Introduction
Gallbladder cancer (GBC) is the most common biliary
malignancy and is characterized by an advanced stage diagnosis,
high recurrence rate and poor prognosis due to no specific clinical
signs, symptoms or reliable sensitive markers (1). Patients with GBC have a poor outcome,
with a 5-year survival rate of only 10–20% (2). Therefore, identifying novel biomarkers
remains essential for the early diagnosis and effective treatment
of patients with GBC (3).
MicroRNAs (miRNAs/miRs) regulate biological
processes, including cell proliferation, differentiation and
migration, by binding to the 3′-untranlsated region (3′-UTR) and
degrading the mRNA of target genes (4). For example, miR-182 has been reported
to promote breast cancer cell motility and invasiveness by
targeting Missing in Metastasis (5).
Furthermore, miR-182 expression is upregulated in colorectal
cancer, which significantly promotes epithelial-mesenchymal
transition (EMT), cell proliferation, invasion and migration by
targeting Special AT-rich Sequence-Binding protein 2 (6). Aberrant miR profiles in GBC have been
reported (7); however, the role of
miR-182 in the pathogenesis of GBC remains unclear.
The EMT event of cancer cells is associated with
cancer invasion, metastasis and recurrence (8). The Wnt/β-catenin pathway has been
implicated in EMT (9). Activation of
β-catenin leads to β-catenin stabilization and translocation to the
nucleus, where it associates with transcription factors, lymphoid
enhancer factor/T-cell (LEF/TCF), to activate the expression of
target genes (10). It has been
demonstrated that the EMT factor, ZEB1, is transcriptionally
regulated by the β-catenin/TCF4 complex pathway (11). Recently, it was reported that
Forkhead box N3 (FOXN3) may interact with β-catenin and suppress
the activity of the Wnt/β-catenin pathway (12). Prediction analysis using TargetScan
7.2 software (http://www.targetscan.org/vert_72) demonstrates that
miR-182 may bind to the 3′-UTR of FOXN3. However, whether miR-182
promotes EMT by negatively regulating FOXN3 remains unclear.
Therefore, the present study aimed to investigate the role of
miR-182 in GBC.
Materials and methods
Tissue collection
A total of 18 GBC tissues and adjacent normal
tissues were collected from patients with GBC (8 males and 10
females; mean age, 70.2±3.6 years; age range, 63–79 years) who
underwent surgical resection at The First Affiliated Hospital of
Gannan Medical University between March 2017 and May 2019.
Inclusion criteria were as follows: i) patients who were diagnosed
by histopathological examinations for the first time and had not
received any therapies and ii) patients who had provided written
informed consent. Exclusion criteria were as follows: i) patients
complicated with other clinical disorders and ii) patients with a
history of malignancies. Tissue samples were collected from all
patients prior to biopsy, which was performed using a fine needle
under the guidance of Magnetic Resonance Imaging. Tissue samples
were stored at −80°C until subsequent experimentation. The present
study was approved by the Ethics Committee of The First Affiliated
Hospital of Gannan Medical University, Ganzhou, China (GMU38972623)
and was performed in accordance with the Declaration of Helsinki
(13). Written informed consent was
provided by all patients prior to the study.
Cell culture
The non-tumorigenic human intrahepatic biliary
epithelial cell line (H69) and the human GBC cell lines (GBC-SD and
SGC-996) were purchased from the American Type Culture Collection.
Cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 100 U/ml penicillin and streptomycin (Thermo
Fisher Scientific, Inc.) at 37°C in 5% CO2.
Cell transfection
miR-182 inhibitors (5′-TTCTACCATTGCCAA-3′) and miRNA
negative control (miR-NC; 5′-ACGTCTATACGCCCA-3′) were purchased
from Guangzhou RiboBio Co., Ltd. To the best of our knowledge, no
effects of miR-NC on cell viability have been confirmed. GBC-SD and
SGC-996 cells were seeded onto 6-well plates at a density of
1×105 cells/well. Once they reached 60% confluence, the
cells were transfected with 50 nM miR-182 inhibitors and miR-NC
using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Inc.)
for 24 h at 37°C, according to the manufacturer's protocol.
The pcDNA3.1-FOXN3 vector (Guangzhou RiboBio Co.,
Ltd) was prepared by cloning the open reading frame of FOXN3 into
the vector. GBC-SD and SGC-996 cells were transfected with the
pcDNA3.1-FOXN3 vectors for 24 h at 37°C, using Lipofectamine 3000
reagent.
MTT assay
GBC-SD and SGC-996 cells transfected with miR-182
inhibitors and miR-NC were seeded onto 96-well plates at a density
of 5×103 cells/well and cultured at 37°C for 48 h. Cells
were subsequently incubated with 0.5 mg/ml MTT reagent at 37°C for
4 h. Following the MTT incubation, the purple formazan crystals
were dissolved using 150 µl dimethyl sulfoxide (Sigma-Aldrich;
Merck KGaA) in the dark and viability was subsequently analyzed at
a wavelength of 490 nm, using a microplate reader (Thermo Fisher
Scientific, Inc.).
Transwell migration and invasion
assays
GBC-SD and SGC-996 cells were collected 24 h
post-transfection and prepared for single-cell suspensions using
serum-free medium, at a density of 3×104 cells/ml. The
single-cell suspensions were transferred into the upper chambers of
Transwell plates, while culture medium supplemented with 20% FBS
was plated into the lower chambers. For the invasion assay,
Transwell membranes were precoated with Matrigel (EMD Millipore)
overnight at room temperature. Following incubation for 2 h at
37°C, the migratory and invasive cells were collected, washed and
stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA) for 15
min at room temperature. Stained cells were counted in five
randomly selected fields under an inverted microscope (Olympus
CK-40; Olympus Corporation) at a magnification of ×100.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from GBC tissues and cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocols. Total RNA (2 µg) was
reverse transcribed into cDNA using M-MLV (Promega Corporation) for
60 min at 42°C. qPCR was subsequently performed using an ABI 7500
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) to
detect the mRNA expression levels of FOXN3, β-catenin, E-cadherin
and vimentin. The procedure was performed as follows: 95°C for 6
min, followed by 40 cycles at 95°C for 40 sec, 65°C for 30 sec, and
finally at 75°C for 8 min. miR-182 primers (Biomics) and EzOmics
SYBR qPCR kits (cat. no. BK2200) (Biomics) were used to detect the
expression levels of miRNAs. The following primer sequences
(Biomics) were used for the qPCR: FOXN3 forward,
5′-TGCCAATCACTCCCATTGGG-3′ and reverse, 5′-CCGCATCCGGCAGCTGG-3′;
Cyclin D1 forward, 5′-TGTTTGCAAGCAGGACTTTG-3′ and reverse,
5′-ACGTCAGCCTCCACACTCTT-3′; and c-Myc forward,
5′-CTCCTGGCAAAAGGTCAGAG-3′ and reverse, 5′-TCGGTTGTTGCTGATCTGTC-3′.
The expression levels of miRNA and mRNA were normalized to the
endogenous reference, U6 (forward, 5′-AGACAATTGATGCGTGCGATC-3′ and
reverse, 5′-GCTGCAACTGCACTACCAAC-3′) and 18S rRNA (forward,
5′-GTAACCCGTTGAACCCCATT-3′ and reverse,
5′-CCATCCAATCGGTAGTAGCG-3′), respectively. Relative expression
levels were calculated using the 2−ΔΔCq method (14).
Western blotting
Cells were lysed using RIPA lysis buffer (Beyotime
Institute of Biotechnology). Total protein was quantified using a
BCA protein assay kit (cat. no. 23250; Pierce Biotechnology; Themo
Fisher Scientific, Inc.) and 25 µg protein/lane was separated via
SDS-PAGE on a 10% gel. The separated proteins were subsequently
transferred onto polyvinylidene difluoride membranes and blocked
with Tris-buffered saline supplemented with 5% skimmed milk for 1 h
at room temperature. The membranes were incubated with primary
antibodies against FOXN3 (cat. no. 95568), β-catenin (cat. no.
8480), TCF4 (cat. no. 2569), E-cadherin (cat. no. 3195), Vimentin
(cat. no. 5741), Cyclin D1 (cat. no. 55506), c-Myc (cat. no. 18583)
and GAPDH (cat. no. 5174S) overnight at 4°C (all 1:1,000 dilutions
and purchased from Cell Signaling Technology, Inc.). Following the
primary incubation, the membranes were incubated with a horseradish
peroxidase-labelled secondary antibody (1:2,000; cat. no. 32935;
Cell Signaling Technology, Inc.) for 1 h at room temperature.
Protein bands were detected using the enhanced chemiluminescence
detection system (Bio-Rad Laboratories, Inc.) and Quantity One
software v4.6.2 (Bio-Rad Laboratories, Inc.).
Dual-luciferase reporter assay
Prediction analysis was conducted using TargetScan
7.2 software (http://www.targetscan.org/vert_72) (15). After inputting miR-182 and clicking
the button ‘submit’, the prediction results were obtained. It
indicated that the position 6109–6116 of FOXN3 3′-UTR was paired
with miR-182. To validate the prediction, the 3′-UTR of FOXN3 mRNA
(both the wild-type and mutant) was cloned into the psiCHECK™-2
vector (Promega Corporation) at the XhoI/NotI sites,
according to the manufacturer's protocol (In-Fusion Advantage PCR
cloning kit; cat. no. 630489; Clontech Laboratories, Inc.). Cells
were cultured for 24 h at 37°C. Subsequently, the psiCHECK-FOXN3
3′-UTR reporter vector (the wild-type or the mutant) was
co-transfected into cells with miR-182 mimics or miR-NC, using
Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Following incubation for 48 h at 37°C, firefly and
Renilla luciferase activities were detected using the Glomax
96 luminometer (Promega Corporation). Firefly luciferase reporter
was normalized to Renilla luciferase activity.
Co-immunoprecipitation (Co-IP)
assay
Cells were harvested using IP lysis buffer (Pierce
Biotechnology; Thermo Fisher Scientific, Inc.), of which, 10% was
used as the input sample. The remaining amount (90% of IP lysate)
was used for IP assays through co-incubation with the goat IgG
(cat. no. 6990) or β-catenin (cat. no. 8480) antibodies (both 1:50
dilutions and purchased from Cell Signaling Technology, Inc.).
Following the primary incubation, the samples were incubated with
protein A-sepharose beads at 4°C for 2 h. IP washing buffer was
used, and the eluted samples (25 µg protein) after centrifugation
at 3,000 × g for 4 min at 4°C were subjected to western
blotting.
Statistical analysis
All experiments were performed in triplicate and
data are presented as the mean ± standard error of the mean. SPSS
20.0 software (IBM Corp.) was used for statistical analysis.
Differences between GBC and normal tissues were analyzed using a
paired t-test. One-way analysis of variance, followed by Tukey's
post hoc test, was used to compare differences between multiple
groups. An unpaired Student's t-test was used to make statistical
comparisons between two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-182 expression is upregulated in
GBC tissues and cells
To investigate the role of miR-182 in GBC, RT-qPCR
analysis was performed to detect relative miR-182 expression in GBC
tissues and cells. As presented in Fig.
1A, miR-182 expression was significantly upregulated in GBC
tissues, compared with adjacent normal tissues. miR-182 expression
was also assessed in GBC cells. As presented in Fig. 1B, miR-182 expression markedly
increased in GBC-SD and SGC-996 cells, compared with H69 cells. To
further investigate the association between miR-182 and EMT
mediated by β-catenin signaling in GBC cells, western blot analysis
was performed to detect protein expression levels of
FOXN3/β-catenin and EMT-related factors, E-cadherin and vimentin.
As presented in Fig. 1C-F, the
protein expression levels of β-catenin and vimentin significantly
increased, while FOXN3 and E-cadherin expression levels were
attenuated in GBC tissues (Fig. 1C and
D) and GBC cells (Fig. 1E and
F), compared with adjacent normal tissues and H69 cells,
respectively. Taken together, these results suggested that
increased miR-182 expression may be involved in the EMT of GBC
cells.
miR-182-knockdown inhibits EMT in GBC
cells
To investigate the role of miR-182 in EMT, GBC-SD
and SGC-996 cells were transfected with miR-182 inhibitors and
miR-NC, respectively. RT-qPCR analysis was performed to detect
miR-182 and the results demonstrated that miR-182 expression was
significantly decreased in cells transfected with miR-182
inhibitors, compared with cells transfected with miR-NC (Fig. 2A). These results demonstrated that
transfection of GBC cells with miR-182 inhibitors was successful.
The results of the MTT assay indicated that miR-182 inhibitors
decreased the viability of GBC cells (Fig. 2B and C). To determine the biological
functions of miR-182 inhibitors on EMT, cell migration and invasion
assays were performed. The results demonstrated that miR-182
inhibitors significantly attenuated the activity of migration and
invasion in GBC-SD and SGC-996 cells (Fig. 2D-F). To further investigate the
potential molecular mechanism of miR-182 inhibitors on EMT, western
blot analysis was performed to detect protein expression levels of
the EMT-related factors, E-cadherin and vimentin. miR-182-knockdown
effectively decreased vimentin expression and increased E-cadherin
expression in GBC-SD (Fig. 2G and H)
and SGC-996 (Fig. 2I and J) cells.
Taken together, these results suggested that miR-182-knockdown may
inhibit EMT in GBC cells.
FOXN3 is a direct target of
miR-182
To investigate the potential molecular mechanism of
miR-182 in promoting EMT in GBC cells, it was hypothesized that
miR-182 targets and degrades FOXN3, which may form a complex with
β-catenin and negatively regulate its nuclear transcriptional
activity in GBC cells. Prediction analysis demonstrated that FOXN3
is a direct target of miR-182. These results were verified via the
dual-luciferase reporter assay. As presented in Fig. 3A, the 3′-UTR of FOXN3 containing a
wild-type or mutant binding site for miR-182 was artificially
cloned into the firefly luciferase reporter system. No significant
differences were observed in the relative luciferase activities
between the NC reporter and the reporter containing the mutant
binding site of FOXN3. By contrast, the reporter containing the
wild-type binding site of FOXN3 exhibited decreased luciferase
activity for >60% (Fig. 3B).
Taken together, these results suggested that miR-182 specifically
aims to degrade FOXN3 by binding to its 3′-UTR.
miR-182/FOXN3 promotes EMT by
regulating the β-catenin pathway
The effects of miR-182/FOXN3 on EMT by mediating the
β-catenin pathway were assessed. FOXN3 has been reported to be
associated with β-catenin, and negatively regulates the expression
of its downstream factors (16). The
present study assessed the expression levels of Cyclin D1 and
c-Myc, which are the downstream factors of the Wnt/β-catenin
signaling pathway (17). As
presented in Fig. 4A-F,
miR-182-knockdown downregulated the mRNA and protein expression
levels of Cyclin D1 and c-Myc in GBC cells transfected with miR-182
inhibitors.
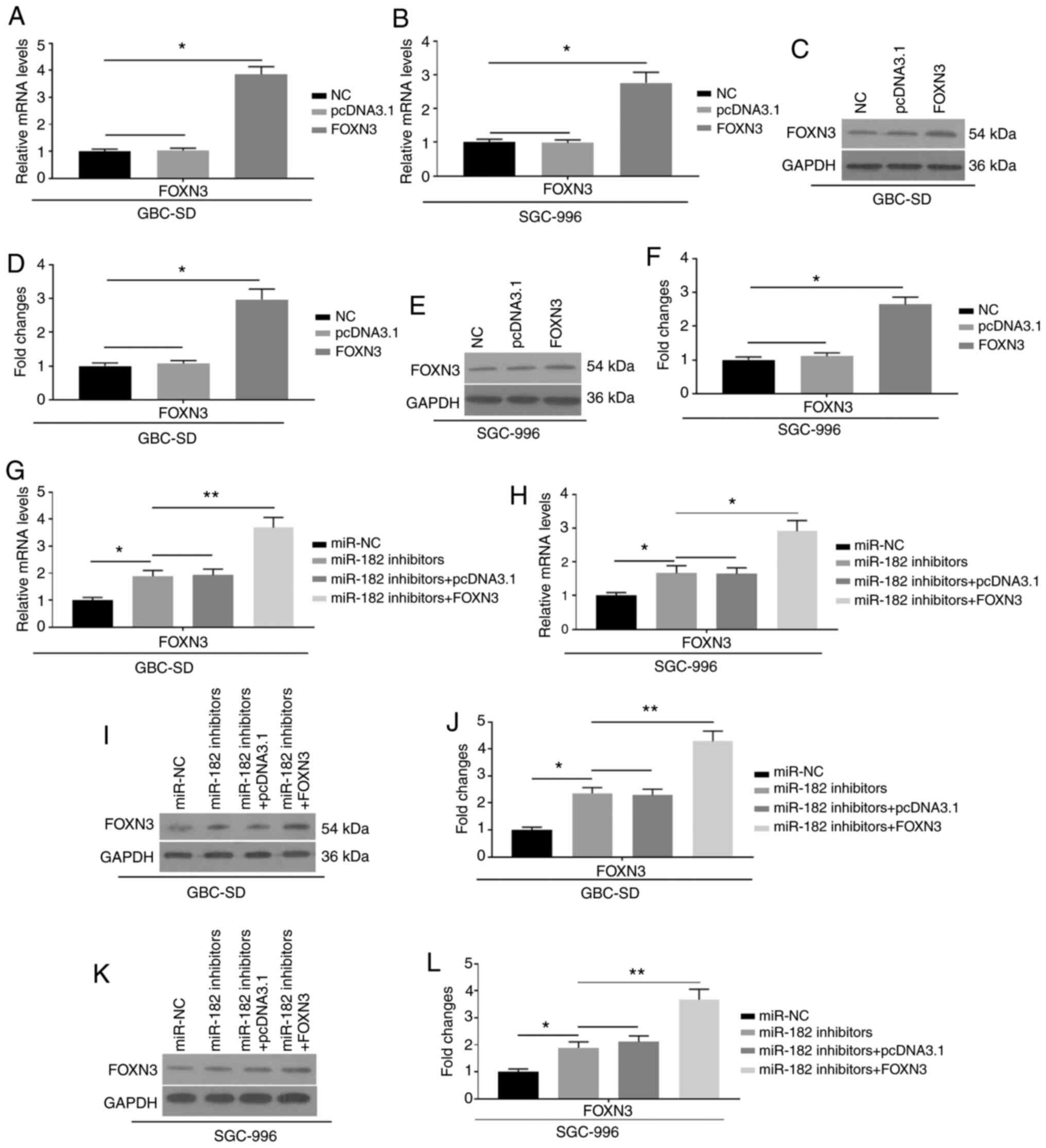
To further investigate the effects of miR-182 on
EMT, FOXN3 was overexpressed by transfection with pcDNA3.1-FOXN3
into GBC cells, which were also co-transfected with miR-182
inhibitors. The mRNA (Fig. 5A and B)
and protein expression (Fig. 5C-F)
levels of FOXN3 were significantly upregulated, indicating
successful transfection of FOXN3 into GBC-SD and SGC-996 cells.
Furthermore, successful co-transfection of FOXN3 and miR-182
inhibitors is indicated in Fig.
5G-L. Notably, the overexpression of FOXN3 was demonstrated to
compromise the promoting activity of miR-182 on EMT, as indicated
by the increased migration and invasion (Fig. 6A-C), increased E-cadherin expression,
and decreased vimentin expression (Fig.
6D-G). In addition, overexpression of FOXN3 ameliorated the
promoting activity of miR-182 on the expression levels of Cyclin D1
and c-Myc. The results of the Co-IP assay demonstrated that FOXN3
may form a complex with β-catenin in GBC cells by competing with
TCF4 (Fig. 6H and I). Taken
together, these results suggested that miR-182 may promote EMT by
targeting FOXN3 and negatively regulating the Wnt/β-catenin pathway
in GBC cells.
 | Figure 6.Overexpression of FOXN3 compromises
the effects of miR-182 on epithelial-mesenchymal transition in GBC
cells co-transfected with miR-182 inhibitors and pcDNA3.1-FOXN3.
(A) The cell migratory and invasive abilities were detected via the
Transwell assays following transfection with miR-NC and FOXN3,
respectively. (B) The summarized staining levels for migration. (C)
The summarized staining levels for invasion. Western blot analysis
was performed to detect the protein expression levels of
E-cadherin, Vimentin, Cyclin D1 and c-Myc in (D) GBC-SD and (E)
SGC-996 cells. (F) The summarized data of (D). (G) The summarized
data of (E). (H and I) The Co-IP assay was performed to assess the
association between FOXN3 and β-catenin. Cell protein extracts
(10%) were used as the input sample, which was subjected to Western
blot analysis. The remaining protein extracts were subjected to IP
using control goat IgG or β-catenin antibodies, followed by IB with
anti-FOXN3 or anti-TCF4. All experiments were performed in
triplicate and data are presented as the mean ± standard deviation.
*P<0.05 and **P<0.01. FOXN3, Forkhead box N3; miR, microRNA;
GBC, gallbladder cancer; NC, negative control; IP,
immunoprecipitation; IB, immunoblotting. |
Discussion
GBC is the most common type of cancer in the biliary
tract, and only 20% of patients with GBC are diagnosed with
non-metastatic GBC (18). Studies of
the molecular mechanisms of GBC pathogenesis are required to
identify and develop effective therapeutic strategies. EMT is
associated with the invasion and metastasis of GBC (19). The results of the present study
demonstrated that miR-182 was significantly upregulated in GBC
tissues and cell lines. miR-182-knockdown was demonstrated to
inhibit EMT. This may be due to the fact that miR-182 targets and
degrades FOXN3, which suppressed the activity of the β-catenin
pathway by competing with TCF4. The rescue assays from
pcDNA3.1-FOXN3 transfection supported that miR-182/FOXN3 promoted
EMT by mediating the β-catenin pathway in GBC cells.
Increasing evidence has demonstrated an association
between miRNA expression profiles and cancer development. Recently,
the pathogenesis of GBC has been indicated to be associated with
the dysregulation of miRNAs (7,20).
miR-181b expression is increased in GBC tissues, and it may promote
cell proliferation and autophagy, and attenuate cell apoptosis in
GBC cells by targeting CREBRF, which is a suppressor of autophagy
(21). miR-7 and miR-29c have been
demonstrated to be downregulated in GBC, while the overexpression
of these two miRNAs may effectively reverse EMT and decrease the
metastatic activity of GBC cells (22). Overexpression of miR-182 promotes
motility and the invasive ability of hepatocellular carcinoma cells
by targeting FOXO3A, which represses the activity of Wnt/β-catenin
signaling (23). Recently, miR-182
has been reported to promote transforming growth factor
(TGF)-β-mediated migration and invasion in GBC cells by targeting
the cell adhesion molecule 1 (24).
Consistent with these findings, the results of the present study
demonstrated the aberrant expression of miR-182 in GBC tissues. In
addition, miR-182-knockdown in GBC cells may effectively attenuate
EMT, as demonstrated by decreased migration and invasion, decreased
vimentin expression and increased E-cadherin expression.
Several studies have reported that activation of the
Wnt/β-catenin signaling pathway may promote the transcription of
EMT-related genes (25–27). β-catenin interacts with SMAD3 and
promotes TGF-β-regulated EMT in lens epithelial cells (25). In addition, β-catenin is required for
TGF-β to induce EMT in lens epithelial cells by interacting with
SMAD3 and translocation into the nucleus (27). The expression of β-catenin and its
downstream factors, Cyclin D1 and c-Myc, are augmented by
H2O2, accompanied by an increase of EMT in
human lens epithelial cells (28).
miR-425 has been demonstrated to promote cell proliferation,
migration, invasion and EMT by activating the β-catenin pathway
(29). miR-23b is involved in
tumor-promoting effects, which are associated with the activation
of the JAK/STAT and Wnt/β-catenin pathways in A549 cells (30). In addition, the key EMT factor, ZEB1,
is transcriptionally regulated by the β-catenin/TCF4 complex
(11). These findings suggested that
activation of the Wnt/β-catenin pathways may promote EMT. The
overexpression of miR-182 has been demonstrated to target SMAD7,
leading to an increase in TGFβ-induced EMT and osteoclastogenesis
for bone metastasis in cancer cells (31). The results of the present study
demonstrated that the transfection with miR-182 inhibitors
decreased migration and invasion, and decreased the expression
levels of Cyclin D1 and c-Myc, which are the two downstream factors
of the Wnt/β-catenin pathway.
FOXN3 is a tumor suppressor, and alternative FOXN3
expression profiles are often observed in different types of
cancer, including melanoma, osteosarcoma and hepatocellular
carcinoma (32). The association
between FOXN3 and tumor development has been reviewed
comprehensively (32). FOXN3 has
been demonstrated to regulate the TGF-β/SMAD signaling pathway,
which modulates the proliferation and differentiation of cancer
cells (33). Overexpression of FOXN3
significantly inhibits tumor growth of papillary thyroid carcinoma,
accompanied by decreased expression of the β-catenin pathway
(12). Furthermore, FOXN3 may
interact with β-catenin and block the interaction between β-catenin
and TCF4 in colon cancer (16). The
present study on the prediction from the system on the website
indicated that miR-182 may bind to the 3′-UTR of FOXN3. The results
of the present study demonstrated that the overexpression of FOXN3,
similar to miR-182 inhibitors transfection, may effectively
attenuate EMT in GBC cells, as indicated by decreased migration and
invasion, decreased vimentin expression, and increased E-cadherin
expression. The present study also demonstrated that FOXN3 may
co-precipitate with β-catenin, interrupting the interaction between
β-catenin and TCF4.
Further studies were still required. The biological
effects of FOXN3 in the pathological development of GBC were not
fully understood. In addition, the positive effects of miR-182 on
the regulation of cellular metabolism in the gallbladder were still
absent, although the deficiency of miR-182 may ameliorate EMT.
However, the miR-182-knockout mice may be further used to confirm
the effects of miR-182 on EMT in GBC. Taken together, the results
of the present study demonstrated that miR-182 expression was
increased in GBC tissues and cells, and miR-182-knockdown
ameliorated EMT. The potential molecular mechanisms may be that
miR-182 targets and degrades FOXN3, which competes with TCF4 for
binding to β-catenin and suppresses the Wnt/β-catenin pathway.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
National Science Foundation of China (grant no. 81960883),
Scientific Research Fund of Jiangxi Provincial Education Department
(grant no. GJJ180820), Science and Technology Plan of Jiangxi
Health and Family Planning Commission (grant no. 20195366), Project
of the First Affiliated Hospital of Gannan Medical University
(grant no. YJZD202001) and Key Research and Development Projects of
Ganzhou Science and Technology Program (Title: Clinical application
of multidisciplinary diagnosis and treatment of primary liver
cancer; 2018).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YX designed the study and wrote the manuscript. JZ,
ZH, CW, QL, BH, JP, XT and ZC performed the experiments and
analyzed the data. QL, BH, JP, XT and ZC revised and finalized the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Gannan Medical
University, Ganzhou, China (GMU38972623) and was performed in
accordance with the Declaration of Helsinki (13). Written informed consent was provided
by all patients prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wennmacker SZ, Lamberts MP, Di Martino M,
Drenth JP, Gurusamy KS and van Laarhoven CJ: Transabdominal
ultrasound and endoscopic ultrasound for diagnosis of gallbladder
polyps. Cochrane Database Syst Rev. 8:CD0122332018.PubMed/NCBI
|
|
2
|
Cubertafond P, Gainant A and Cucchiaro G:
Surgical treatment of 724 carcinomas of the gallbladder. Results of
the French Surgical Association Survey. Ann Surg. 219:275–280.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zali MR, Zamanian Azodi M, Razzaghi Z and
Heydari MH: Gallbladder cancer integrated bioinformatics analysis
of protein profile data. Gastroenterol Hepatol Bed Bench. 12 (Suppl
1):S66–S73. 2019.PubMed/NCBI
|
|
4
|
Lee HE, Huh JW and Kim HS: Bioinformatics
analysis of evolution and human disease related transposable
element-derived microRNAs. Life (Basel). 10:952020.
|
|
5
|
Lei R, Tang J, Zhuang X, Deng R, Li G, Yu
J, Liang Y, Xiao J, Wang HY, Yang Q and Hu G: Suppression of MIM by
microRNA-182 activates RhoA and promotes breast cancer metastasis.
Oncogene. 33:1287–1296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang MH, Yu J, Jiang DM, Li WL, Wang S and
Ding YQ: microRNA-182 targets special AT-rich sequence-binding
protein 2 to promote colorectal cancer proliferation and
metastasis. J Transl Med. 12:1092014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chandra V, Kim JJ, Mittal B and Rai R:
MicroRNA aberrations: An emerging field for gallbladder cancer
management. World J Gastroenterol. 22:1787–1799. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao M, Ang L, Huang J and Wang J:
MicroRNAs regulate the epithelial-mesenchymal transition and
influence breast cancer invasion and metastasis. Tumour Biol.
39:10104283176916822017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li S, Liu F, Xu L, Li C, Yang X, Guo B, Gu
J and Wang L: Wnt/β-catenin signaling axis is required for
TFEB-mediated gastric cancer metastasis and epithelial-mesenchymal
transition. Mol Cancer Res. 18:1650–1659. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu L, Huang X, Li L, Huang H, Xu R and
Luyten W: Insights on biology and pathology of HIF-1α/-2α,
TGFβ/BMP, Wnt/β-catenin, and NF-κB pathways in osteoarthritis. Curr
Pharm Des. 18:3293–3312. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sánchez-Tilló E, de Barrios O, Siles L,
Cuatrecasas M, Castells A and Postigo A: β-catenin/TCF4 complex
induces the epithelial-to-mesenchymal transition (EMT)-activator
ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci USA.
108:19204–19209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao C, Mo L, Li C, Han S, Zhao W and Liu
L: FOXN3 suppresses the growth and invasion of papillary thyroid
cancer through the inactivation of Wnt/β-catenin pathway. Mol Cell
Endocrinol. 515:1109252020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y,
Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes
tumor progression by regulating the miR-143/HK2 axis in gallbladder
cancer. Mol Cancer. 18:332019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
16
|
Dai Y, Wang M, Wu H, Xiao M, Liu H and
Zhang D: Loss of FOXN3 in colon cancer activates beta-catenin/TCF
signaling and promotes the growth and migration of cancer cells.
Oncotarget. 8:9783–9793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim GH, Fang XQ, Lim WJ, Park J, Kang TB,
Kim JH and Lim JH: Cinobufagin suppresses melanoma cell growth by
inhibiting LEF1. Int J Mol Sci. 21:67062020. View Article : Google Scholar
|
|
18
|
Mishra SK, Kumari N and Krishnani N:
Molecular pathogenesis of gallbladder cancer: An update. Mutat Res.
816-818:1116742019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakada S, Kuboki S, Nojima H, Yoshitomi H,
Furukawa K, Takayashiki T, Takano S, Miyazaki M and Ohtsuka M:
Roles of Pin1 as a key molecule for EMT induction by activation of
STAT3 and NF-κB in human gallbladder cancer. Ann Surg Oncol.
26:907–917. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Jin Y, Li S, Song Q and Tang P:
Identification of microRNAs associated with the survival of
patients with gallbladder carcinoma. J Int Med Res.
48:3000605209180612020.PubMed/NCBI
|
|
21
|
Wu K, Huang J, Xu T, Ye Z, Jin F, Li N and
Lv B: MicroRNA-181b blocks gensenoside Rg3-mediated tumor
suppression of gallbladder carcinoma by promoting autophagy flux
via CREBRF/CREB3 pathway. Am J Transl Res. 11:5776–5787.
2019.PubMed/NCBI
|
|
22
|
Lu K, Feng F, Yang Y, Liu K, Duan J, Liu
H, Yang J, Wu M, Liu C and Chang Y: High-throughput screening
identified miR-7-2-3p and miR-29c-3p as metastasis suppressors in
gallbladder carcinoma. J Gastroenterol. 55:51–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY,
Zhang SZ, Zhang KW, Cai H, Shi WK, Li XL, et al: miR-182-5p
promotes hepatocellular carcinoma progression by repressing FOXO3a.
J Hematol Oncol. 11:122018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qiu Y, Luo X, Kan T, Zhang Y, Yu W, Wei Y,
Shen N, Yi B and Jiang X: TGF-β upregulates miR-182 expression to
promote gallbladder cancer metastasis by targeting CADM1. Mol
Biosyst. 10:679–685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Charbonney E, Speight P, Masszi A, Nakano
H and Kapus A: β-catenin and Smad3 regulate the activity and
stability of myocardin-related transcription factor during
epithelial-myofibroblast transition. Mol Biol Cell. 22:4472–4485.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou B, Liu Y, Kahn M, Ann DK, Han A, Wang
H, Nguyen C, Flodby P, Zhong Q, Krishnaveni MS, et al: Interactions
between β-catenin and transforming growth factor-β signaling
pathways mediate epithelial-mesenchymal transition and are
dependent on the transcriptional co-activator cAMP-response
element-binding protein (CREB)-binding protein (CBP). J Biol Chem.
287:7026–7038. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taiyab A, Holms J and West-Mays JA:
β-catenin/Smad3 interaction regulates transforming growth
factor-β-induced epithelial to mesenchymal transition in the lens.
Int J Mol Sci. 20:20782019. View Article : Google Scholar
|
|
28
|
Li J, Chen Y, Han C, Huang S, Chen S, Luo
L and Liu Y: JNK1/β-catenin axis regulates
H2O2-induced epithelial-to-mesenchymal
transition in human lens epithelial cells. Biochem Biophys Res
Commun. 511:336–342. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu D, Zhang H, Cui M, Chen C and Feng Y:
Hsa-miR-425-5p promotes tumor growth and metastasis by activating
the CTNND1-mediated β-catenin pathway and EMT in colorectal cancer.
Cell Cycle. 19:1917–1927. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang L, Hu Z, Guo Q, Yang L, Pang Y and
Wang W: miR-23b functions as an oncogenic miRNA by downregulating
Mcl-1S in lung cancer cell line A549. J Biochem Mol Toxicol.
34:e224942020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu J, Lei R, Zhuang X, Li X, Li G, Lev S,
Segura MF, Zhang X and Hu G: MicroRNA-182 targets SMAD7 to
potentiate TGFβ-induced epithelial-mesenchymal transition and
metastasis of cancer cells. Nat Commun. 7:138842016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kong X, Zhai J, Yan C, Song Y, Wang J, Bai
X, Brown JAL and Fang Y: Recent advances in understanding FOXN3 in
breast cancer, and other malignancies. Front Oncol. 9:2342019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kirmizitas A, Meiklejohn S, Ciau-Uitz A,
Stephenson R and Patient R: Dissecting BMP signaling input into the
gene regulatory networks driving specification of the blood stem
cell lineage. Proc Natl Acad Sci USA. 114:5814–5821. 2017.
View Article : Google Scholar : PubMed/NCBI
|