).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WH contributed to the conceptualization and
investigation of the study, data curation and writing of the
original draft. HS contributed to the conceptualization of the
study, funding acquisition and review and editing of the
manuscript. TH contributed to the analysis of experimental data.
DZ, XL and HG performed the experiments. QL contributed to the
conceptualization and supervision of the study. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All experimental procedures involving animals were
approved by the Animal Ethics and Use Committee of the Second
Affiliated Hospital of Chongqing Medical University (Chongqing,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ALR
|
augmenter of liver regeneration
|
|
MM
|
multiple myeloma
|
|
McAb
|
monoclonal antibody
|
|
GO
|
Gene Ontology
|
References
|
1
|
Kazandjian D: Multiple myeloma
epidemiology and survival: A unique malignancy. Semin Oncol.
43:676–681. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kristinsson SY, Anderson WF and Landgren
O: Improved long-term survival in multiple myeloma up to the age of
80 years. Leukemia. 28:1346–1348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Landgren O and Iskander K: Modern multiple
myeloma therapy: Deep, sustained treatment response and good
clinical outcomes. J Intern Med. 281:365–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sonneveld P and Broijl A: Treatment of
relapsed and refractory multiple myeloma. Haematologica.
101:396–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hagiya M, Francavilla A, Polimeno L, Ihara
I, Sakai H, Seki T, Shimonishi M, Porter KA and Starzl TE: Cloning
and sequence analysis of the rat augmenter of liver regeneration
(ALR) gene: Expression of biologically active recombinant ALR and
demonstration of tissue distribution. Proc Natl Acad Sci USA.
91:8142–8146. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu J, Xu WX, Zhan YQ, Cui XL, Cai WM, He
FC and Yang XM: Identification and characterization of a novel
isoform of hepatopoietin. World J Gastroenterol. 8:353–356. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hofhaus G, Lee JE, Tews I, Rosenberg B and
Lisowsky T: The N-terminal cysteine pair of yeast sulfhydryl
oxidase Erv1p is essential for in vivo activity and interacts with
the primary redox centre. Eur J Biochem. 270:1528–1535. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng J, Zhong YW, Liu Y, Dong J, Yang JZ
and Chen JM: Cloning and sequence analysis of human genomic DNA of
augmenter of liver regeneration. World J Gastroenterol. 6:275–277.
2000.PubMed/NCBI
|
|
9
|
Gandhi CR, Kuddus R, Subbotin VM, Prelich
J, Murase N, Rao AS, Nalesnik MA, Watkins SC, DeLeo A, Trucco M, et
al: A fresh look at augmenter of liver regeneration in rats.
Hepatology. 29:1435–1445. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fass D: The Erv family of sulfhydryl
oxidases. Biochim Biophys Acta. 1783:557–566. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jung YS, Kim SJ, Kwon DY, Jun DS and Kim
YC: Significance of alterations in the metabolomics of
sulfur-containing amino acids during liver regeneration. Biochimie.
95:1605–1610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferecatu I, Gonçalves S, Golinelli-Cohen
MP, Clémancey M, Martelli A, Riquier S, Guittet E, Latour JM,
Puccio H, Drapier JC, et al: The diabetes drug target MitoNEET
governs a novel trafficking pathway to rebuild an Fe-S cluster into
cytosolic aconitase/iron regulatory protein 1. J Biol Chem.
289:28070–28086. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Todd LR, Damin MN, Gomathinayagam R, Horn
SR, Means AR and Sankar U: Growth factor erv1-like modulates Drp1
to preserve mitochondrial dynamics and function in mouse embryonic
stem cells. Mol Biol Cell. 21:1225–1236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ibrahim S and Weiss TS: Augmenter of liver
regeneration: Essential for growth and beyond. Cytokine Growth
Factor Rev. 45:65–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zeng HQ, Luo Y, Lou SF, Liu Q, Zhang L and
Deng JC: Silencing of augmenter of liver regeneration inhibited
cell proliferation and triggered apoptosis in U266 human multiple
myeloma cells. Braz J Med Biol Res. 50:e61392017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim HY, Stojadinovic A and Izadjoo MJ:
Immunization, hybridoma generation, and selection for monoclonal
antibody production. Methods Mol Biol. 1131:33–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering
C, et al: STRING v9.1: Protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41D:D808–D815. 2013.
|
|
19
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vodovotz Y, Prelich J, Lagoa C, Barclay D,
Zamora R, Murase N and Gandhi CR: Augmenter of liver regeneration
(ALR) is a novel biomarker of hepatocellular stress/inflammation:
In vitro, in vivo and in silico studies. Mol Med. 18:1421–1429.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao Y, Fu YL, Yu M, Yue PB, Ge CH, Xu WX,
Zhan YQ, Li CY, Li W and Wang XH: Human augmenter of liver
regeneration is important for hepatoma cell viability and
resistance to radiation-induced oxidative stress. Free Radic Biol
Med. 47:1057–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45D:D353–D361. 2017.
View Article : Google Scholar
|
|
24
|
Sun Y, Liu WZ, Liu T, Feng X, Yang N and
Zhou HF: Signaling pathway of MAPK/ERK in cell proliferation,
differentiation, migration, senescence and apoptosis. J Recept
Signal Transduct Res. 35:600–604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Giorda R, Hagiya M, Seki T, Shimonishi M,
Sakai H, Michaelson J, Francavilla A, Starzl TE and Trucco M:
Analysis of the structure and expression of the augmenter of liver
regeneration (ALR) gene. Mol Med. 2:97–108. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu HY, Xiang DR, Huang HJ, Li J and Sheng
JF: Expression level of augmenter of liver regeneration in patients
with hepatic failure and hepatocellular carcinoma. Hepatobiliary
Pancreat Dis Int. 9:492–498. 2010.PubMed/NCBI
|
|
27
|
Gatzidou E, Mantzourani M, Giaginis C,
Giagini A, Patsouris E, Kouraklis G and Theocharis S: Augmenter of
liver regeneration gene expression in human colon cancer cell lines
and clinical tissue samples. J BUON. 20:84–91. 2015.PubMed/NCBI
|
|
28
|
Shen Y, Liu Q, Sun H, Li X, Wang N and Guo
H: Protective effect of augmenter of liver regeneration on
vincristine-induced cell death in Jurkat T leukemia cells. Int
Immunopharmacol. 17:162–167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Polimeno L, Pesetti B, Lisowsky T, Iannone
F, Resta L, Giorgio F, Mallamaci R, Buttiglione M, Santovito D,
Vitiello F, et al: Protective effect of augmenter of liver
regeneration on hydrogen peroxide-induced apoptosis in SH-SY5Y
human neuroblastoma cells. Free Radic Res. 43:865–875. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Polimeno L, Pesetti B, De Santis F, Resta
L, Rossi R, De Palma A, Girardi B, Amoruso A and Francavilla A:
Decreased expression of the augmenter of liver regeneration results
in increased apoptosis and oxidative damage in human-derived glioma
cells. Cell Death Dis. 3:e2892012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang LM, Liu DW, Liu JB, Zhang XL, Wang
XB, Tang LM and Wang LQ: Effect of naked eukaryotic expression
plasmid encoding rat augmenter of liver regeneration on acute
hepatic injury and hepatic failure in rats. World J Gastroenterol.
11:3680–3685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ilowski M, Kleespies A, de Toni EN,
Donabauer B, Jauch KW, Hengstler JG and Thasler WE: Augmenter of
liver regeneration (ALR) protects human hepatocytes against
apoptosis. Biochem Biophys Res Commun. 404:148–152. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dayoub R, Thasler WE, Bosserhoff AK,
Singer T, Jauch KW, Schlitt HJ and Weiss TS: Regulation of
polyamine synthesis in human hepatocytes by hepatotrophic factor
augmenter of liver regeneration. Biochem Biophys Res Commun.
345:181–187. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Polimeno L, Pesetti B, Annoscia E, Giorgio
F, Francavilla R, Lisowsky T, Gentile A, Rossi R, Bucci A and
Francavilla A: Alrp, a survival factor that controls the apoptotic
process of regenerating liver after partial hepatectomy in rats.
Free Radic Res. 45:534–549. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao XH, Zhang L, Liu Q, Sun H, Peng CM
and Guo H: Augmenter of liver regeneration protects kidneys from
ischaemia/reperfusion injury in rats. Nephrol Dial Transplant.
25:2921–2929. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liao XH, Chen GT, Li Y, Zhang L, Liu Q,
Sun H and Guo H: Augmenter of liver regeneration attenuates tubular
cell apoptosis in acute kidney injury in rats: The possible
mechanisms. Ren Fail. 34:590–599. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang N, Sun H, Shen Y, Li XF, Pan T, Liu
GL and Liu Q: Augmenter of liver regeneration inhibits apoptosis of
activated human peripheral blood lymphocytes in vitro.
Immunopharmacol Immunotoxicol. 35:257–263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Francavilla A, Vujanovic NL, Polimeno L,
Azzarone A, Iacobellis A, Deleo A, Hagiya M, Whiteside TL and
Starzl TE: The in vivo effect of hepatotrophic factors augmenter of
liver regeneration, hepatocyte growth factor, and insulin-like
growth factor-II on liver natural killer cell functions.
Hepatology. 25:411–415. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Weiss TS, Lupke M, Ibrahim S, Buechler C,
Lorenz J, Ruemmele P, Hofmann U, Melter M and Dayoub R: Attenuated
lipotoxicity and apoptosis is linked to exogenous and endogenous
augmenter of liver regeneration by different pathways. Plos One.
12:e01842822017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang TC, Chiu PR, Chang WT, Hsieh BS,
Huang YC, Cheng HL, Huang LW, Hu YC and Chang KL: Epirubicin
induces apoptosis in osteoblasts through death-receptor and
mitochondrial pathways. Apoptosis. 23:226–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y, Li M, Xing G, Hu Z, Wang Q, Dong C,
Wei H, Fan G, Chen J, Yang X, et al: Stimulation of the
mitogen-activated protein kinase cascade and tyrosine
phosphorylation of the epidermal growth factor receptor by
hepatopoietin. J Biol Chem. 275:37443–37447. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu J, Bing C and Wilding JPH:
1α,25(OH)2D3 attenuates IL-6 and IL-1β-mediated inflammatory
responses in macrophage conditioned medium-stimulated human white
preadipocytes by modulating p44/42 MAPK and NF-κB signaling
pathways. Diabetol Metab Syndr. 11:92019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu Y, Wei SG, Zhang ZH, Weiss RM and
Felder RB: ERK1/2 MAPK signaling in hypothalamic paraventricular
nucleus contributes to sympathetic excitation in rats with heart
failure after myocardial infarction. Am J Physiol Heart Circ
Physiol. 310:H732–H739. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
von Kriegsheim A, Baiocchi D, Birtwistle
M, Sumpton D, Bienvenut W, Morrice N, Yamada K, Lamond A, Kalna G,
Orton R, et al: Cell fate decisions are specified by the dynamic
ERK interactome. Nat Cell Biol. 11:1458–1464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sławińska-Brych A, Zdzisińska B, Czerwonka
A, Mizerska-Kowalska M, Dmoszyńska-Graniczka M, Stepulak A and
Gagoś M: Xanthohumol exhibits anti-myeloma activity in vitro
through inhibition of cell proliferation, induction of apoptosis
via the ERK and JNK-dependent mechanism, and suppression of sIL-6R
and VEGF production. Biochim Biophys Acta, Gen Subj.
1863:1294082019. View Article : Google Scholar
|
|
46
|
Park S, Lee HJ, Jeong SJ, Song HS, Kim M,
Lee HJ, Lee EO, Kim DH, Ahn KS and Kim SH: Inhibition of JAK1/STAT3
signaling mediates compound K-induced apoptosis in human multiple
myeloma U266 cells. Food Chem Toxicol. 49:1367–1372. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dayoub R, Buerger L, Ibrahim S, Melter M
and Weiss TS: Augmenter of liver regeneration (ALR) exhibits a dual
signaling impact on hepatic acute-phase response. Exp Mol Pathol.
102:428–433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Teng EC, Todd LR, Ribar TJ, Lento W,
Dimascio L, Means AR and Sankar U: Gfer inhibits Jab1-mediated
degradation of p27kip1 to restrict proliferation of hematopoietic
stem cells. Mol Biol Cell. 22:1312–1320. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang J, Chen L, Yan Y, Qiu J, Chen J, Song
J, Rao Q, Ben-David Y, Li Y and Hao X: BW18, a C-21 steroidal
glycoside, exerts an excellent anti-leukemia activity through
inducing S phase cell cycle arrest and apoptosis via MAPK pathway
in K562 cells. Biomed Pharmacother. 112:1086032019. View Article : Google Scholar : PubMed/NCBI
|