Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly
aggressive malignancy with an overall 5-year survival rate of ~9%
(1). Globally, PDAC-associated
deaths are predicted to increase by 0.5% in 2030 (2). Due to the lack of specific symptoms for
early diagnosis and a tendency to metastasize quickly, the majority
of patients present with late stage PDAC at diagnosis (3). Consequently, even after
pancreaticoduodenectomy, the 5-year survival rate remains <20%
(3). Additional diagnostic and
prognostic markers are required to improve early detection, the
assessment of disease activity and prognosis, and therapeutic
decisions and monitoring, as well as for the development of novel
treatment strategies.
Numerous biomarkers have been identified and
developed to improve the early diagnosis of PDAC (4). Among the genetic factors, BRCA1 and
BRCA2 are two well-characterized tumor suppressor genes implicated
in gene transcription and DNA repair (5). Gene mutations in BRCA1 and BRCA2 are
associated with breast, ovarian, colorectal and prostate cancer,
and genetic variants in these genes have also been associated with
the risk of PDAC (6,7).
Partner and localizer of BRCA2 (PALB2) was first
identified as a protein that co-localized with BRCA2 in the
nucleus; PALB2 serves as a linker between BRCA1 and BRCA2 in the
repair of DNA breaks (8). Mutations
in PALB2 are responsible for Fanconi anemia complementation group N
and are associated with childhood cancers, such as solid tumors of
the kidney (Wilms' tumor), medulloblastoma and neuroblastoma
(9,10). Additionally, single nucleotide
polymorphisms in the PALB2 gene have been associated with the risk
of breast cancer (11,12). The association among hereditary
mutations in PALB2, PDAC risk and incidence rates exhibits
population-specific characteristics (13–16).
Compared with no or mild mutations (replacement of a single amino
acid with a structurally related one), inactivating mutations in
BRCA1, BRCA2 or PALB2 confer a more favorable prognosis in patients
with PDAC (17), potentially due to
higher genomic instability and an improved response to
platinum-based chemotherapy (18–20). In
addition, carbohydrate antigen 19-9 displays some prognostic value,
but standardized predictive biomarkers to assess treatment success
have not yet been identified (21).
The mRNA and protein expression levels of PALB2, as
well as the role of PALB2 in PDAC tissues and its potential
prognostic value, are not completely understood. Therefore, the
present study investigated PALB2 expression in human PDAC cell
lines and PDAC tumor and peritumoral tissue sections. Moreover, the
association between PALB2 expression and disease characteristics or
patient survival was further assessed in the present study.
Materials and methods
Patients and sample collection
Human PDAC tissue microarrays, as well as human
breast and gastric cancer tissue microarrays (including 15
surgically resected breast/gastric tumor tissues), were obtained
from Shanghai BioChip Co., Ltd. PDAC tissue microarray chips
contained 157 tumor tissue samples (Table I) and 121 peritumoral tissues, which
were free of signs of malignant transformation and distant from the
tumor. The information on patient survival ranged from 1.2 to 7.0
years. TNM staging (7th edition) and American Joint Committee on
Cancer staging data (7th edition) (22) were available for the majority of PDAC
samples (the data were not available for 23 samples). Expression
levels of tumor-associated markers, including Ki-67, p53, CD8 and
programmed death ligand 1 (PDL1), were obtained from the database
of the pathology department of the National Engineering Center of
Shanghai BioChip Co., Ltd. All patients with PDAC were diagnosed by
positive histology of invasive ductal carcinoma. Overall survival
(OS) was calculated as the time interval between the date of
surgery and the date of death or last follow-up visit. Not all
deceased subjects were subjected to pathological analysis, and
therefore the rate of PDAC-associated death is unknown.
 | Table I.Clinicopathological characteristics
of patients with PDAC (n=157) and PALB2 protein expression in PDAC
tissues. |
Table I.
Clinicopathological characteristics
of patients with PDAC (n=157) and PALB2 protein expression in PDAC
tissues.
| Features | PALB2 negative,
n=93 | PALB2 positive,
n=64 | P-value |
|---|
| Mean age ± SD,
years | 64.1±11.0 | 62.6±10.1 | 0.3851 |
| Age range,
years | 41–85 | 34–81 |
|
| Sex, n (%) |
|
| 0.9325 |
|
Male | 56 (58.3) | 40 (41.7) |
|
|
Female | 36 (59.0) | 25 (41.0) |
|
| T stage, n
(%)a |
|
| 0.0554 |
|
T1-T2 | 55 (66.7) | 27 (33.3) |
|
| T3 | 26 (50.0) | 26 (50.0) |
|
| N stage, n
(%)a |
|
| 0.4895 |
| N0 | 47 (63.5) | 27 (36.5) |
|
| N1 | 34 (57.6) | 25 (42.4) |
|
| AJCC, n
(%)a |
|
| 0.3948 |
| I | 28 (68.2) | 13 (31.8) |
|
|
IIA | 18 (58.1) | 13 (41.9) |
|
|
IIB | 33 (60.0) | 22 (40.0) |
|
| IV | 2
(33.3) | 4
(66.7) |
|
| Tumor location |
|
| 0.6776 |
|
Head | 54 (57.6) | 39 (42.4) |
|
|
Body/Tail | 39 (60.9) | 25 (39.1) |
|
| Ki-67 expression,
nb |
|
| 0.3718 |
|
Positive | 47 (75.8) | 15 (24.2) |
|
|
Negative | 18 (66.7) | 9
(33.3) |
|
| P53 expression,
nb |
|
| 0.1810 |
|
Positive | 48 (69.5) | 21 (30.5) |
|
|
Negative | 17 (85.0) | 3
(15.0) |
|
| PDL1 expression,
nc |
|
| 0.1795 |
|
Positive | 19 (48.7) | 20 (51.3) |
|
|
Negative | 5
(29.4) | 12 (70.6) |
|
| CD8 expression,
nc |
|
| 0.8460 |
|
Positive | 19 (42.2) | 26 (57.8) |
|
|
Negative | 5
(45.5) | 6
(54.5) |
|
Public data resource analysis
The level 3 information (aggregated, normalized
and/or segmented data) of The Cancer Genome Atlas (TCGA, http://portal.gdc.cancer.gov/) containing
pancreatic adenocarcinoma datasets of 178 individual tumors were
downloaded for expression analysis (survival data were available
for 177 patients). The Root Mean Squared Error normalized mRNA
count (‘count’), which represents PALB2 gene expression, was
evaluated. For Gene Set Enrichment Analysis (GSEA), the latest
official tool was downloaded from software.broadinstitute.org/gsea (version 3.0). The
cBio Cancer Genomics Portal (cbioportal.org) was used to analyze PALB2 mRNA
expression across different types of human cancer, based on TCGA
public database.
Immunohistochemistry (IHC) and
evaluation of immunostaining
Thin slices (4-µm-thick sections) from 10%
formalin-fixed (24 h at room temperature), paraffin-embedded tissue
specimens were used. The sections were deparaffinized in xylene and
rehydrated in a descending alcohol series (100, 95, 90, 75 and
70%), using routine procedures. Antigen retrieval was achieved by
digesting the sections with proteinase K (0.2 mg/ml at 37°C for 10
min) before IHC, followed by repeated washing steps and blocking
with 3% H2O2 for 30 min at room temperature.
Sections were incubated with a rabbit polyclonal anti-PALB2
antibody (1:2,000; cat. no. ab202970; Abcam) for 12 h at 4°C and
then with an HRP-conjugated anti-rabbit secondary antibody
(1:5,000; cat. no. P044801; Dako; Agilent Technologies, Inc.) for
30 min at room temperature. Tissue microarray slides were scanned
using a Leica Aperio digital slide scanner (Leica Microsystems
GmbH). Tumor cells displaying staining in the nucleus were
categorized as positively stained. The percentage of
PALB2+ tumor cells was calculated as the ratio of
stained to unstained tumor cells. The percentages of positive cells
were classified into five scores: 0, 0%; 1, 1–5%; 2, 6–30%; 3,
31–60%; and 4, 61–100% positively stained cells. The scoring of
each tissue section was conducted independently by two
pathologists. In cases of inconsistent results (~15% of samples),
the pathologists re-evaluated the sample in question together to
achieve a consensus. Tissue sections with scores 0–1 were
considered as low expression, whereas scores of 2–4 were considered
as high expression. According to the information from TCGA, gastric
cancer tissues are known to express low PALB2 protein levels, while
breast cancer tissues are known for high PALB2 expression;
therefore, the reliability of staining with the commercial PALB2
antibody was tested in a gastric cancer tissue as a negative
control, and a breast cancer tissue as a positive control.
Cell lines, cell culture and small
interfering (si)RNA
SW1990, PANC1 and CFPAC1 cell lines were purchased
from The Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences. SW1990, PANC1 and CFPAC1 cells were maintained
at 37°C with 5% CO2 and cultured in DMEM supplemented
with 10% FBS (both Gibco; Thermo Fisher Scientific, Inc.). A siRNA
targeting PALB2 (si-PALB2) and a scrambled negative control siRNA
(si-NC) were purchased from Santa Cruz Biotechnology, Inc. (cat.
nos. sc-93396 and sc-37007). Transfection was performed using
standard protocols of Lipofectamine® 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Lipofectamine 3000 reagent and
siRNAs were diluted separately with OPTI-MEM (Gibco; Thermo Fisher
Scientific, Inc.) in a centrifuge tube, then Lipofectamine 3000 and
siRNAs were mixed and incubated for 15 min at room temperature.
Subsequently, DNA-lipid complex was added to the cells and
incubated for 48 h at 37°C. The transfected cells were used for
subsequent experiments.
Cell proliferation and migration via
wound healing assay
Cell proliferation was assessed by performing the
Cell Counting Kit-8 assay (Dojindo Molecular Technologies, Inc.).
Briefly, cells were seeded into 96-well plates. After 0, 24 or 48
h, the number of viable cells was quantified after incubation with
the CCK-solution for 2 h by measuring the optical density at a
wavelength of 450 nm using a microplate reader. For the wound
healing assay, cells were seeded into 6-well plates and transfected
with si-PALB2 or si-NC. At 24 h post-transfection, a scratch wound
was made in the confluent cell monolayer using a 200-µl pipette
tip. Cells were cultured in serum-free medium. The wound was
observed at 0 and 48 h with a light microscope (×100 magnification)
after scratching. Cell migration area was measured using ImageJ
(v1.52q; National Institutes of Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from cells was isolated using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
Total RNA (1 µg) was reverse transcribed into cDNA using a ReverTra
Ace qPCR RT kit (Toyobo Life Science) according to the
manufacturer's protocol. Subsequently, qPCR was performed using
SYBR Green Gene Expression Assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.) and an AB7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Thermocycling
conditions included denaturation at 95°C for 15 sec, followed by
annealing at 60°C for 10 sec and elongation at 72°C for 20 sec, for
40 cycles. The following primers were used for qPCR: Snail family
transcriptional repressor 1 (Snail) forward,
5′-ACCACTATGCCGCGCTCTT-3′ and reverse, 5′-GGTCGTAGGGCTGCTGGAA-3′;
snail family transcriptional repressor 2 (Slug) forward,
5′-ATGAGGAATCTGGCTGCTGT-3′ and reverse, 5′-CAGGAGAAAATGCCTTTGGA-3′;
zinc finger E-box binding homeobox 1 (Zeb1) forward,
5′-GCACCTGAAGAGGACCAGAG-3′ and reverse, 5′-TGCATCTGGTGTTCCATTTT-3′;
Vimentin forward, 5′-GACGCCATCAACACCGAGTT-3′ and reverse,
5′-CTTTGTCGTTGGTTAGCTGGT-3′; and GAPDH forward,
5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′.
mRNA expression levels were quantified using the 2−∆∆Cq
method (23) and normalized to the
internal reference gene GAPDH.
Western blotting
Total protein was extracted from pancreatic cancer
cell lines using RIPA lysis buffer (cat. no. 89900; Thermo Fisher
Scientific, Inc.). Protein concentration was measured using BCA kit
and western blotting was performed according to standard protocols.
Briefly, 30 µg protein/lane was separated via 8% SDS-PAGE and
transferred to a PVDF membrane. The membrane was blocked with 5%
BSA (cat. no. 9998; Cell Signaling Technology, Inc.) for 2 h at
room temperature. Subsequently, the membrane was incubated with the
following primary antibodies: Anti-PALB2 (1:1,000; cat. no.
ab202970; Abcam) and anti-β-actin (1:2,000; cat. no. sc-130656;
Santa Cruz Biotechnology, Inc.) for 12 h at 4°C. A mouse
anti-rabbit IgG HRP-conjugated secondary antibody (1:2,000; cat.
no. sc-2357; Santa Cruz Biotechnology, Inc.) was used and the
membrane was incubated for 30 min at room temperature. TBS-Tween
(0.5% Tween) was used for membrane washing, and an ECL kit (Bio-Rad
Laboratories, Inc.) was used for visualization. Protein expression
levels were semi-quantified using ImageJ software v1.52q (National
Institutes of Health) with β-actin as the loading control.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism (v8.0; GraphPad Software, Inc.) or SPSS (v24.0; IBM Corp.)
softwares. Continuous variables were compared using the unpaired
Student's t-test or Mann-Whitney U test. The χ2 test was
used to analyze the distribution of categorical variables between
PALB2− and PALB2+ groups. OS was plotted as a
Kaplan-Meier survival curve with 95% CIs, and differences between
subgroups were compared using log-rank tests. Cox regression
analysis was performed to identify independent prognostic factors.
P<0.05 was considered to indicate a statistically significant
difference.
Results
PALB2 IHC
To establish the IHC method and test whether the
selected antibody selected was capable of yielding congruent data
in agreement with current knowledge, stomach and breast carcinoma
tissues were measured for PALB2 expression as negative and positive
controls, respectively. Weak staining signals were observed for the
negative control stomach cancer tissue (Fig. S1; upper panels), while strong
staining signals were observed for the positive control breast
carcinoma tissues (Fig. S1; lower
panels). The results supported the suitability of the antibody and
the IHC protocol, and the staining pattern was consistent with the
results obtained using the public TCGA database.
PALB2 expression in PDAC tissues
Subsequently, PALB2 expression in PDAC samples was
analyzed using characterized tissue samples arranged in an array
format. The staining patterns were evaluated by two independent
pathologists and a consensus was reached on the resulting data. The
number of PALB2− samples was slightly higher compared
with that of PALB2+ samples within the array of 157
tumor samples analyzed (Table I). In
PALB2+ samples, compared with in the para-tumor normal
tissues, staining was stronger in PDAC tissues (Fig. 1A). PALB2+ cell nuclei in
PDAC samples and peritumoral tissues were identified and counted in
relation to unlabeled cells. Overall, the positive rates for PALB2
were 24.8% in para-tumor healthy tissues and 40.8% in PDAC tissues.
The distribution of the samples with respect to the IHC score of
0–4 for the PDAC and peritumor tissue samples are presented in
Table SI. Representative images of
PALB2 IHC labeling for each score are shown in Fig. 1B.
Association between PALB2 and
classical cancer markers
To assess whether PALB2 expression was associated
with classical cancer markers, the expression pattern of PALB2
expression in tumor and peritumoral tissues was assessed. The
results indicated that PALB2 expression was significantly lower in
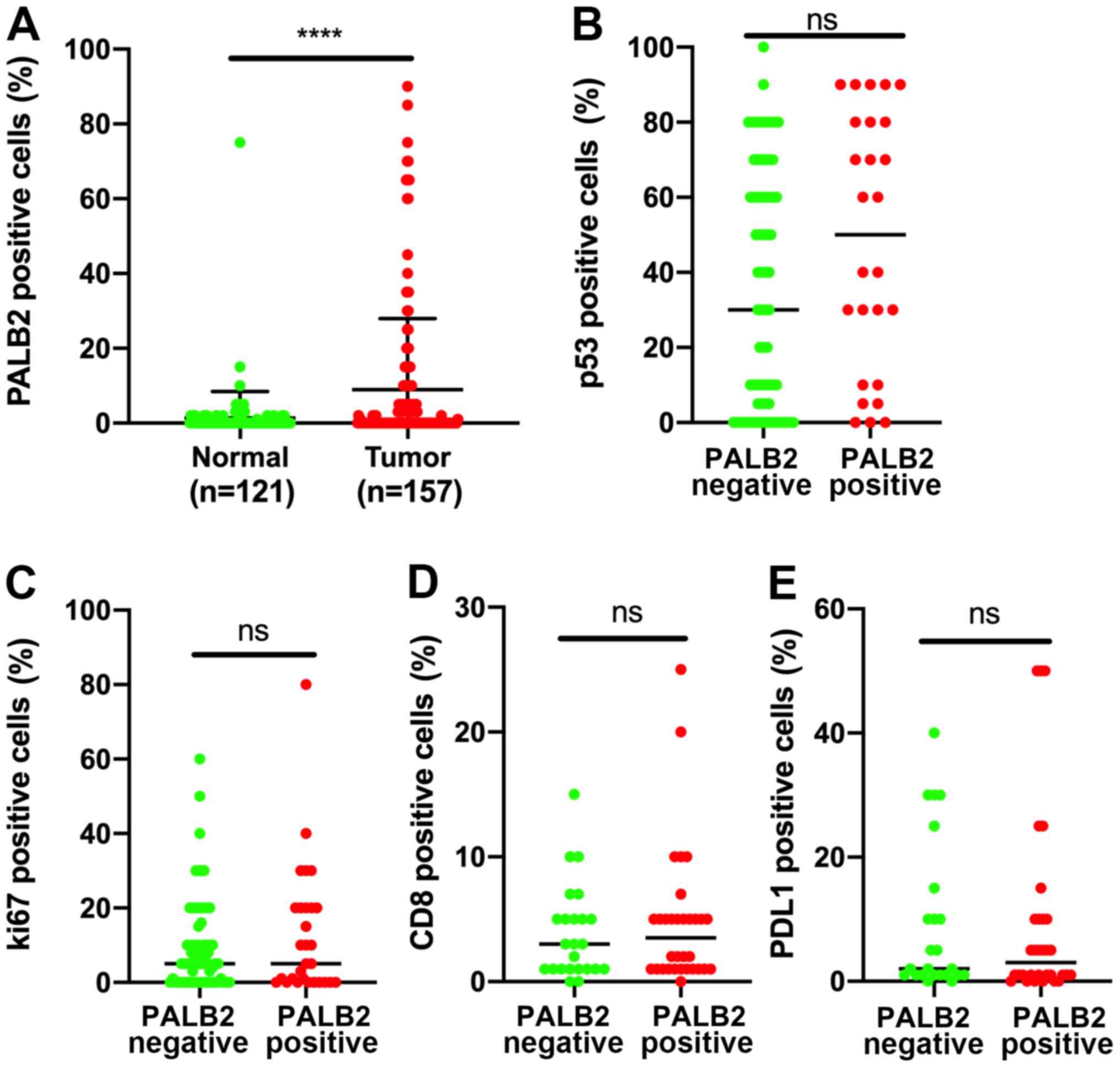
peritumoral tissues compared with in PDAC tissues (Fig. 2A). A direct comparison of Ki-67, p53,
CD8 and PDL1 staining patterns in tissues with positive or negative
PALB2 expression indicated no significant differences, suggesting
that the expression levels of Ki-67, p53, CD8 and PDL1 were
independent from PALB2 expression (Fig.
2B-E),
Association of PALB2 and other
parameters with OS in patients with PDAC
The association between patient clinical
characteristics, including tumor PALB2 expression, and OS was
analyzed in 157 patients. Multivariate and univariate Cox
proportional hazard regression analyses identified that N stage,
age and PALB2 expression were significantly associated with OS in
patients with PDAC (Table II).
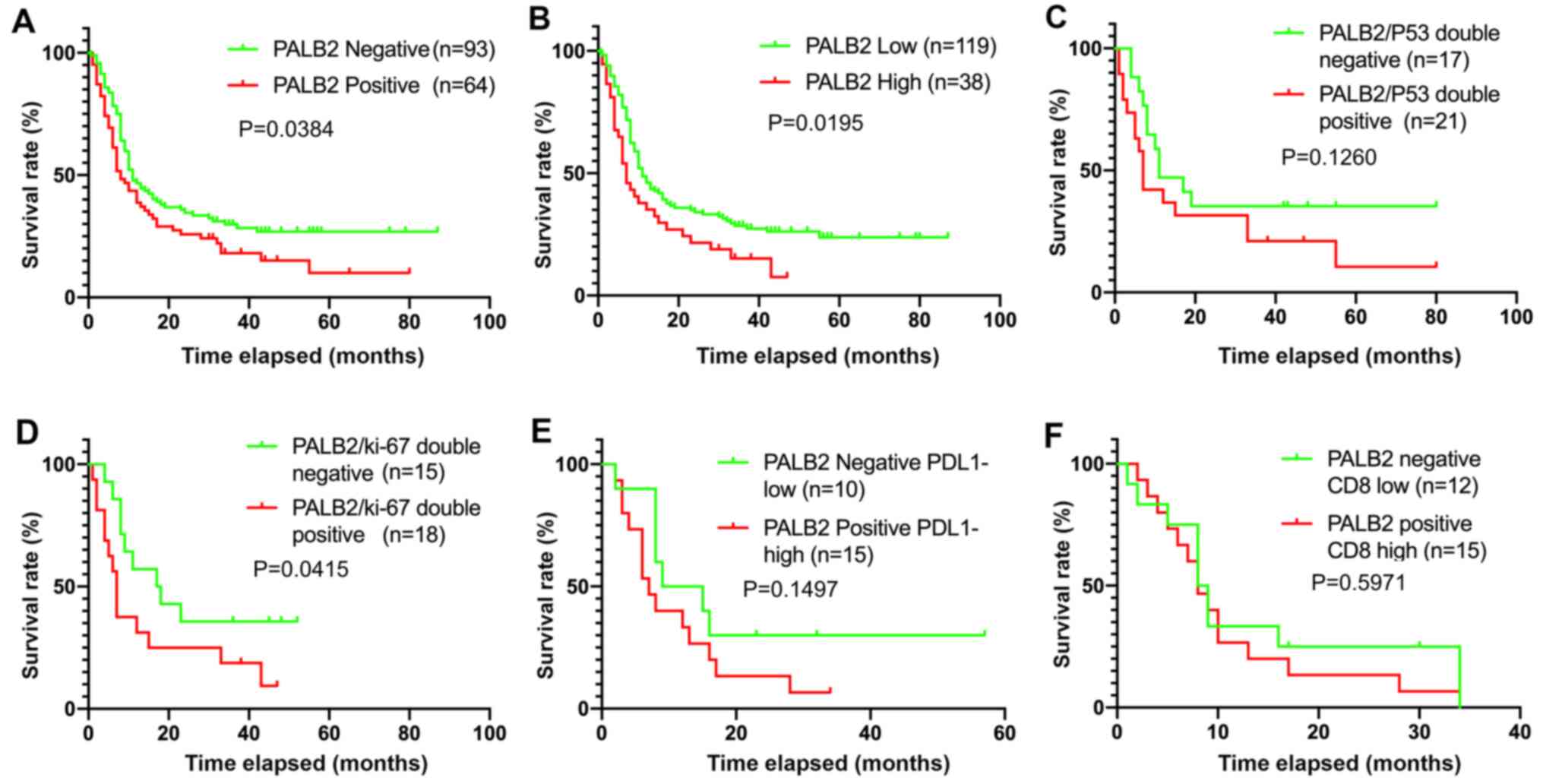
Kaplan-Meier analysis revealed that patients with positive PALB2
expression had a poorer OS rate compared with patients with
negative PALB2 expression (P=0.0384; Fig. 3A). Furthermore, patients were divided
into two groups: Low PALB2 expression (positive rate ≤5%; n=119)
and high PALB2 expression (positive rate >5%; n=38).
Kaplan-Meier curves demonstrated a significant negative association
between high PALB2 expression and OS (P=0.0195; Fig. 3B). Therefore, high PALB2 expression
may indicate a poor prognosis in patients with surgically
resectable PDAC. By contrast, co-expression of p53 and PALB2 was
not significantly associated with survival (P=0.1260; Fig. 3C), whereas co-expression of PALB2 and
Ki-67 indicated a poor prognosis in patients with PDAC (P=0.0415;
Fig. 3D). In addition, co-expression
of PALB2 and the cancer immune markers CD8 and PDL1 were not
associated with survival (P=0.1497; Fig.
3E and P=0.5971; Fig. 3F).
 | Table II.Multivariate Cox regression analyses
of the overall survival of 157 patients with PDAC and PDAC samples
represented in the TCGA database (May 2019). |
Table II.
Multivariate Cox regression analyses
of the overall survival of 157 patients with PDAC and PDAC samples
represented in the TCGA database (May 2019).
| A, Patients with
PDAC (n=157) |
|---|
|
|---|
|
| Multivariate
analysis | Univariate
analysis |
|---|
|
|
|
|
|---|
| Factors | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex
(Male/Female) | 1.375
(0.913–2.071) | 0.128 | 1.182
(0.816–1.712) | 0.377 |
| Age (≥64/<64
years) | 0.645
(0.423–0.982) | 0.041a | 0.773
(0.541–1.106) | 0.159 |
| Tumor location
(head and body/tail) | 1.129
(0.731–1.744) | 0.584 | 1.172
(0.813–1.689) | 0.395 |
| T classification(T1
and 2/T3) | 1.097
(0.722–1.665) | 0.665 | 1.077
(0.730–1.589) | 0.708 |
| N classification
(N0/N1) | 2.267
(1.480–3.472) |
<0.001a | 1.720
(1.182–2.504) | 0.005a |
| PALB2 (high/low
expression) | 1.789
(1.142–2.803) | 0.011a | 1.598
(1.068–2.389) | 0.022a |
|
| B, TCGA PDAC
samples (n=177) |
|
|
| Multivariate
analysis | Univariate
analysis |
|
|
|
|
| Factors | HR (95%
CI) | P-value | HR (95%
CI) | P-value |
|
| Sex
(Male/Female) | 0.963
(0.548–1.693) | 0.896 | 0.868
(0.577–1.306) | 0.497 |
| Age (≥65/<65
years) | 1.005
(0.549–1.841) | 0.986 | 1.084
(0.698–1.684) | 0.720 |
| Tumor location
(head and body/tail) | 1.970
(1.083–3.585) | 0.026a | 2.108
(1.170–3.801) | 0.013a |
| T classification
(T1 and 2/T3 and 4) | 1.032
(0.508–2.097) | 0.930 | 2.052
(1.088–3.870) | 0.026a |
| N classification
(N0/N1) | 1.894
(1.109–3.234) | 0.019a | 2.048
(1.243–3.374) | 0.005a |
| PALB2 (high/low
expression) | 1.980
(1.245–3.249) | 0.004a | 1.996
(1.302–3.060) | 0.002a |
Relative PALB2 gene expression across
different types of cancer
To investigate the spectrum of cancers associated
with PALB2 expression on a larger scale, TCGA public database was
analyzed. The mRNA expression levels of PALB2 in each type of
cancer compared with their respective para-tumor tissues were
presented in the log scale format (Fig.
4A). Most types of cancer in the database displayed upregulated
PALB2 mRNA expression, including breast, cervical and pancreatic
cancer, which is consistent with the detection of PALB2 protein
expression in the present study. The 177 patients with PDAC from
TCGA database were divided into two groups: Relatively high PALB2
expression group (n=88) and relatively low PALB2 expression group
(n=89). The Kaplan-Meier curves demonstrated a significant negative
association between high PALB2 expression and long-term survival
rate (P<0.001; Fig. 4B).
Multivariate Cox regression analysis were conducted on the PALB2
mRNA expression data extracted from TCGA database (Table II). The multivariate analysis
identified tumor location, N stage and PALB2 mRNA expression as
tumor parameters with a significant association with OS (Table II). The summary of
clinicopathological information of patients with PDAC from TCGA
database are shown in Table
SII.
Functional analysis of PALB2 in PDAC
cells in vitro
To investigate the potential biological role of
PALB2 in PDAC, PALB2 expression was knocked down using a siRNA in
PDAC cells. The western blotting results indicated successful
PALB2-knockdown by si-PALB2 compared with si-NC in SW1990 PDAC
cells (Fig. 5A). Cell proliferation
assays indicated similar proliferation rates in SW1990 cells
regardless of PALB2-knockdown (Fig.
5B). Migration was assessed by performing wound healing
experiments. Cell migration area was measured using ImageJ
software. The results indicated that PALB2-knockdown inhibited PDAC
cell migration depending on cell line. Migration was significantly
decreased by PALB2-knockdown in SW1990 cells and CFPAC1 cells, but
it was independent of PALB2 expression in PANC1 cells. (Figs. 5C and S2). Additionally, PALB2-knockdown
significantly decreased the cell proliferation rate of CFPAC1
cells, but had no effect on PANC1 cells (Fig. S2).
Identification of biological signaling
pathways associated with PALB2 expression
To identify biological signaling pathways associated
with PALB2 expression, a GSEA was conducted on TCGA gene expression
datasets from patients with different levels of PALB2 mRNA
expression. Among the 189 available datasets, the top and bottom 20
datasets were selected. The most significantly enriched signaling
pathways were selected based on the normalized enrichment scores.
The results indicated that samples with relatively high PALB2 mRNA
expression were enriched for genes associated with the ‘epithelial
mesenchymal transition’ (EMT) signaling pathway, along with genes
involved in the TNF-α, TGF-β, p53, NOTCH and mTORC1 signaling
pathways (Fig. 5D). To further
validate the GSEA results, the expression levels of characteristic
EMT-associated genes were quantified via RT-qPCR in PALB2-knockdown
PDAC cells in vitro. SW1990 cells displayed PALB2-dependent
migration effects. PALB2-knockdown significantly decreased Slug,
Zeb1 and vimentin gene expression, but induced a small but
non-significant increase on Snail expression. (Fig. 5E).
Discussion
In the present study, PALB2 expression in PDAC and
peritumoral tissue samples was analyzed, and the effect of PALB2 on
PDAC cell proliferation and migration in vitro was assessed.
PALB2 protein expression was increased in tumor tissues compared
with surrounding healthy tissues, and PALB2 mRNA expression levels
were relatively high in PDAC compared with in other types of
cancer. Upregulated PALB2 expression was not observed in stomach
cancer in TCGA database and in the present IHC analysis. In PDAC,
high mRNA and protein expression levels of PALB2 were negatively
associated with OS, suggesting a potential positive effect of PALB2
on tumor cell migration and EMT signaling pathway-associated genes.
The results suggested that elevated PALB2 expression may represent
a diagnostic and prognostic marker in PDAC. Furthermore, N
classification and age were significantly associated with OS in
patients with PDAC; therefore, PALB2 expression, N classification
and age may be combined to obtain a reliable marker for mortality
risk. As PDAC is a rising and leading cause of cancer-associated
mortality, additional markers are required to improve early and
specific diagnosis of PDAC, and to aid with the development of
treatment strategies (24).
The molecular mechanisms underlying the role of
PALB2 in tumorigenesis are not completely understood (25). PALB2 co-localizes with BRCA1 and
BRCA2, and contributes to error-free homologous recombination
repair (26). Dysfunctions in BRCA1
and BRCA2 promote carcinogenesis (27). Upregulated BRCA2 expression predicts
a poor prognosis in patients with breast carcinoma (28). Similarly, elevated PALB2 expression
is associated with poor clinical outcomes in patients with advanced
breast carcinoma (29). However,
independent research has indicated no association between PALB2
expression and breast cancer prognosis (30), and therefore additional studies are
required to clarify this interaction. PALB2 mutations are
frequently described for some types of tumor, such as lung
(31), breast and ovarian cancer
(32). PALB2 mutations have been
reported to predispose men and women to breast cancer (33), with the highest risk resulting from
protein truncation mutations (34).
Missense polymorphisms in BRCA1 exert a moderate
effect on the prognosis of patients with PDAC (35). Certain mutations in other BRCA
signaling pathway genes predict an improved prognosis in patients
with PDAC (17). Germline truncating
mutations in PALB2 have been detected in patients with PDAC,
indicating that PALB2 may serve as a potential susceptibility gene
for pancreatic tumorigenesis (36).
Another study suggested that truncating mutations of PALB2 may
predispose individuals to breast carcinoma, as well as pancreatic
carcinoma (37). Although the
association between PALB2 and tumorigenesis has been reported, the
relative importance of PALB2 dysregulation for the course of the
disease, mortality risk and tumor cell characteristics are not
completely understood. The identified role of PALB2 in PDAC cell
migration is consistent with the inverse association between PALB2
expression and OS observed in the present study. Therefore, it may
be hypothesized that elevated PALB2 expression may be consistent
with improved protection against DNA damage and low mutation rates
in tumor cells as part of the BRCA-complex without conferring an
increased risk of cancer-associated mortality. However, the
aforementioned hypothesis was not in accordance with the present
data, and it remains to be analyzed whether metastases are
characterized by particularly high PALB2 expression supporting
migration, and relatively low mutation rates compared with primary
PDAC cells, potentially suggesting new therapeutic approaches for
PDAC treatment and metastasis control (38). Furthermore, the integrity of the
PALB2 gene, and whether overexpressed PALB2 is functional or
mutated, requires further investigation.
The results of the present study indicated the
potential prognostic significance of PALB2 expression in PDAC. The
Kaplan-Meier analysis identified a strong association between PALB2
expression and poor OS. PALB2 expression was independent from other
clinicopathological parameters, including Ki-67, p53, CD8 and PDL1
expression. Therefore, the results of the present study suggested
that PALB2 may serve as an independent prognostic factor of OS in
patients with surgically resectable PDAC and may be an additional
biomarker to the classical parameters of PDAC staging (39).
In the present study, the in vitro
experiments demonstrated that cell migration, but not cell
proliferation, was altered by PALB2-knockdown. The corresponding
GSEA highlighted several signaling pathways associated with PALB2,
including EMT, TNF-α, TGF-β, p53, NOTCH and mTORC1. As EMT was the
most prominent signaling pathway associated with decreased OS, the
expression levels of certain EMT genes in PDAC cells were assessed.
Consistent with the GSEA results, PALB2-knockdown affected the
expression levels of Slug, Zeb1 and vimentin, which was also
consistent with a previous study (29). However, the extent to which the other
significantly associated signaling pathways contribute to the
decreased OS of patients with high PALB2 expression requires
further investigation. PALB2 mutations are frequently observed in
tumor metastases, as indicated by comparisons between localized and
metastasized breast and prostate cancer, respectively (40,41).
Therefore, comparing sequence and expression level information on
localized and metastasized PDAC should be conducted in future
studies to further identify the potential functional role of
PDAC.
In the present study, the association between PALB2
expression and OS was relatively strong. The present study used a
relatively large sample size to identify and characterize the
association between PALB2 expression and OS. However, the present
study had a number of limitations. For example, as the study was a
retrospective study, not all data on patients with PDAC and their
treatments were available. Due to the observational type of the
present study, mechanistic explanation could not be drawn, and the
potential impact of varying expression levels of PALB2 on DNA
damage was not studied. Additional functional studies are required
to elucidate the biological role of elevated PALB2 protein
expression in tumor cells and to determine the effect of PALB2 on
cell proliferation, migration and tumor susceptibility to
oncostatic medication, especially in association with the integrity
of the gene. In conclusion, the present study demonstrated that
PALB2 expression was increased in PDAC tissues, and an association
between high PALB2 mRNA or protein expression and decreased OS was
identified. The results suggested that PALB2 may serve as an
additional diagnostic marker and may aid in predicting the risk of
mortality in patients with PDAC. Moreover, the present results may
aid in the management of patients in clinical practice and
potentially in the development of personalized treatment
strategies.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Waldemar Minich
and Dr Eddy Rijntjes (Charité-Universitätsmedizin Berlin, Berlin,
Germany) for constructive discussions.
Funding
The present study was supported by Deutsche
Forschungsgemeinschaft (Research Unit 2558 TraceAge, Scho 849/6-1).
OG received a stipend from the China National Scholarship for
Abroad.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
OG and LS were responsible for the conceptualization
of the study. OG and XW conducted the in silico analysis of
The Cancer Genome Atlas database. YC and YY collected and
translated patients' clinical data, and identified and selected the
tissue samples for immunohistochemistry. OG and AH conducted the
in vitro experiments. OG, YC and XW formally analyzed the
data and conducted the statistical analyses. YY and LS supervised
the project. OG, XW, YC, YY and LS contributed to drafting and
writing the final manuscript. All authors read and approved the
final manuscript. OG, YY and LS confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The use of the pancreas, gastric and breast tumor
tissues was approved by the Research Ethics Committees of the
National Engineering Center of Shanghai BioChip Co., Ltd. The
research has been performed according to the ethical standards of
the Declaration of Helsinki. Oral informed consent was obtained
from all patients prior to analysis according to the committee's
regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang X, Shi S, Zhang B, Ni Q, Yu X and Xu
J: Circulating biomarkers for early diagnosis of pancreatic cancer:
Facts and hopes. Am J Cancer Res. 8:332–353. 2018.PubMed/NCBI
|
|
5
|
Venkitaraman AR: Cancer susceptibility and
the functions of BRCA1 and BRCA2. Cell. 108:171–182. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang L, Wu C, Yu D, Wang C, Che X, Miao
X, Zhai K, Chang J, Jiang G, Yang X, et al: Identification of
common variants in BRCA2 and MAP2K4 for susceptibility to sporadic
pancreatic cancer. Carcinogenesis. 34:1001–1015. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sikdar N, Saha G, Dutta A, Ghosh S,
Shrikhande SV and Banerjee S: Genetic alterations of periampullary
and pancreatic ductal adenocarcinoma: An overview. Curr Genomics.
19:444–463. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B
and Yu X: PALB2 links BRCA1 and BRCA2 in the DNA-damage response.
Curr Biol. 19:524–529. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reid S, Schindler D, Hanenberg H, Barker
K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al:
Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and
predispose to childhood cancer. Nat Genet. 39:162–164. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia B, Dorsman JC, Ameziane N, de Vries Y,
Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al:
Fanconi anemia is associated with a defect in the BRCA2 partner
PALB2. Nat Genet. 39:159–161. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen P, Liang J, Wang Z, Zhou X, Chen L,
Li M, Xie D, Hu Z, Shen H and Wang H: Association of common PALB2
polymorphisms with breast cancer risk: A case-control study. Clin
Cancer Res. 14:5931–5937. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Southey MC, Teo ZL and Winship I: PALB2
and breast cancer: Ready for clinical translation! Appl Clin Genet.
6:43–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Borecka M, Zemankova P, Vocka M, Soucek P,
Soukupova J, Kleiblova P, Sevcik J, Kleibl Z and Janatova M:
Mutation analysis of the PALB2 gene in unselected pancreatic cancer
patients in the Czech Republic. Cancer Genet. 209:199–204. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blanco A, de la Hoya M, Osorio A, Diez O,
Miramar MD, Infante M, Martinez-Bouzas C, Torres A, Lasa A, Llort
G, et al: Analysis of PALB2 gene in BRCA1/BRCA2 negative Spanish
hereditary breast/ovarian cancer families with pancreatic cancer
cases. PLoS One. 8:e675382013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Harinck F, Kluijt I, van Mil SE, Waisfisz
Q, van Os TA, Aalfs CM, Wagner A, Olderode-Berends M, Sijmons RH,
Kuipers EJ, et al: Routine testing for PALB2 mutations in familial
pancreatic cancer families and breast cancer families with
pancreatic cancer is not indicated. Eur J Hum Genet. 20:577–579.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hofstatter EW, Domchek SM, Miron A, Garber
J, Wang M, Componeschi K, Boghossian L, Miron PL, Nathanson KL and
Tung N: PALB2 mutations in familial breast and pancreatic cancer.
Fam Cancer. 10:225–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takeuchi S, Doi M, Ikari N, Yamamoto M and
Furukawa T: Mutations in BRCA1, BRCA2, and PALB2, and a panel of 50
cancer-associated genes in pancreatic ductal adenocarcinoma. Sci
Rep. 8:81052018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Waddell N, Pajic M, Patch AM, Chang DK,
Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al:
Whole genomes redefine the mutational landscape of pancreatic
cancer. Nature. 518:495–501. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Golan T, Kanji ZS, Epelbaum R, Devaud N,
Dagan E, Holter S, Aderka D, Paluch-Shimon S, Kaufman B,
Gershoni-Baruch R, et al: Overall survival and clinical
characteristics of pancreatic cancer in BRCA mutation carriers. Br
J Cancer. 111:1132–1138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wattenberg MM, Asch D, Yu S, O'Dwyer PJ,
Domchek SM, Nathanson KL, Rosen MA, Beatty GL, Siegelman ES and
Reiss KA: Platinum response characteristics of patients with
pancreatic ductal adenocarcinoma and a germline BRCA1, BRCA2 or
PALB2 mutation. Br J Cancer. 122:333–339. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dell'Aquila E, Fulgenzi CAM, Minelli A,
Citarella F, Stellato M, Pantano F, Russano M, Cursano MC,
Napolitano A, Zeppola T, et al: Prognostic and predictive factors
in pancreatic cancer. Oncotarget. 11:924–941. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Edge S, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer-Verlag; New York, NY: 2009
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ducy M, Sesma-Sanz L, Guitton-Sert L,
Lashgari A, Gao Y, Brahiti N, Rodrigue A, Margaillan G, Caron MC,
Côté J, et al: The tumor suppressor PALB2: Inside out. Trends
Biochem Sci. 44:226–240. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sy SM, Huen MS and Chen J: PALB2 is an
integral component of the BRCA complex required for homologous
recombination repair. Proc Natl Acad Sci USA. 106:7155–7160. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang G, Mercado-Uribe I, Multani AS, Sen
S, Shih Ie M, Wong KK, Gershenson DM and Liu J: RAS promotes
tumorigenesis through genomic instability induced by imbalanced
expression of Aurora-A and BRCA2 in midbody during cytokinesis. Int
J Cancer. 133:275–285. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Egawa C, Miyoshi Y, Taguchi T, Tamaki Y
and Noguchi S: High BRCA2 mRNA expression predicts poor prognosis
in breast cancer patients. Int J Cancer. 98:879–882. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li J, Li M, Chen P and Ba Q: High
expression of PALB2 predicts poor prognosis in patients with
advanced breast cancer. FEBS Open Bio. 8:56–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Poumpouridou N, Acha-Sagredo A, Goutas N,
Vlachodimitropoulos D, Chatziioannidou I, Lianidou E, Liloglou T
and Kroupis C: Development and validation of molecular
methodologies to assess PALB2 expression in sporadic breast cancer.
Clin Biochem. 49:253–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bleuyard JY, Butler RM and Esashi F:
Perturbation of PALB2 function by the T413S mutation found in small
cell lung cancer. Wellcome Open Res. 2:1102017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakagomi H, Sakamoto I, Hirotsu Y, Amemiya
K, Mochiduki H and Omata M: Analysis of PALB2 mutations in 155
Japanese patients with breast and/or ovarian cancer. Int J Clin
Oncol. 21:270–275. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rahman N, Seal S, Thompson D, Kelly P,
Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, et
al: PALB2, which encodes a BRCA2-interacting protein, is a breast
cancer susceptibility gene. Nat Genet. 39:165–167. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Casadei S, Norquist BM, Walsh T, Stray S,
Mandell JB, Lee MK, Stamatoyannopoulos JA and King MC: Contribution
of inherited mutations in the BRCA2-interacting protein PALB2 to
familial breast cancer. Cancer Res. 71:2222–2229. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu Y, Zhai K, Ke J, Li J, Gong Y, Yang Y,
Tian J, Zhang Y, Zou D, Peng X, et al: BRCA1 missense polymorphisms
are associated with poor prognosis of pancreatic cancer patients in
a Chinese population. Oncotarget. 8:36033–36039. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jones S, Hruban RH, Kamiyama M, Borges M,
Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, et
al: Exomic sequencing identifies PALB2 as a pancreatic cancer
susceptibility gene. Science. 324:2172009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Slater EP, Langer P, Niemczyk E, Strauch
K, Butler J, Habbe N, Neoptolemos JP, Greenhalf W and Bartsch DK:
PALB2 mutations in European familial pancreatic cancer families.
Clin Genet. 78:490–494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giovannetti E, van der Borden CL, Frampton
AE, Ali A, Firuzi O and Peters GJ: Never let it go: Stopping key
mechanisms underlying metastasis to fight pancreatic cancer. Semin
Cancer Biol. 44:43–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Garces-Descovich A, Beker K,
Jaramillo-Cardoso A, James Moser A and Mortele KJ: Applicability of
current NCCN Guidelines for pancreatic adenocarcinoma
resectability: Analysis and pitfalls. Abdom Radiol (NY).
43:314–322. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lefebvre C, Bachelot T, Filleron T,
Pedrero M, Campone M, Soria JC, Massard C, Lévy C, Arnedos M,
Lacroix-Triki M, et al: Mutational profile of metastatic breast
cancers: A retrospective analysis. PLoS Med. 13:e10022012016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pritchard CC, Mateo J, Walsh MF, De Sarkar
N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R,
et al: Inherited DNA-Repair gene mutations in men with metastatic
prostate cancer. New Engl J Med. 375:443–453. 2016. View Article : Google Scholar : PubMed/NCBI
|