Introduction
In Korean women, breast cancer (BC) is one of the
most common type of cancer (1).
Despite the significant improvements in the overall survival and
quality of life for women with BC, BC remains a leading cause of
cancer-associated deaths (2).
Therefore, it is necessary to develop new prognostic and
therapeutic markers for patients with BC.
Cancer cells frequently exhibit altered cellular
metabolism, which can mediate tumor progression and can be used for
therapeutic purposes (3).
Cholesterol is a unique lipid that is crucial for membrane
formation, cell proliferation and cell differentiation (4). The critical role of cholesterol in the
pathogenesis of various types of cancer, such as prostate and
breast cancer, has been recognized in tumor cell proliferation,
survival and treatment resistance (4–6).
Moreover, several studies have revealed that inhibition of
cholesterol synthesis at different steps results in human cancer
cell death in both in vitro and in vivo models
(3,4,7).
Overall, dysregulation of cholesterol metabolism may be a promising
new therapeutic target for cancer treatment.
Squalene epoxidase (SQLE) is one of the
rate-limiting enzymes in cholesterol synthesis by catalyzing the
first step of squalene oxygenation (8). Previous studies have revealed that
dysregulation of SQLE expression is involved in the molecular
pathogenesis of various types of cancer, such as prostate cancer
(9,10), hepatocellular carcinoma (11), pancreatic cancer (12), esophageal squamous cell carcinoma
(13) and squamous lung cancer
(14). SQLE has been proposed as a
new molecular marker to predict a poor prognosis in the
aforementioned types of cancer (9–14).
Several studies have already been conducted with the
aim of exploring the potential effect of the dysregulation of SQLE
expression in BC (15–26). SQLE is involved in the maintenance of
lipid droplet homeostasis in BC (15). Additionally, SQLE is involved in the
process by which normal mammary fibroblasts induce a reversion of
the malignant phenotype in primary BC (16). SQLE overexpression is more prevalent
in groups with an unfavorable prognosis of stage I/II estrogen
receptor-positive (ER+) BC (17), early-onset BC (18) and African-American patients with
luminal A BC (19). Increased SQLE
expression in patients with ER+ BC has been associated
with poor response to endocrine therapy (20). Furthermore, high SQLE expression has
been associated with increased risk of BC recurrence (21–23).
SQLE copy number amplification has been associated with its
overexpression and poor prognosis (24). MYC gene amplification and aberrant
SQLE methylation have also been observed in aggressive BC (25). Brown et al (26) have confirmed that SQLE is a true
oncogene by clinically relevant amplification in BC. Brown et
al (26) have demonstrated that
SQLE overexpression represents an independent adverse prognostic
factor and that SQLE inhibition may be a novel therapeutic target
for BC. Most of the aforementioned SQLE studies in BC have been
based on a molecular approach. Although ductal carcinoma in
situ (DCIS) is a non-invasive cancerous lesion of the breast
(27), to the best of our knowledge,
there are no studies on the profile of SQLE expression during BC
progression, including DCIS.
We have previously performed massive parallel RNA
sequencing (RNA-Seq) analysis in 21 samples (normal, cancer and
nodal metastases) from 7 patients with ER+,
HER2− BC, revealing SQLE as one of the differentially
expressed genes (DEGs) (28).
Therefore, the present study evaluated the potential involvement of
SQLE in the tumorigenic process of BC. Additionally, whether SQLE
detection by immunohistochemistry may predict the prognosis in
patients with BC was investigated. SQLE expression was examined in
10 pairs of DCIS and BC tissues and their adjacent normal tissues
at the mRNA level by reverse transcription-quantitative PCR
(RT-qPCR). In addition, immunohistochemical staining of SQLE
expression on tissue microarray (TMA) was performed in 26 normal
breast, 79 DCIS and 198 BC samples. The role of SQLE as a
prognostic biomarker in patients with BC was then verified using
BreastMark (29).
Materials and methods
Validation of SQLE RNA-Seq data by
RT-qPCR
We have previously generated comprehensive gene
expression profiles of matched normal, cancer and lymph node
metastatic tissues from 7 patients with ER+,
HER2− BC using RNA-Seq analysis (28). To validate the RNA-Seq data, SQLE
mRNA expression was analyzed by RT-qPCR, as previously described
(30). The isolated RNA used for
RNA-Seq in our previous study (28)
was used for RT-qPCR. The qPCR reaction was performed in a 7500
Fast Real-Time PCR System using TaqMan® Gene Expression
Master Mix (Thermo Fisher Scientific, Inc.) using the following
thermocycling conditions: Initial denaturation for 30 sec at 95°C,
followed by 40 cycles at 95°C for 15 sec and 60°C for 60 sec. The
following probes (Thermo Fisher Scientific, Inc.) were used:
Hs01123768_m1 (SQLE) and Hs02758991_g1 (GAPDH). The
2−ΔΔCq method (31) was
used for data analysis and the value of 2−ΔΔCq indicated
the fold change in SQLE expression normalized to GAPDH
expression.
Samples for SQLE mRNA expression in
DCIS and BC tissues and their adjacent normal tissues
New frozen samples of DCIS and BC tissues and their
adjacent normal tissues (≥2 cm from DCIS/BC tissues) were provided
by the Biobank of Chonnam National University Hwasun Hospital
(Jeollanam, Republic of Korea), which is a member of the Korea
Biobank Network, and were used for RT-qPCR, as aforementioned. The
present study included 10 female patients each with DCIS or BC. The
mean age in patients with DCIS was 57.2 years (median age, 57.6
years; age range, 48–73 years), while the mean age in patients with
BC was 56.6 years (median age, 53.0 years; age range, 39–86
years).
SQLE expression in normal, DCIS and BC
tissues
Patients and tissues
Tissues were fixed in 10% formalin for 12–24 h at
room temperature, cut into <3-mm-thick sections and embedded in
paraffin. Formalin-fixed paraffin-embedded (FFPE) samples of normal
breast tissues with no pathological lesions (n=26), DCIS (n=79) and
BC (n=198) were selected from female patients within Chonnam
National University Hospital (Gwangju, Republic of Korea) and
Chonnam National University Hwasun Hospital (Jeollanam, Republic of
Korea). The mean age in patients with no pathological lesions was
44.3 years (median age, 45.5 years; age range, 25–69 years), the
mean age in patients with DCIS was 49.3 years (median age, 48.0
years; age range, 27–73 years) and the mean age in patients with BC
was 46.6 years (median age, 45.0 years; age range, 26–89 years).
Patients diagnosed with BC between January 1997 and December 2002
were included in the present study and had a follow-up of ≥10
years. After surgery, the patients underwent standard radiation
therapy or adjuvant systemic therapy (hormone therapy or
chemotherapy), according to the medical insurance program
controlled by the Ministry of Health and Welfare of Korea. Medical
records were reviewed to obtain clinicopathological information
including patient outcome. ER, progesterone receptor (PR) and HER2
expression was assessed according to the American Society of
Clinical Oncology/College of American Pathologists guidelines
(32–34). DCIS samples between February 2005 and
December 2011 were selected based on the availability of FFPE
samples.
TMA construction
TMA blocks were constructed using one representative
FFPE block in each case. Three cores of 1-mm diameter for BC and
two cores of 2-mm diameter for normal breast and DCIS were punched
from the donor block.
Immunohistochemistry and evaluation of
immunostaining
SQLE expression was examined by immunohistochemical
staining using the Bond-max automatic device (Leica Microsystems,
Inc.), as previously described (35). Mouse polyclonal antibody against SQLE
(1:200; cat. no. 042278; United States Biological) was used.
According to a previous study (36),
SQLE immunoreactivity was scored as 0 (negative), 1 (weak), 2
(moderate) or 3 (strong), based on the intensity of cytoplasmic
staining. SQLE expression was evaluated by light microscopy
(magnification, ×200). Samples with intensity staining scores of
0–2 were considered as low SQLE expression, while scores of 3 were
defined as high SQLE expression.
Validation of SQLE as a prognostic
biomarker using BreastMark
SQLE was further analyzed to validate its prognostic
value in patients with BC using the BreastMark database, as
previously described (29).
Statistical analysis
The data of SQLE expression in matched normal,
cancer and lymph node metastatic tissues from seven patients were
analyzed using one-way repeated measures ANOVA followed by post-hoc
analysis with Fisher's Least Significant Difference adjustment for
multiple comparisons. SQLE mRNA expression according to RT-qPCR in
the DCIS and BC tissues and their adjacent normal tissues were
compared using a paired Student's t-test (two-sided). Categorical
nominal variables were tested using the χ2 test or
Fisher's exact test. Linear-by-linear association was added to test
for the trend. Univariate survival analysis was performed according
to the Kaplan-Meier method and the differences in survival curves
were assessed with the log-rank test. Cox's proportional hazard
model was used for multivariate analysis. SPSS (v25 for windows;
IBM Corp.) was used for all statistical analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
Validation of SQLE RNA-Seq data by
RT-qPCR
SQLE expression was assessed by RNA-Seq (Fig. 1A) and RT-qPCR (Fig. 1B) in matched normal, cancer and nodal
metastatic tissues in seven patients with BC. In the RNA-Seq
results, SQLE expression (mean ± SD) was upregulated in BC tissues
compared with in adjacent normal breast tissues (22.8±10.4 vs.
5.3±3.9 for BC vs. normal, respectively; fold change, 3.915;
P=0.007) and subsequently unchanged when comparing nodal metastatic
tissues with corresponding BC tissues (17.4±13.5 vs. 22.8±10.4 for
nodal vs. BC; fold change, −1.3014; P=0.354) (data not shown). For
RT-qPCR, the unamplified total RNA (from the same batch used for
RNA-Seq) was used as the template. Although no significant
differences among groups were observed, SQLE expression was mainly
upregulated in BC and nodal metastatic tissues compared with in
adjacent normal breast tissues (data not shown). Overall, the
results obtained with the two different techniques were consistent
for ~70% (5/7) of the samples tested.
SQLE mRNA in DCIS and BC tissues and
their adjacent normal breast tissues
SQLE expression was assessed by RT-qPCR in 10 frozen
DCIS tissues (Fig. 2A) and 10 BC
tissues (Fig. 2B) and their
respective adjacent normal breast tissues. The expression levels of
SQLE mRNA (mean ± SD) were significantly increased in DCIS and BC
tissues compared with in their adjacent normal breast tissues
(6.27±7.54 vs. 1.00±0.00 for DCIS vs. normal, P<0.05;
14.02±22.95 vs. 1.00±0.00 for BC vs. normal, P<0.05) (data not
shown). No significant differences in SQLE mRNA expression (mean ±
SD) were observed between DCIS and BC (6.27±7.54 vs. 14.02±22.95,
respectively; P=0.323) (data not shown).
SQLE expression in normal, DCIS and BC
tissues
Normal breast tissues exhibited weak cytoplasmic
SQLE expression (Fig. 3A and B). In
DCIS and BC tissues, the carcinoma cells displayed variable SQLE
expression (Fig. 3C-F). Table I summarizes SQLE expression in normal
breast, DCIS and BC tissues. High SQLE expression was detected in
0/26 (0%) of normal breast tissues, 38/79 (48.1%) of DCIS samples,
and 80/198 (40.4%) of BC samples. Differential SQLE expression was
noted in the DCIS and BC groups compared with in the normal breast
group (P<0.05, linear-by-linear association for trend). SQLE
expression in DCIS and BC tissues was significantly higher compared
with in normal breast tissues (DCIS vs. normal, 48.1 vs. 0%,
P<0.001; BC vs. normal, 40.4 vs. 0%, P<0.001). There was no
significant difference in the expression levels of SQLE between
DCIS and BC (DCIS vs. BC, 48.1 vs. 40.4%, P=0.242).
 | Figure 3.SQLE expression in normal breast,
DCIS and BC tissues. (A and B) Normal breast tissue exhibited weak
cytoplasmic SQLE expression (A: magnification, ×4; scale bar, 500
µm; B: magnification, ×400; scale bar, 60 µm). (C) High and (D) low
SQLE expression in the cytoplasm of DCIS tissues. (E) High and (F)
low SQLE expression in the cytoplasm of BC tissues. Magnification,
×4; scale bar, 500 µm (inlet magnification, ×400). DCIS, ductal
carcinoma in situ; BC, breast cancer; SQLE, squalene
epoxidase. |
 | Table I.SQLE expression in normal breast,
ductal carcinoma in situ and breast cancer tissues. |
Table I.
SQLE expression in normal breast,
ductal carcinoma in situ and breast cancer tissues.
| Type of tissue | High SQLE
expression, n/total n (%) | P-value |
|---|
| Normal | 0/26 (0.0) | 0.018a |
| Ductal carcinoma
in situ | 38/79 (48.1) |
|
| Breast cancer | 80/198 (40.4) |
|
Tables II and
III summarize the associations
between SQLE expression and the clinicopathological features of
patients with DCIS and BC, respectively. High SQLE expression in
DCIS tissues was associated with nuclear grade (P<0.01),
comedo-type necrosis (P<0.05) and HER2 status (P<0.01)
(Table II). Although not
statistically significant, the percentage of high SQLE expression
was higher in patients with recurrence (5/7; 71.4%) than in
patients without recurrence (33/72; 45.8%) (Table II). High SQLE expression in BC
tissues was associated with tumor size (P<0.05), nodal
metastases (P<0.001), stage (P<0.01), molecular subtype
(P<0.05) and distant metastatic relapse (P<0.05) (Table III).
 | Table II.Association between SQLE expression
and clinicopathological parameters of patients with ductal
carcinoma in situ (n=79). |
Table II.
Association between SQLE expression
and clinicopathological parameters of patients with ductal
carcinoma in situ (n=79).
|
Characteristics | High SQLE
expression, n/total n (%) |
P-valuea |
|---|
| Age, years |
| >0.05 |
|
≤50 | 21/45 (46.7) |
|
|
>50 | 17/34 (50.0) |
|
| Size, cm |
| >0.05 |
|
≤2.5 | 21/45 (46.7) |
|
|
>2.5 | 17/34 (50.0) |
|
| Nuclear grade |
| <0.01 |
| 1 | 1/7 (14.3) |
|
| 2 | 19/46 (41.3) |
|
| 3 | 18/26 (69.2) |
|
| Comedo-type
necrosis |
| <0.05 |
| No | 11/32 (34.4) |
|
|
Yes | 27/47 (57.4) |
|
| Estrogen
receptor-α |
| >0.05 |
|
Negative | 17/28 (60.7) |
|
|
Positive | 21/51 (41.2) |
|
| HER2 |
| <0.01 |
|
Negative | 18/51 (35.3) |
|
|
Positive | 20/28 (71.4) |
|
| Recurrence |
|
>0.05b |
| No | 33/72 (45.8) |
|
|
Yes | 5/7 (71.4) |
|
 | Table III.Association between SQLE expression
and clinicopathological parameters of patients with breast cancer
(n=198). |
Table III.
Association between SQLE expression
and clinicopathological parameters of patients with breast cancer
(n=198).
|
Characteristics | High SQLE
expression, n/total n (%) |
P-valuea |
|---|
| Age, years |
| >0.05 |
|
≤46 | 40/112 (35.7) |
|
|
>46 | 41/86 (47.7) |
|
| Histopathologic
type |
| >0.05 |
|
Invasive ductal carcinoma,
NOS | 67/171 (39.2) |
|
|
Invasive lobular
carcinoma | 14/25 (56.0) |
|
|
Mucinous carcinoma | 0/2 (0.0) |
|
| Tumor size, cm |
| <0.05 |
| ≤2 | 11/35 (31.4) |
|
|
2–5 | 51/133(38.3) |
|
|
>5 | 19/30 (63.3) |
|
| Number of nodal
metastasis |
| <0.001 |
| 0 | 29/105 (27.6) |
|
|
1–3 | 25/51 (49.0) |
|
|
4–9 | 13/25 (52.0) |
|
|
≥10 | 14/17 (82.4) |
|
| Histological
grade |
| >0.05 |
| 1 | 4/26 (15.4) |
|
| 2 | 47/101 (46.5) |
|
| 3 | 30/71 (42.3) |
|
| Stage |
| <0.01 |
| I | 11/35 (31.4) |
|
| II | 37/112 (33.0) |
|
|
III | 33/51 (64.7) |
|
| Estrogen
receptor-α |
| >0.05 |
|
Negative | 40/87 (46.0) |
|
|
Positive | 41/111 (36.9) |
|
| Progesterone
receptor |
| >0.05 |
|
Negative | 39/87 (44.8) |
|
|
Positive | 42/111 (37.8) |
|
| HER2 |
| >0.05 |
|
Negative | 64/162 (39.5) |
|
|
Positive | 17/36 (47.2) |
|
| Molecular
subtypes |
| <0.05 |
|
Luminal | 50/135 (37.0) |
|
|
HER2 | 14/21 (66.7) |
|
| Triple
negative | 17/42 (40.5) |
|
| Distant metastatic
relapse |
| <0.05 |
| No | 52/146 (35.6) |
|
|
Yes | 29/52 (55.8) |
|
Summary of survival analysis in
patients with BC
Table IV summarizes
the results of univariate survival analysis according to log-rank
test for Kaplan-Meier survival analysis. Patients with high SQLE
expression exhibited a poor prognosis for disease-free survival and
overall survival compared with those with low expression (P=0.001
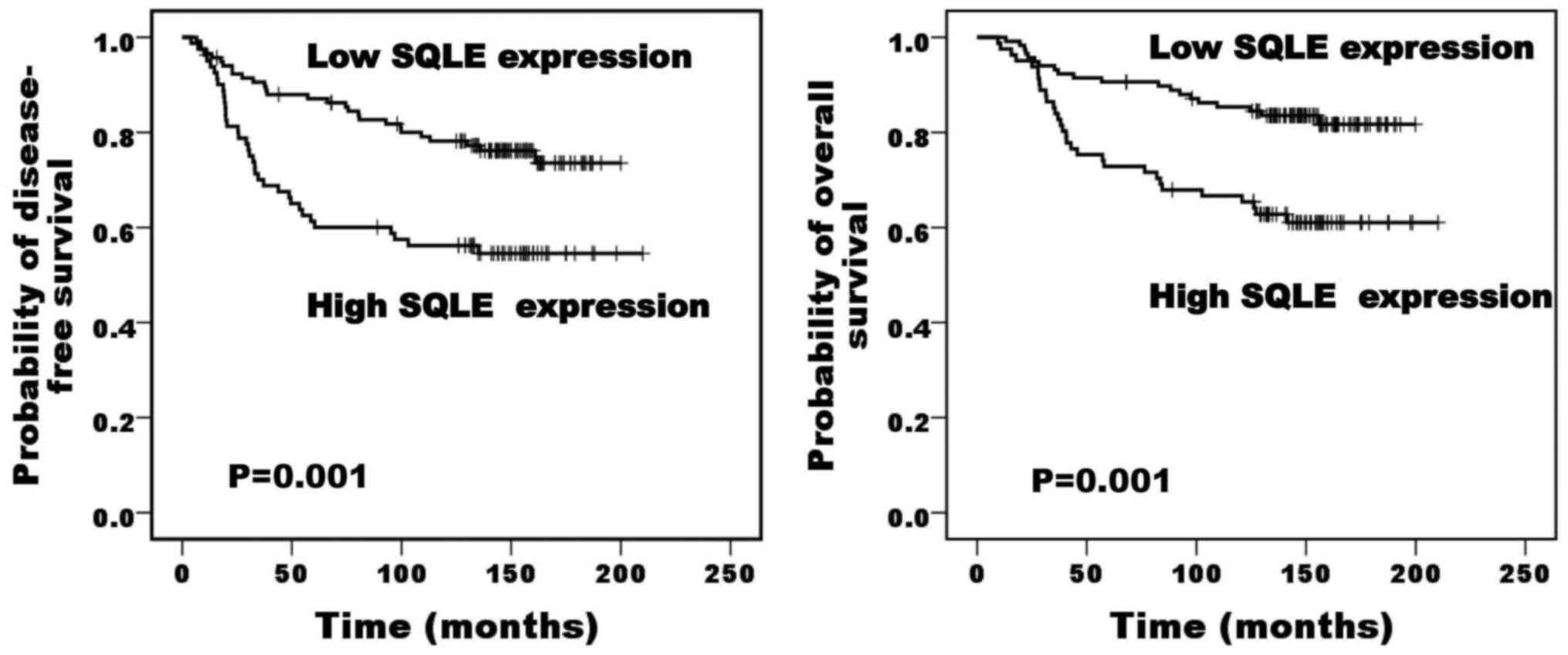
and P=0.001, respectively; Fig. 4).
Lymph node status and SQLE expression were independent poor
prognostic factors for disease-free survival, while lymph node
status was the only independent poor prognostic factor for overall
survival (Table V).
 | Table IV.Univariate analysis of prognostic
factors in patients with breast cancer. |
Table IV.
Univariate analysis of prognostic
factors in patients with breast cancer.
|
Characteristics | Disease-free
survival (P-value) | Overall survival
(P-value) |
|---|
| Age (≤46 vs. >46
years) | 0.972 | 0.522 |
| Histological type
(invasive carcinoma of no special type vs. other types) | 0.076 | 0.409 |
| Tumor size (pT1 vs.
pT2 vs. pT3) | <0.001 | <0.001 |
| Lymph node status
(pN0 vs. pN1 vs. pN2 vs. pN3) | <0.001 | <0.001 |
| Histological grade
(1 vs. 2 vs. 3) | 0.513 | 0.105 |
| Stage (I vs. II vs.
III vs. IV) | <0.001 | <0.001 |
| Hormonal therapy
(no vs. yes) | 0.395 | 0.295 |
|
Chemotherapy/radiotherapy (no vs.
yes) | 0.026 | 0.017 |
| Estrogen receptor-α
status (negative vs. positive) | 0.430 | 0.424 |
| Progesterone
receptor status (negative vs. positive) | 0.527 | 0.625 |
| HER-2 status
(negative vs. positive) | 0.966 | 0.627 |
| Squalene epoxidase
expression (low vs. high) | 0.001 | 0.001 |
 | Table V.Multivariate analysis with Coxs
proportional hazard model for prognostic factors in patients with
breast cancer. |
Table V.
Multivariate analysis with Coxs
proportional hazard model for prognostic factors in patients with
breast cancer.
|
| Disease-free
survival | Overall
survival |
|---|
|
|
|
|
|---|
|
Characteristics | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (≤46 vs. >46
years) | 0.072 | 0.566–1.539 | 0.788 | 0.038 | 0.597–1.877 | 0.845 |
| Tumor size (≤5 vs.
>5 cm) | 2.885 | 0.291–1.092 | 0.089 | 1.492 | 0.320–1.303 | 0.222 |
| Lymph node status
(negative vs. positive) | 6.443 | 0.212–0.819 | 0.011 | 4.922 | 0.170–0.896 | 0.027 |
| Stage (I/II vs.
III) | 0.440 | 0.394–1.584 | 0.507 | 1.885 | 0.265–1.263 | 0.170 |
|
Chemotherapy/radiotherapy (no vs.
yes) | 1.285 | 0.193–1.551 | 0.257 | 1.484 | 0.096–1.730 | 0.223 |
| Squalene epoxidase
expression (low vs. high) | 4.079 | 0.354–0.985 | 0.043 | 3.561 | 0.318–1.022 | 0.059 |
Validation of SQLE as a prognostic
biomarker using BreastMark
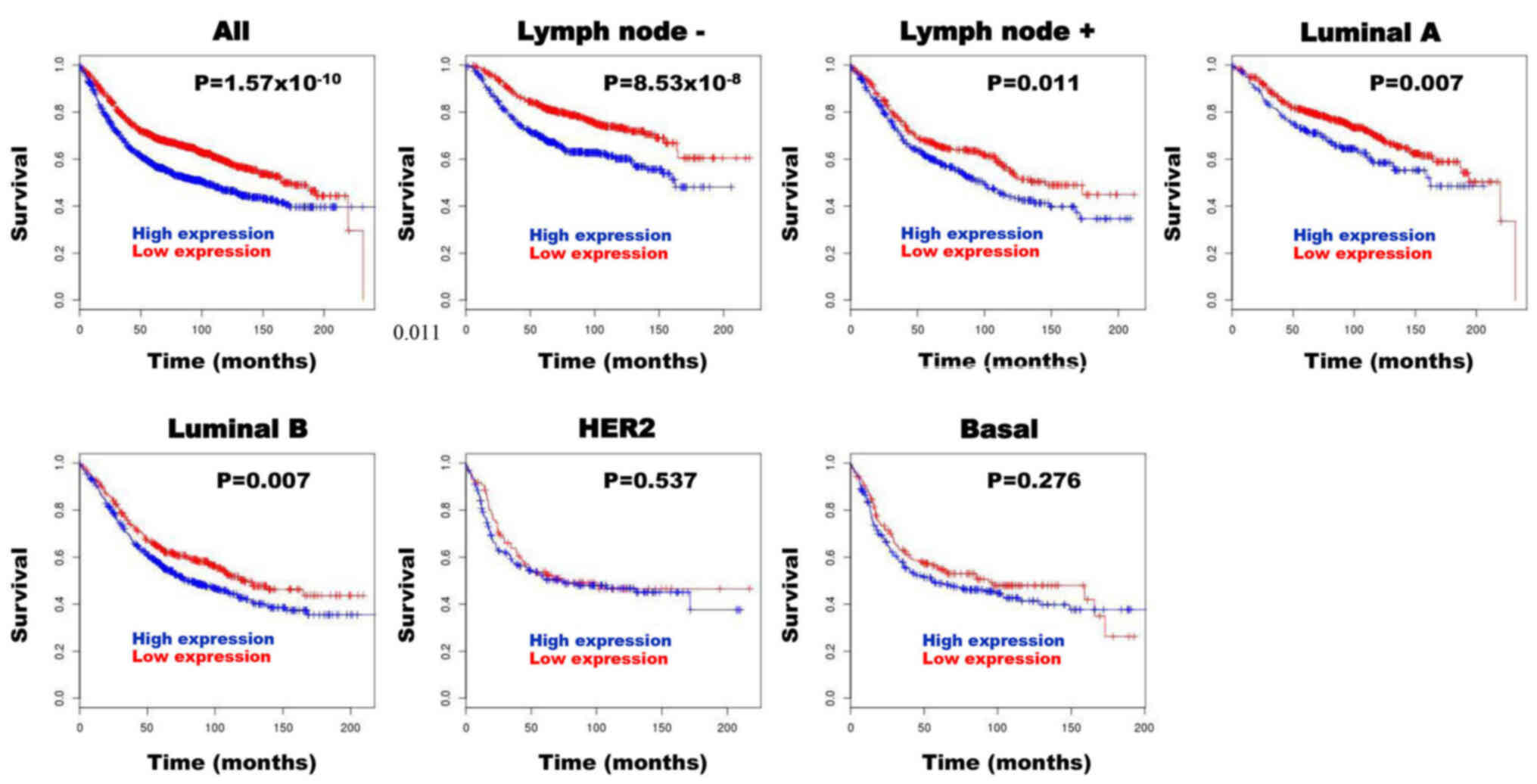
Following BreastMark analysis, high SQLE mRNA
expression in patients with BC was significantly associated with
poor prognosis in all groups [hazard ratio (HR), 1.467;
P=1.57×10−10; n=2,652), lymph node negative group (HR,
1.781; P=8.53×10−8; n=1,183), lymph node positive group
(HR, 1.337; P=0.011; n=744), luminal A subtype (HR, 1.427; P=0.007;
n=823) and luminal B subtype (HR, 1.284; P=0.007; n=1,013)
(Fig. 5). However, high SQLE mRNA
expression was not associated with prognosis in patients with HER
subtype (HR, 1.117; P=0.537; n=286) and basal subtype (HR, 1.169;
P=0.276; n=424) (Fig. 5).
Discussion
SQLE expression is associated with pathogenesis and
clinical outcome in various types of cancer (9–14). The
present study demonstrated the upregulation of SQLE mRNA expression
in DCIS and BC tissues compared with in their adjacent normal
breast tissues. SQLE expression was associated with breast tumor
progression and with a poor clinical outcome in patients with
BC.
Cholesterol is the main sterol that is synthesized
in all animal cells and is an essential structural component of
cell membrane (4). An association
between cholesterol and cancer has been previously recognized
(6). Moreover, previous studies have
indicated that inhibition at different stages of cholesterol
synthesis contributes to inhibition of tumor cell proliferation,
cell death and resistance to therapies in cancer (3,4,7). Overall, dysregulation of cholesterol
metabolism may be a new therapeutic target in cancer treatment.
SQLE encodes squalene epoxidase, one of the major
enzymes in the late stages of cholesterol synthesis (8). SQLE catalyzes the oxidation of squalene
to 2,3-oxidosqualene (squalene epoxide), and its inhibition can
affect the synthesis of sterols and of the cell membrane, or even
cell growth (3). Previous studies
have indicated that dysregulation of SQLE is involved in the
molecular pathogenesis of various types of cancer and has also been
associated with poor prognosis (9–14). SQLE
has been associated with radiation resistance in pancreatic cancer
(12) and metastasis in esophageal
squamous cell carcinoma (13), as
well as with poor outcomes in patients with prostate cancer
(9,10), hepatocellular carcinoma (11) and squamous lung cancer (14).
Several studies have already been conducted to
explore the potential role of the dysregulation of SQLE in BC
(15–26). Polycarpou-Schwarz et al
(16) identified a new microprotein
gene, Cancer-Associated Small Integral Membrane Open reading frame
1 (CASIMO1), utilizing a microarray approach. CASIMO1 was found to
be overexpressed and important for proper proliferation of breast
cancer cells, and exerted its effects through interactions with
SQLE maintaining lipid droplet homeostasis (16). Römer et al (15) assessed the ability of normal human
mammary fibroblasts (HMFs) to induce reversion of the malignant
phenotype of primary breast carcinoma cells in a three-dimensional
cell culture model. The reversion of the malignant phenotype was
detected in 5/13 primary breast carcinoma cell co-cultures with
HMFs (15). Gene expression analysis
revealed that SQLE expression was downregulated in the reverted
cases compared with in the non-reverted cases (15). These findings were consistent with
the role of SQLE in regulating BC progression and inhibition of
differentiation (15). cDNA
microarrays reported that SQLE was differentially expressed in BC
and that SQLE expression was increased dramatically in cancer
tissues compared with in normal and cancer-adjacent normal tissues
(18,19). In the present study, SQLE expression
was directly compared in sets of matched normal breast, BC and
nodal metastatic tissues using RNA-Seq and RT-qPCR. In the RNA-Seq
data, SQLE expression was upregulated in BC tissues compared with
in matched normal breast tissues, and was unchanged when comparing
nodal metastatic tissues with matched BC tissues. Based on changes
in gene expression during progression from normal tissue to cancer
tissue and then to metastatic node of BC, SQLE may belong to genes
associated with tumorigenesis (normal-cancer transition) rather
than metastasis (cancer-metastasis transition).
Although BC is believed to develop from DCIS
(27), to the best of our knowledge,
there are no studies that have examined the association between
SQLE and tumor progression in BC, including DCIS lesions. In the
present study, mRNA expression levels of SQLE in frozen DCIS and BC
tissues and their adjacent normal breast tissues were assessed
using RT-qPCR. The present data revealed that SQLE mRNA expression
was significantly increased in DCIS and BC tissues compared with in
adjacent normal tissues. This result demonstrated the oncogenic
properties of SQLE in BC as well as in DCIS. However, there were no
significant differences in SQLE mRNA expression between DCIS and
BC. In accordance with the RT-qPCR findings, immunohistochemical
analysis revealed that SQLE expression in DCIS and BC was
significantly higher than that in normal breast tissues, but there
was no significant difference in SQLE expression between DCIS and
BC tissues. These results suggest that SQLE may serve an essential
role in BC progression and may be especially activated during the
process of DCIS development.
Although the underlying mechanism of SQLE
dysregulation in BC remains unclear, SQLE is frequently altered by
copy number gains in BC, and SQLE expression appears to be tightly
regulated by increases in the copy dosage of its gene locus
(17,24,26).
SQLE methylation has also been associated with SQLE overexpression
in BC (25).
Recently, it has been suggested that the
dysregulation of SQLE may be associated with the prognosis in
patients with BC. Helms et al (17) compared SQLE mRNA expression of BC
cases with and without chromosomal 7p and 8q gains by suppression
subtractive hybridization PCR. SQLE mRNA expression was upregulated
in BC with ER+ 7p/8q gains (17). Although SQLE expression was not
associated with tumor size, grade and ER or HER2 status, SQLE mRNA
expression was an independent risk factor for the early onset of
distant metastasis among early stage I/II BC cases (17). Simigdala et al (20) found that the cholesterol biosynthesis
pathway was the common adaptive mechanism associated with acquired
resistance to aromatase inhibitors in the ER+ long-term
estrogen-deprived cell lines. The aforementioned study analyzed
in-silico data from primary patients with ER+ BC with
neoadjuvant aromatase inhibitor therapy and revealed that increased
on-treatment SQLE expression was significantly associated with poor
response to endocrine therapy (20).
Kim et al (21) found that
SQLE was one of the important genes for BC metastasis based on a
standardized pathway-based approach. Shkurnikov et al
(22) searched for novel parameters
predicting the risk of relapse in patients with BC using public
microarray datasets, revealing that SQLE expression was
significantly associated with the risk of relapse in patients with
BC. Parada et al (23)
examined racial differences in the expression levels of eight
genes, including SQLE, and their associations with the risk of BC
recurrence among white and black women with BC. Compared with white
women, black women exhibited higher expression levels of SQLE, and
high SQLE expression was associated with increased risk of BC
recurrence (23). Chin et al
(24) performed a high-resolution
comparative genomic hybridization analysis in BC, revealing that
8q24 locus, where the SQLE gene resides, is one of the hotspot
genomic regions exhibiting the strongest association between copy
number gain and aberrant gene expression in high-grade,
ER− BC. Furthermore, amplification of 8q24 was
associated with a poor prognosis independently of standard
prognostic factors (24). Brown
et al (26) independently
confirmed that SQLE is a bona fide oncogene by amplification
with clinical relevance in BC. SQLE overexpression was more
prevalent in high-grade, HER2+ and hormone
receptor-negative invasive BC, and was an independently significant
unfavorable prognostic biomarker in BC (26). Yu et al (18) evaluated gene expression profiles from
early-onset BC tissues (age of patients, <40 years) and their
adjacent normal tissues to explore the genes and prognostic factors
associated with BC, revealing that SQLE expression was upregulated
in BC and that high SQLE expression was associated with a poor
prognosis.
Most studies on SQLE in BC have been based on
molecular approaches. The present study investigated whether SQLE
detection by immunohistochemistry could predict the prognosis in
patients with BC. High SQLE expression was more prevalent in
aggressive BC, such as larger tumor size, nodal metastases, higher
stage, HER2 subtype and distant metastatic relapse. High SQLE
expression was associated with poor disease-free survival and
overall survival, and independently predicted unfavorable
disease-free survival in patients with BC. SQLE was subsequently
verified as a prognostic biomarker for BC using the public
BreastMark database. High SQLE mRNA expression was significantly
associated with a poor prognosis in the ‘all’, lymph node negative,
lymph node positive, luminal A subtype and luminal B subtype
groups. The present results support the findings of previous
studies (17,18,20–24,26) and
suggest that high SQLE expression assessed by immunohistochemistry
may be associated with a more aggressive phenotype in BC and may be
used as a prognostic marker in patients with BC.
In the present study, high SQLE expression in DCIS
was significantly associated with high nuclear grade, comedo-type
necrosis and HER2 positivity, which are risk factors for DCIS
recurrence or progression (27).
However, high SQLE expression was not associated with the
recurrence of DCIS. Considering the potential prognostic value of
SQLE expression in patients with DCIS, further studies in a large
cohort of DCIS cases with long-term follow-up period are
required.
Several studies have demonstrated that SQLE promotes
cancer cell proliferation and migration, and the presence of SQLE
inhibitors in both in vitro and in vivo models causes
cancer cell death (11,13). Brown et al (26) demonstrated that SQLE inhibition
decreased the viability of a BC cell line in a
copy-dosage-dependent way and increased the doubling time only in
BC cell lines with high SQLE expression. In the present study, SQLE
mRNA expression was significantly increased in BC tissues compared
with in adjacent normal breast tissues. The current findings may
pave the way for the development of novel therapeutic strategies
aimed at SQLE in BC. Further studies in vivo, such as animal
models of BC, are required to elucidate the mechanism of action of
SQLE in promoting BC and to evaluate the therapeutic efficacy of
SQLE inhibitors for the treatment of BC.
The selection of appropriate internal control genes
is crucial for proper interpretation of RT-qPCR data. In the
present study, only the GAPDH gene was used as an internal
reference control because the mRNA expression levels were constant
in different tissue samples. It is recommended to use at least two
reference genes to increase the resolution and accuracy of the
RT-qPCR analysis (37). Therefore,
the accuracy of the RT-qPCR results in the present study may be
limited.
In summary, the current results suggested that
upregulation of SQLE expression serves an important role in BC
progression. Analysis of SQLE expression by immunohistochemistry
may be a useful biomarker to predict the prognosis in patients with
BC. Therefore, the present findings may open the way for further
research in clinical settings to assess the relevance of SQLE
inhibition as a new treatment option in patients with BC.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Chonnam National University Hwasun Hospital Institute for
Biomedical Science (grant no. HCRI20013).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to privacy and other
restrictions, but are available from the corresponding author on
reasonable request.
Authors' contributions
NIK conceived the experiments and wrote the
manuscript. NC designed the experiments. MHP prepared the samples.
NIK and MHP performed the experiments and confirmed the
authenticity of all the raw data. NIK and NC processed and analyzed
the data. SSK designed the study, performed statistical analysis
and edited the manuscript. JSL developed the project, collected and
analyzed the data, and wrote and revised the manuscript with input
from all authors. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was a retrospective study that
utilized archived materials of normal, DCIS and BC tissues and did
not impact patient care; hence, approval was granted by the
Institutional Review Board of Chonnam National University Hwasun
Hospital (Jeollanam, Republic of Korea) without patient consent
(reference no. CNUHH-2020-056).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Park EH, Min SY, Kim Z, Yoon CS, Jung KW,
Nam SJ, Oh SJ, Lee S, Park BW, Lim W, et al Korean Breast Cancer
Society, : Basic facts of breast cancer in Korea in 2014: The
10-year overall survival progress. J Breast Cancer. 20:1–11. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akram M, Iqbal M, Daniyal M and Khan AU:
Awareness and current knowledge of breast cancer. Biol Res.
50:332017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cirmena G, Franceschelli P, Isnaldi E,
Ferrando L, De Mariano M, Ballestrero A and Zoppoli G: Squalene
epoxidase as a promising metabolic target in cancer treatment.
Cancer Lett. 425:13–20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Silvente-Poirot S and Poirot M:
Cholesterol metabolism and cancer: The good, the bad and the ugly.
Curr Opin Pharmacol. 12:673–676. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu J, Locasale JW, Bielas JH, O'Sullivan
J, Sheahan K, Cantley LC, Vander Heiden MG and Vitkup D:
Heterogeneity of tumor-induced gene expression changes in the human
metabolic network. Nat Biotechnol. 31:522–529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Silvente-Poirot S and Poirot M: Cancer.
Cholesterol and cancer, in the balance. Science. 343:1445–1446.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Silvente-Poirot S and Poirot M:
Cholesterol epoxide hydrolase and cancer. Curr Opin Pharmacol.
12:696–703. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagai M, Sakakibara J, Wakui K, Fukushima
Y, Igarashi S, Tsuji S, Arakawa M and Ono T; Nagai M1, ; Sakakibara
J, Wakui K, Fukushima Y, Igarashi S, Tsuji S, Arakawa M and Ono T:
Localization of the squalene epoxidase gene (SQLE) to human
chromosome region 8q24.1. Genomics. 44:141–143. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stopsack KH, Gerke TA, Sinnott JA, Penney
KL, Tyekucheva S, Sesso HD, Andersson SO, Andrén O, Cerhan JR,
Giovannucci EL, et al: Cholesterol metabolism and prostate cancer
lethality. Cancer Res. 76:4785–4790. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stopsack KH, Gerke TA, Andrén O, Andersson
SO, Giovannucci EL, Mucci LA and Rider JR: Cholesterol uptake and
regulation in high-grade and lethal prostate cancers.
Carcinogenesis. 38:806–811. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu D, Wong CC, Fu L, Chen H, Zhao L, Li
C, Zhou Y, Zhang Y, Xu W, Yang Y, et al: Squalene epoxidase drives
NAFLD-induced hepatocellular carcinoma and is a pharmaceutical
target. Sci Transl Med. 10:4372018. View Article : Google Scholar
|
|
12
|
Souchek JJ, Baine MJ, Lin C, Rachagani S,
Gupta S, Kaur S, Lester K, Zheng D, Chen S, Smith L, et al:
Unbiased analysis of pancreatic cancer radiation resistance reveals
cholesterol biosynthesis as a novel target for radiosensitisation.
Br J Cancer. 111:1139–1149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qin Y, Zhang Y, Tang Q, Jin L and Chen Y:
SQLE induces epithelial-to-mesenchymal transition by regulating of
miR-133b in esophageal squamous cell carcinoma. Acta Biochim
Biophys Sin (Shanghai). 49:138–148. 2017.PubMed/NCBI
|
|
14
|
Zhang HY, Li HM, Yu Z, Yu XY and Guo K:
Expression and significance of squalene epoxidase in squamous lung
cancerous tissues and pericarcinoma tissues. Thorac Cancer.
5:275–280. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Römer AM, Lühr I, Klein A, Friedl A,
Sebens S, Rösel F, Arnold N, Strauss A, Jonat W and Bauer M: Normal
mammary fibroblasts induce reversion of the malignant phenotype in
human primary breast cancer. Anticancer Res. 33:1525–1536.
2013.PubMed/NCBI
|
|
16
|
Polycarpou-Schwarz M, Gross M, Mestdagh P,
Schott J, Grund SE, Hildenbrand C, Rom J, Aulmann S, Sinn HP,
Vandesompele J, et al: The cancer-associated microprotein CASIMO1
controls cell proliferation and interacts with squalene epoxidase
modulating lipid droplet formation. Oncogene. 37:4750–4768. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Helms MW, Kemming D, Pospisil H, Vogt U,
Buerger H, Korsching E, Liedtke C, Schlotter CM, Wang A, Chan SY,
et al: Squalene epoxidase, located on chromosome 8q24.1, is
upregulated in 8q+ breast cancer and indicates poor clinical
outcome in stage I and II disease. Br J Cancer. 99:774–780. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu Z, He Q and Xu G: Screening of
prognostic factors in early-onset breast cancer. Technol Cancer Res
Treat. Feb 7–2020.(Epub ahead of print). doi:
10.1177/1533033819893670. View Article : Google Scholar
|
|
19
|
D'Arcy M, Fleming J, Robinson WR, Kirk EL,
Perou CM and Troester MA: Race-associated biological differences
among Luminal A breast tumors. Breast Cancer Res Treat.
152:437–448. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Simigdala N, Gao Q, Pancholi S,
Roberg-Larsen H, Zvelebil M, Ribas R, Folkerd E, Thompson A, Bhamra
A, Dowsett M, et al: Cholesterol biosynthesis pathway as a novel
mechanism of resistance to estrogen deprivation in estrogen
receptor-positive breast cancer. Breast Cancer Res. 18:582016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim S, Kon M and DeLisi C: Pathway-based
classification of cancer subtypes. Biol Direct. 7:212012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shkurnikov MY, Galatenko VV, Lebedev AE,
Podol'skii VE, Tonevitskii EA and Mal'tseva DV: On statistical
relationship between ADRA2A expression and the risk of breast
cancer relapse. Bull Exp Biol Med. 157:454–458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parada H Jr, Sun X, Fleming JM,
Williams-DeVane CR, Kirk EL, Olsson LT, Perou CM, Olshan AF and
Troester MA: Race-associated biological differences among luminal A
and basal-like breast cancers in the Carolina Breast Cancer Study.
Breast Cancer Res. 19:1312017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chin SF, Teschendorff AE, Marioni JC, Wang
Y, Barbosa-Morais NL, Thorne NP, Costa JL, Pinder SE, van de Wiel
MA, Green AR, et al: High-resolution aCGH and expression profiling
identifies a novel genomic subtype of ER negative breast cancer.
Genome Biol. 8:R2152007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parris TZ, Kovács A, Hajizadeh S, Nemes S,
Semaan M, Levin M, Karlsson P and Helou K: Frequent MYC
coamplification and DNA hypomethylation of multiple genes on 8q in
8p11-p12-amplified breast carcinomas. Oncogenesis. 3:e952014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brown DN, Caffa I, Cirmena G, Piras D,
Garuti A, Gallo M, Alberti S, Nencioni A, Ballestrero A and Zoppoli
G: Squalene epoxidase is a bona fide oncogene by amplification with
clinical relevance in breast cancer. Sci Rep. 6:194352016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schnitt SJ, Allred C, Britton P, Ellis IO,
Lakhani SR, Morrow M, Palazzo J, Reynolds C, Rutgers E, Simpson J,
et al: Ductal carcinoma in situ. WHO Classification of Tumours of
the Breast. Lakhani SR, Ellis IO, Tan PH and van de Vijver MJ: IARC
Press; Lyon: pp. 90–94. 2012
|
|
28
|
Kim GE, Kim NI, Lee JS, Park MH and Kang
K: Differentially expressed genes in matched normal, cancer, and
lymph node metastases predict clinical outcomes in patients with
breast cancer. Appl Immunohistochem Mol Morphol. 28:111–122. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Madden SF, Clarke C, Gaule P, Aherne ST,
O'Donovan N, Clynes M, Crown J and Gallagher WM: BreastMark: An
integrated approach to mining publicly available transcriptomic
datasets relating to breast cancer outcome. Breast Cancer Res.
15:R522013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu YF, Yi Y, Qiu SJ, Gao Q, Li YW, Dai CX,
Cai MY, Ju MJ, Zhou J, Zhang BH, et al: PEBP1 downregulation is
associated to poor prognosis in HCC related to hepatitis B
infection. J Hepatol. 53:872–879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hammond ME, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS,
Hayes M, et al: American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer. Arch Pathol Lab Med. 134:907–922. 2010.PubMed/NCBI
|
|
33
|
Wolff AC, Hammond ME, Schwartz JN, Hagerty
KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer
A, et al American Society of Clinical Oncology; College of American
Pathologists, : American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for human epidermal
growth factor receptor 2 testing in breast cancer. J Clin Oncol.
25:118–145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bae YK, Gong G, Kang J, Lee A, Cho EY, Lee
JS, Suh KS, Lee DW and Jung WH; Breast Pathology Study Group of
Korean Society of Pathologists, : HER2 status by standardized
immunohistochemistry and silver-enhanced in situ hybridization in
Korean breast cancer. J Breast Cancer. 15:381–387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim GE, Kim JH, Lee KH, Choi YD, Lee JS,
Lee JH, Nam JH, Choi C, Park MH and Yoon JH: Stromal matrix
metalloproteinase-14 expression correlates with the grade and
biological behavior of mammary phyllodes tumors. Appl
Immunohistochem Mol Morphol. 20:298–303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jardel P, Debiais C, Godet J, Irani J and
Fromont G: Ductal carcinoma of the prostate shows a different
immunophenotype from high grade acinar cancer. Histopathology.
63:57–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kozera B and Rapacz M: Reference genes in
real-time PCR. J Appl Genet. 54:391–406. 2013. View Article : Google Scholar : PubMed/NCBI
|