Introduction
Hepatocellular carcinoma (HCC) is the most commonly
occurring form of primary liver cancer and is one of the leading
causes of cancer-associated death worldwide (1). HCC has poor prognosis and accounts for
75–85% of all primary liver cancer cases according to global cancer
statistics in 2018 (2). Current
therapies against HCC include ablation, liver transplantation and
radical resection, which is the main treatment (3). However, HCC shows a high recurrence.
Roayaie et al (4) reported
that the recurrence rate of HCC is 60% at 5 years after surgery
(4). Another study reported HCC
recurrence, including true recurrence due to dissemination and
de novo tumors within the oncogenic liver, makes 70% of
cases worsen within 5 years after surgery (5). These have become the main problems that
limit the 5-year survival rate of patients with liver cancer. This
highlights the need to determine molecular mechanisms involved in
the development of HCC and identify molecules that can be used in
anti-HCC therapy. Developing new methods for predicting the
prognosis of this disease is also important for improving the
prognosis prediction of patients with HCC may help inform treatment
choices.
Peroxisome proliferator activated receptor γ
(PPARγ), a ligand-activated transcription factor, is a member of
the nuclear receptor superfamily of PPAR proteins. PPARγ has two
subtypes: PPARα And PPARβ (6). PPARα
is involved in regulating various processes, from inflammation and
immunity to nutrient metabolism and energy balance (7). PPARβ has been shown to be involved in
metabolism, angiogenesis and inflammatory responses (8). PPARγ plays an important role in the
occurrence and development of various diseases, including obesity,
inflammation, diabetes, atherosclerosis and cancer (9). This protein is also a major regulator
of fat cell formation. In adipocytes, PPARγ regulates the
expression of genes controlling the uptake and storage of free
fatty acids and mediates the endocrine function of adipose tissue
(10). A previous study showed that
PPARγ may be involved in the occurrence and development of liver
cancer (11). However, the specific
mechanisms involved are unclear. It is also undetermined whether
PPARγ acts as an antitumor or tumor-promoting factor in liver
cancer (12).
Patients with HCC usually have a background of
chronic liver disease such as non-alcoholic fatty liver disease
(NAFLD), and chronic hepatitis B and C (13). NAFLD, a disease closely associated
with metabolic syndrome, includes chronic liver diseases ranging
from simple steatosis to non-alcoholic steatohepatitis (NASH)
(14). Increasing epidemiological
evidence indicates that NAFLD is a major cause of HCC (15). Numerous studies have shown that PPARγ
is involved in the pathogenesis of NAFLD (10,15–20).
Elevated PPARγ levels can increase the expression of genes
responsible for lipid metabolism in the liver, which induces liver
steatosis and leads to NAFLD (21–23).
Therefore, based on the previously identified role of PPARγ in
liver cancer (24–26), the present study hypothesized that
PPARγ may be involved in the development of HCC by affecting the
metabolism of liver fat cells.
The present study examined the role of PPARγ in HCC
development. Databases including The Cancer Genome Atlas (TCGA),
Gene Expression Omnibus (GEO) and Kaplan-Meier plotter were used to
predict the expression and function of PPARγ using bioinformatics.
Then, the differences between the results obtained via
bioinformatics analysis and those obtained by analysis of clinical
data of 125 patients with HCC were compared. It was explored
whether PPARγ participates in the development and progression of
HCC by affecting lipid metabolism. Finally, PPARγ expression
profiles and other selected prognostic clinical indicators were
used to build a model for predicting the prognosis of patients with
HCC.
Materials and methods
Differential analysis of PPARG mRNA
expression in patients with HCC conducted using TCGA and GEO
databases
The data on PPARγ expression were downloaded
from TCGA database (https://genome-cancer.ucsc.edu/) and GEO database
(https://www.ncbi.nlm.nih.gov/geo/),
then analyzed using the limma package of R software (version
3.5.2.) (27). The Wilcoxon
signed-rank test was used to evaluate PPARG expression in
HCC tissues and normal adjacent tissues of patients with HCC.
Interactive gene expression
profiling
Gene Expression Profiling Interactive Analysis
(GEPIA) (http://gepia.cancer-pku.cn/) is an
interactive web application based on 9,736 tumors and 8,587 samples
of normal tissue from TCGA and Genotype-Tissue Expression
databases. GEPIA can be used for the profiling of cancerous and
normal gene expression, and for interactive analyses (28). This database was used to explore the
relationship between PPARγ expression levels and prognosis
in patients with HCC.
Kaplan-Meier plotter analysis
The Kaplan-Meier plotter (http://kmplot.com/analysis/) database contains gene
expression and clinical data for 21 different types of cancer
including liver, lung, ovarian, gastric and breast cancer (29). Kaplan-Meier plotter was used to
evaluate the prognostic value of PPARγ in patients with HCC.
The median PPARγ expression (score of 1; Table I) was used as the cut-off value,
which was used to divide patients into high and low expression
groups and the prognosis of the two groups was compared.
 | Table I.Specific scoring criteria for
staining intensity and proportion of stained cells. |
Table I.
Specific scoring criteria for
staining intensity and proportion of stained cells.
|
| Points |
|---|
|
|
|
|---|
| Specific
criteria | 0 | 1 | 2 | 3 | 4 |
|---|
| Dyeing
intensity | Absent | Weak positive | Moderately
positive | Strong
positive | Strong
positive |
| Percentage range of
positively stained cells, % | 0 | 1-25 | 26-50 | 51-75 | 76-100 |
Analysis of the human protein
atlas
The Human Protein Atlas (https://www.proteinatlas.org), an open-access
resource, was used to map all the human proteins in cells, tissues
and organs. This mapping was performed to explore the distribution
of PPARγ protein in tumor and adjacent normal tissues of patients
with HCC.
Analysis of kyoto encyclopedia of
genes and genomes (KEGG) and gene set enrichment analysis
(GSEA)
GSEA (http://www.broadinstitute.org/gsea/index.jsp) is a
powerful analytical method that can interpret genome-wide
expression profiles (30). KEGG
enrichment analysis (31) for
PPARγ was performed using The Database for Annotation,
Visualization and Integrated Discovery (https://david.ncifcrf.gov), and evaluated the
PPARγ pathways using GSEA. These analyses were performed
using the clusterProfiler package of R software (version 3.5.2
(32).
Clinical factors affecting HCC
development and progression
The study was approved by The Ethics Committee of
the Zhejiang Provincial People's Hospital (Hangzhou, China).
Clinical data on 125 patients with HCC were collected from the
Zhejiang Provincial People's Hospital. The diagnosis of HCC in all
the patients included was confirmed using histopathological
examination by independent pathologists. Paraffin samples of HCC
were collected in March 2020. These patients underwent surgical
operations at Zhejiang Provincial People's Hospital from January
2008 to December 2015, and patients provided written informed
consent at the time of tissue collection. The patients had complete
follow-up data from the day of surgical resection of the primary
tumor to death or the last follow-up. The last follow-up date was
March 2016. The Tumor-Node-Metastasis (TNM) stage was defined
according to the criteria described in the 8th American Joint
Committee on Cancer guidelines (33). The inclusion criteria were as
follows: i) Patients who underwent surgery, ii) patients with the
histological type hepatocellular carcinoma and iii) patients with
complete follow-up information. The exclusion criteria included: i)
patients with other malignant diseases and (II) those without
detailed clinical information. In addition, 43 clinical factors,
including PPARγ protein expression, were collected for all the
patients (Table SI). Lasso
regression was then performed on these clinical factors using the
glmnet package (version 4.0–2.) in R software (http://www.jstatsoft.org/v33/i01/) (34). Univariate Cox regression analysis was
also performed using R software on the selected clinical factors.
Finally, the selected factors were used to establish a nomogram
model using the rms, foreign and survival package in R software.
The area under the curve (AUC) of receiver operating characteristic
(ROC) and concordance index (C-index) were used to evaluate the
accuracy of the model. Meanwhile, in order to validate that PPARγ
did play a role in the established nomogram, other clinical factors
besides PPARγ were used to establish another nomogram to obtain its
C-index and AUC for comparison.
Tissue microarray and
immunohistochemical (IHC) analyses
Specimens that included tumor tissue and
corresponding adjacent normal tissues were collected from 125
patients with HCC. A tissue microarray was constructed using paired
tumor tissue and adjacent normal tissue collected 0.5–1.0 cm from
the margin of the tumor (35).
Immunolabeling of the tissue microarray was performed using an
anti-PPARγ polyclonal antibody (1:200; cat. no. ab209350; Abcam).
Briefly, sections from the tissue microarray were baked at 70°C for
2 h and then deparaffinized using xylene (cat. no. 534596;
Sigma-Aldrich; Merck KGaA). A gradient of ethanol concentrations
(100, 95 and 80%) was used to rehydrate the sections, and antigen
retrieval was performed by boiling the sections using a
high-pressure cooker for 3 min in 1 mM Tris-EDTA buffer. The
sections were blocked with 3% hydrogen peroxide at room temperature
for 15 min to inhibit endogenous peroxidase activity. Sections were
then incubated with 10% goat non-immune serum (cat. no. ZLI-9022;
Zsbio Store) for 20 min to reduce non-specific staining.
Consequently, the sections were incubated with the primary antibody
at 4°C overnight, and then with the Biotin-SP-AffiniPure Goat
Anti-Rabbit IgG (H+L) (cat. no. 111-065-144; 1:500; Jackson
ImmunoResearch Europe, Ltd.) at room temperature for 15 min.
Afterwards, the sections were incubated with HRP-conjugated
streptavidin (cat. no. 3999s; CST Biological Reagents Co., Ltd.) at
room temperature for another 15 min. Protein expression was
visualized using a diaminobenzidine substrate kit (cat. no.
ab64238; 1:50; Abcam) and hematoxylin was used for 5 min at room
temperature to counterstain the sections.
Evaluation of IHC labeling
Two experienced pathologists, blinded to the
patients' pathology reports, independently scored the results of
IHC labeling based on intensity and proportion of positively
stained cells by light microscopy (cat. no. NI-SS 933679; Nikon
Corporation). Signal intensity was expressed as: 0 for absent, 1
for weak positive, 2 for moderately positive and 3 for strong
positive. The proportion of positively stained cells was also
quantified as: 0 for 0% positively stained cells, 1 for 1–25%
positively stained cells, 2 for 26–50% positively stained cells, 3
for 51–75% positively stained cells and 4 for 76–100% positively
stained cells. Specific evaluation criteria are shown in Table I. The final score, obtained by
multiplying the intensity score by the percentage of positively
labeled cells, was used to indicate the expression of the PPARγ
protein. A score of ≤1 was defined as low PPARγ expression, while
that of >1 was defined as high PPARγ expression.
Statistical analysis
Kaplan-Meier analysis was performed using log-rank
tests. Lasso regression was used to assess the prognosis of
patients with HCC and identify potential risk factors, and Lasso
analysis sampled and analyzed the sample 1,000 times. Univariate
Cox regression analysis was used to analyze the prognosis of
clinical factors screened by Lasso analysis. The χ2 test
was used to examine the association between PPARγ and other
clinical factors in categorical variables. When variables with ≥20%
of the cells had a count of ≤5, the data were analyzed using
Fisher's exact test. Additionally, the ROC curve was plotted and
the AUC was calculated. The C-index and AUC were used to analyze
the accuracy of the nomogram. All statistical analyses were
performed using the R software package. Confidence interval (CI)
was set to 95%. P<0.05 was considered to indicate a
statistically significant difference.
Results
Bioinformatics analysis of PPARγ mRNA
and protein expression
The results of the bioinformatics analysis showed
that PPARγ mRNA expression in HCC tumor tissues was higher
compared with that in normal tissues obtained from TCGA database
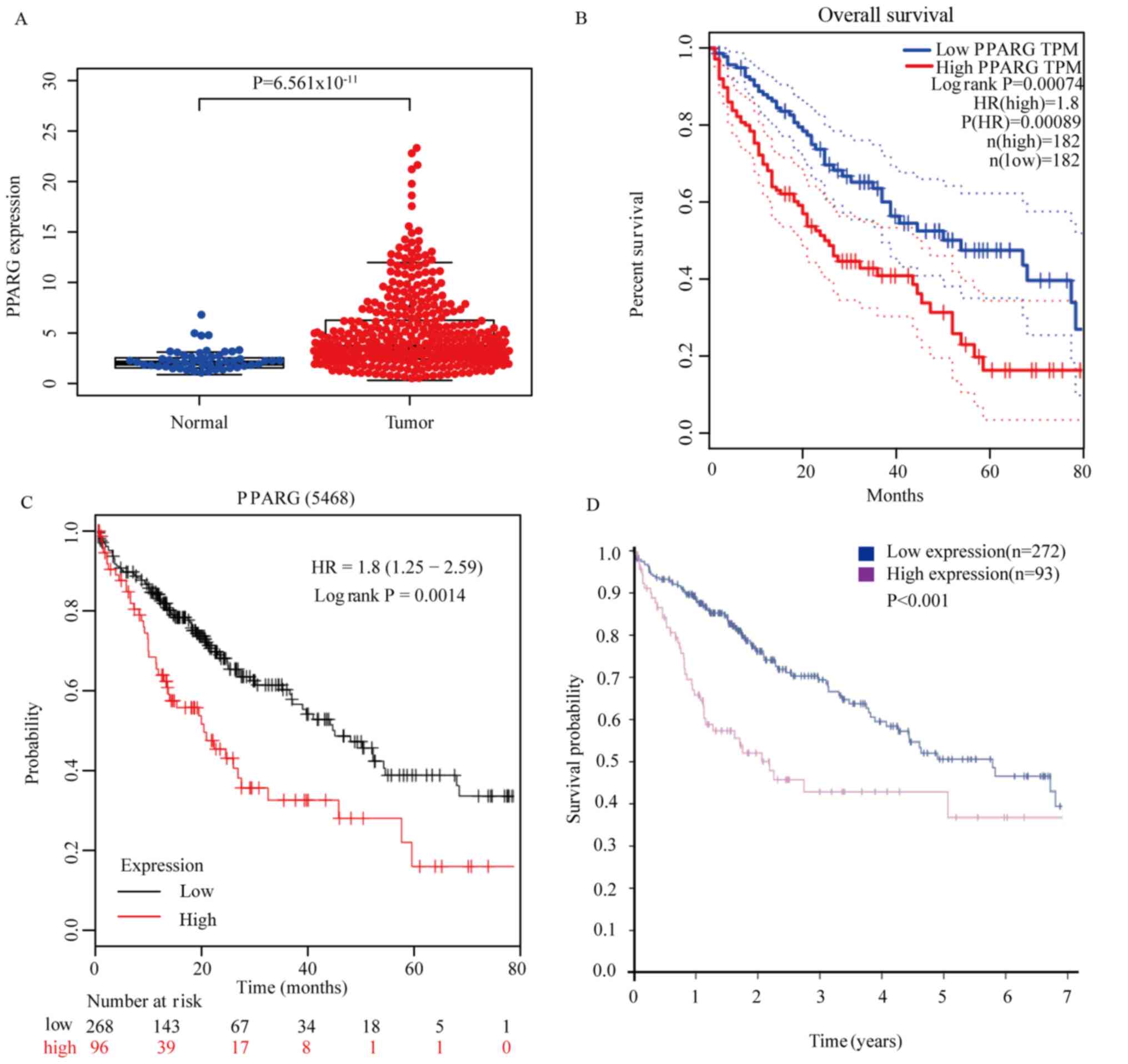
(P=6.561×10−11; Fig. 1A).
High PPARG expression was indicative of poor prognosis in
patients with HCC as assessed using GEPIA (P=0.00074; Fig. 1B). It was concluded that high
PPARγ expression was detrimental to patient prognosis by
using Kaplan-Meier plotter analysis (P=0.0014; Fig. 1C). Overexpression of PPARγ protein in
HCC tissues was predictive of unfavorable prognosis in patients
with HCC as assessed using analysis of the Human Protein Atlas
(P<0.001; Fig. 1D). In summary,
bioinformatics analysis reported that high PPARγ mRNA or
protein level in tumor tissues compared with normal tissues
indicated a poor prognosis.
Relationship between PPARγ expression
in HCC tissues and prognosis of patients with HCC
PPARγ expression in tumor tissues was higher
compared with that in normal liver tissues by analysis of PPARγ
expression in 125 patients with HCC (P<0.001; Fig. 2A), and the high expression of PPARγ
was beneficial to the prognosis of patients (P=0.015; Fig. 2B). IHC was used to evaluate PPARγ
expression in tumor tissues and corresponding adjacent normal
tissues. PPARγ expression in tissues was graded according to the
criteria aforementioned.
Evaluation of IHC labeling
As shown in Fig. 3,
PPARγ levels in tumor tissues (Fig. 3A
and B) were higher compared with those in the adjacent normal
tissues (Fig. 3C and D) of patients
with HCC (P<0.001; Fig. 3A).
Increased expression of PPARγ in HCC tumor tissues was indicative
of an improved prognosis in patients with HCC (P=0.015; Fig. 2B).
Association between PPARγ expression
and clinical features in patients with HCC
The associations between PPARγ expression levels and
key clinical characteristics of patients with HCC were examined
(Table II). The results indicated
that three clinical features including age (P=0.019), tumor
diameter (P=0.041) and vascular invasion (P=0.026) were
significantly associated with PPARγ expression. However, PPARγ
expression was not significantly associated with factors such as
hepatitis B, cirrhosis and metabolism-related indicators (Table II).
 | Table II.Association between PPARγ and the
clinical characteristics. |
Table II.
Association between PPARγ and the
clinical characteristics.
|
| PPARγ
expression |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Factors | Low | High | Total, n | χ2 | P-value |
|---|
| Sex |
|
|
| 0.629 | 0.428 |
|
Male | 46 | 52 | 98 |
|
|
|
Female | 15 | 12 | 27 |
|
|
| Age, years |
|
|
| 5.464 | 0.019 |
|
≤60 | 43 | 32 | 75 |
|
|
|
>60 | 18 | 32 | 50 |
|
|
| Tumor diameter,
cm |
|
|
| 4.163 | 0.041 |
| ≤4 | 29 | 42 | 71 |
|
|
|
>4 | 32 | 22 | 54 |
|
|
| Grade |
|
|
| 2.132 | 0.144 |
|
I+II | 45 | 54 | 99 |
|
|
|
III | 16 | 10 | 26 |
|
|
| TNM stage |
|
|
| 0.642 | 0.663 |
|
IA+IB+II | 56 | 61 | 117 |
|
|
|
IIIA+IIIB+IVA+IVB | 5 | 3 | 8 |
|
|
| AFP, µg/l |
|
|
| 0.321 | 0.571 |
|
≤200 | 41 | 46 | 87 |
|
|
|
>200 | 20 | 18 | 38 |
|
|
| Vascular
invasion |
|
|
| 4.979 | 0.026 |
|
Negative | 31 | 45 | 76 |
|
|
|
Positive | 30 | 19 | 49 |
|
|
| Hepatitis B |
|
|
| 0.261 | 0.609 |
|
Negative | 12 | 15 | 27 |
|
|
|
Positive | 49 | 49 | 98 |
|
|
| Cirrhosis |
|
|
| 0.913 | 0.339 |
|
Negative | 21 | 17 | 38 |
|
|
|
Positive | 40 | 47 | 87 |
|
|
| TG, mmol/l |
|
|
| 2.243 | 0.134 |
| ≤1 | 31 | 41 | 72 |
|
|
|
>1 | 30 | 23 | 53 |
|
|
| HDL, mmol/l |
|
|
| 0.001 | 0.990 |
|
≤1.04 | 42 | 44 | 86 |
|
|
|
>1.04 | 19 | 20 | 39 |
|
|
| LDL, mmol/l |
|
|
| 0.003 | 0.954 |
|
≤2.1 | 34 | 36 | 70 |
|
|
|
>2.1 | 27 | 28 | 55 |
|
|
| TCHO, mmol/l |
|
|
| 0.473 | 0.492 |
|
≤3.11 | 23 | 28 | 51 |
|
|
|
>3.11 | 38 | 36 | 74 |
|
|
Functional enrichment analyses for
PPARγ
KEGG pathway enrichment was conducted through GSEA
in high-expression group in the entire set by R software
(clusterProfiler package). It revealed that patients with low PPARγ
expression were associated with some pathways, including ‘mitotic
spindle,’ ‘G2M checkpoint,’ ‘E2F targets’,
‘spermatogenesis’, ‘mammalian target of rapamycin complex 1
signaling’, ‘fatty acid metabolism’, ‘TNFA signaling via NFκB’ and
‘xenobiotic metabolism’ (Fig. 4A and
B). Adjusted P<0.05 for a gene set were considered
statistically significant. Then a GSEA pathway analysis was
performed with respect to fatty acid metabolism pathways (Fig. 4C). This indicated that the high
expression of PPARγ gene in liver cancer is closely related
to fatty acid metabolism.
Screening for clinical factors
associated with prognosis
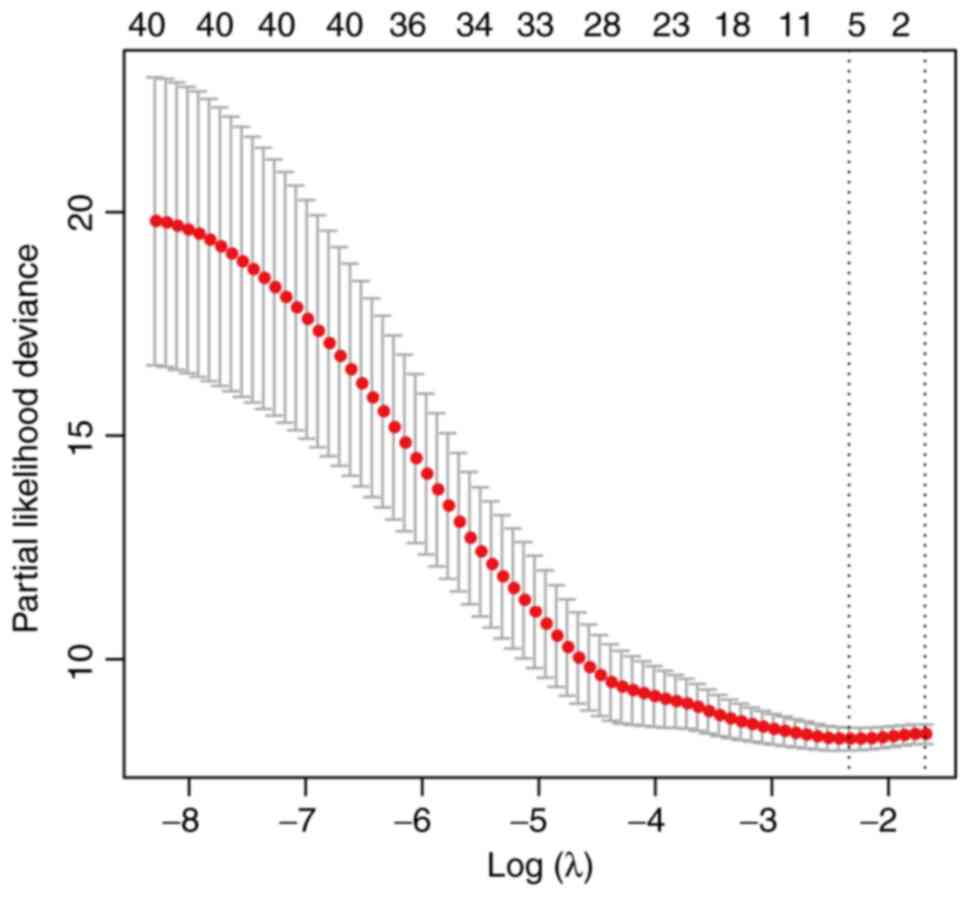
Lasso regression was used to analyze the 43 clinical
factors aforementioned (Table SI),
and identified the following seven clinical factors associated with
prognosis: Tumor-Node-Metastasis (TNM) stage, type of
differentiation, vascular invasion, α fetoprotein (APF),
carbohydrate antigen 199 (CA199), γ-glutamyl transpeptidase (γ-GT)
and PPARγ protein expression (Fig.
5).
Univariate analysis of the seven
clinical factors associated with prognosis
Univariate analysis was performed on the seven
clinical factors of TNM stage, grade, vascular invasion, APF,
CA199, γ-GT and PPARγ protein expression that were screened
previously using Lasso regression analysis. The results indicated
that TNM stage (P=0.005; Table
III), grade (P<0.001; Table
III), vascular invasion (P<0.001; Table III), APF (P=0.035; Table III), CA199 (P=0.024; Table III), γ-GT (P=0.002; Table III) and expression of PPARγ protein
(P=0.015; Table III) were
statistically significant factors affecting survival of patients
with HCC (Table III). These
results indicated that that these seven factors were independent
risk factors for the prognosis of patients with HCC.
 | Table III.Univariate analysis of survival in
125 patients with hepatocellular carcinoma. |
Table III.
Univariate analysis of survival in
125 patients with hepatocellular carcinoma.
| Factors | Value, n | P-value | HR | P-value (HR) |
|---|
| PPARγ |
|
|
|
|
| ≤1 | 61 | 0.015 | Reference | 0.018 |
|
>1 | 64 |
| 0.459
(0.240–0.875) |
|
| Grade |
|
|
|
|
|
I+II | 99 | <0.001 | Reference | 0.001 |
|
III | 26 |
| 2.848
(1.519–5.341) |
|
| TNM stage |
|
|
|
|
|
IA+IB+II | 117 | 0.005 | Reference | 0.009 |
|
IIIA+IIIB+IVA+IVB | 8 |
| 3.537
(1.372–9.116) |
|
| Vascular
invasion |
|
|
|
|
|
Negative | 76 | <0.001 | Reference | 0.001 |
|
Positive | 49 |
| 2.748
(1.475–5.121) |
|
| AFP, µg/l |
|
|
|
|
|
≤200 | 87 | 0.035 | Reference | 0.039 |
|
>200 | 38 |
| 1.928
(1.035–3.593) |
|
| CA199, µg/ml |
|
|
|
|
|
≤37 | 65 | 0.024 | Reference | 0.027 |
|
>37 | 60 |
| 2.141
(1.09–4.025) |
|
| γ-GT, U/l |
|
|
|
|
|
≤51 | 66 | 0.002 | Reference | 0.003 |
|
>51 | 59 |
| 2.707
(1.417–5.17) |
|
Construction of an OS prognostic
nomogram
The aforementioned seven clinical factors were used
to build an effective OS prognostic nomogram to predict the
prognosis of patients with HCC (Fig.
6). The C-index used to evaluate the accuracy of the nomogram
using these clinical factors was 0.755 (95% CI, 0.591 to 0.919;
P<0.001; data not shown) and the AUC values predicting the 1-,
3- and 5-year OS of the nomogram were 0.744, 0.780 and 0.780,
respectively (Fig. 7A-C). As for the
nomogram established by clinical factors that did not include
PPARγ, the C-index and AUC values were both lower. The C-index was
0.748 (95% CI, 0.584 to 0.912; P<0.001; data not shown). The AUC
values predicting the 1-, 3- and 5-year OS of the nomogram were
0.706, 0.751 and 0.751, respectively (Fig. 7D-F).
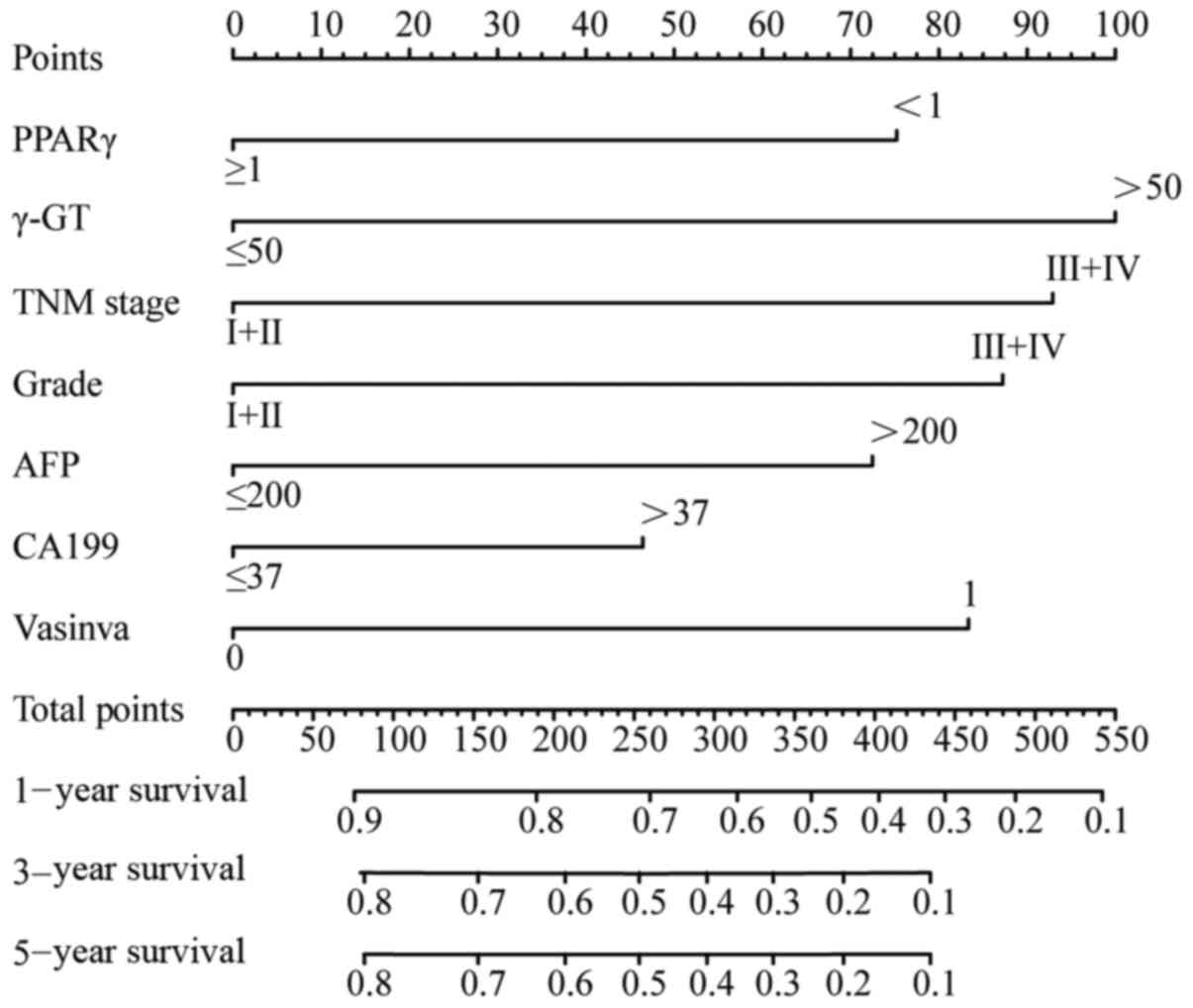 | Figure 6.In total, seven clinical factors,
including TNM stage, grade, vascular invasion, APF, CA199, γ-GT and
PPARγ protein expression, were used to establish an overall
survival prognostic nomogram. PPARγ, peroxisome
proliferator-activated receptor γ; TNM, Tumor-Node-Metastasis; AFP,
α fetoprotein; CA199, carbohydrate antigen 199; γ-GT, γ-glutamyl
transpeptidase. |
Discussion
HCC is a major human health concern because of its
high rates of recurrence and metastasis (36). Elucidating the molecular regulatory
mechanisms involved in HCC is necessary for improving diagnostic
methods and anti-HCC therapies. This requires detailed
understanding of the molecular regulatory mechanisms involved in
HCC (37). PPARγ is potentially
involved in mediation of HCC-related mechanisms (38,39).
However, the role of PPARγ expression in HCC remains controversial.
Some studies have shown high expression of PPARγ protein in HCC
tissues (40–42), while another report indicated that
PPARγ protein expression is decreased in HCC (43). Presently, PPARγ is considered to
exert an antitumor effect in HCC because PPARγ has been
identified as a tumor suppressor gene (44). PPARγ plays a key role in the
proliferation, migration, invasion and apoptosis of HCC cells. Cao
et al (45) have demonstrated
that PPARγ activation can inhibit the proliferation of liver
hepatic cancer cells by downregulating septin 2 expression. Lee
et al (46) reported that
PPARγ may be involved in modulating E-cadherin expression and
motility of HepG2 cells. Another study demonstrated that PPARγ can
directly inhibit the migration of HCC cells and is negatively
correlated with macrovascular invasion observed in HCC (47). Han et al (48) showed that hispidulin inhibits the
growth and metastasis of HCC via PPARγ activation, mediated by
AMPK and ERK signaling. In summary, previous studies
indicate that high expression of PPARγ in liver cancer exerts a
robust tumor-suppressive effect.
The present study first employed bioinformatics
analyses to establish that PPARγ expression in tumor tissues of
patients with HCC was higher compared with that in adjacent normal
liver tissues. PPARγ expression was examined in 125 clinical
samples collected from patients with HCC by performing tissue
microarray and IHC analyses and concluded that PPARγ was highly
expressed in liver tumor tissues. These results showed that PPARγ
can be used as a tumor biomarker in HCC.
The relationship between PPARγ expression levels and
several key clinical characteristics in patients with HCC were also
evaluated. The results indicated that PPARγ expression is
associated with age, tumor diameter and vascular invasion. However,
PPARγ expression was not significantly associated with factors such
as hepatitis B, cirrhosis and metabolism-related indicators. These
results concurred with the results of a previous study by Hsu et
al (47).
Bioinformatics analyses using multiple databases was
used to evaluate the relationship between PPARγ expression and
prognosis of patients with HCC. The results showed that increased
PPARγ protein or gene expression was indicative of a less favorable
prognosis, which contradicted the results from previous studies
(11,36,44). To
explain this discrepancy, the relationship between expression
levels of PPARγ and prognosis of 125 patients with HCC was
evaluated. The results indicated that high expression of PPARγ was
indicative of improved prognosis. The inconsistency between these
results and those obtained using bioinformatics analysis may be due
to the following reasons. The data used for the bioinformatics
analyses was collected on patients in Western countries, while the
present patient cohort included patients residing in China, which
may leads to bias in prognostic results. Second, China has a high
incidence of hepatitis B, and most cases of liver cancer in China
are caused by hepatitis B (49).
However, in Western countries, most liver cancer cases are caused
by over-consumption of alcohol (50). Therefore, the different pathogenesis
of liver cancer may account for the differences in prognosis.
Third, due to differences in eating habits, Westerners weigh more
than East Asians (51). PPARγ
expression may be affected by differences in body weights. These
differences in eating habits and weight likely contribute to
differences in prognosis. In addition, as the two sets of data to
be analyzed have different sources, other factors (such as
ethnicity, weight and treatment method) for the two sets of data
cannot be controlled. At the same time, there is research showing
that PPARγ mRNA and protein levels do not have the same
expression level, which indicates that there may be
post-transcriptional modifications in HCC (11).
Although the present study did not observe an
association between PPARγ expression and related indices of lipid
metabolism, it was reported that PPARγ was indeed enriched
in the ‘fatty acid metabolism’ pathway. Changes in liver metabolism
are critical to the development of liver disease. PPARγ is involved
in mitochondrial oxidative phosphorylation, gluconeogenesis and
fatty acid synthesis. Since modifications that affect mitochondria
and lipid metabolism all contribute to the occurrence and/or
development of liver steatosis, NAFLD, NASH and HCC, the function
of PPARγ in lipid metabolism is closely related to the occurrence
and development of liver cancer (25). This observation will be investigated
further in our future study into the role of PPARγ in liver cancer
pathogenesis.
To make the present study more comprehensive and
accurate, data on a total of 43 clinical factors for patients with
HCC were collected. First, Lasso regression was used to perform a
dimensionality reduction analysis on these factors. Consequently,
seven indicators were identified, including PPARγ expression, that
were closely associated with the prognosis of patients with HCC. A
univariate analysis was then performed on these indicators to
verify the results obtained using Lasso analysis. The results of
univariate analysis confirmed that these seven factors were indeed
independent risk factors associated with the prognosis of patients
with HCC. Finally, these prognosis-associated factors were used to
establish an OS prognostic nomogram for rapidly and accurately
predicting HCC prognosis. The C-index and AUC values of the
nomogram established with PPARγ were higher compared with those of
the nomogram established without PPARγ, which further demonstrated
that the expression of PPARγ may play a role in predicting the
prognosis of patients with HCC.
The present study used several innovative
approaches. First, PPARγ protein and gene expression was assessed
using bioinformatics analyses. Then Lasso analysis was used to
identify clinical and comprehensive factors associated with patient
prognosis. These data demonstrated that PPARγ was associated with
prognosis in patients with HCC. As single prognostic factors play
limited roles in prognosis (52), a
OS prognostic nomogram based on independent factors, such as PPARγ
expression, was constructed to predict the OS rate in patients with
HCC. This type of nomogram, which has been used previously as a
prognostic prediction model, integrates all prognostic factors to
achieve an individualized prediction (53). Additionally, this nomogram
incorporates an enhanced visual interface and is straightforward to
operate, which is advantageous in clinical practice (54). The limitations of the present study
were as follows. First, semi-quantitative analysis was used to
evaluate the results of PPARγ immunolabeling assay, which may have
led to statistical bias. Second, the insufficient number of samples
collected could not rule out the possibility of sampling error.
Additionally, single-center data were used to build the prognostic
model and was not verified using external data. Lastly, the role of
PPARγ in HCC pathogenesis needs to be explored in greater detail.
PPARγ should be knocked down or overexpressed in cells to observe
its effect on tumor progression and specific mechanisms in in
vivo and in vitro experiments, which will be the subject
of our future studies.
The present study reported that PPARγ was highly
expressed in the tumor tissues of patients with HCC, and its
expression level was associated with age, tumor diameter and
vascular invasion. These results indicated that PPARγ expression
can be used as a biomarker for predicting the prognosis of HCC. The
OS prognostic nomogram, established using clinical factors and
PPARγ expression levels, can be used to rapidly and accurately
predict the prognosis of patients with HCC, leading to improved
monitoring of the present patient population.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The study was funded by The Program on the Funds of
Science Technology Department of Zhejiang province (grant. no.
LGF19H160027) and The General Project Funds from the Health
Department of Zhejiang Province (grant. no. 2018259783).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request. The additional datasets analyzed during the current study
are available in The Cancer Genome Atlas database (https://genome-cancer.ucsc.edu/), Gene Expression
Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) and The Human
Protein Atlas (https://www.proteinatlas.org).
Authors' contributions
XLZ designed the study. WP provided administrative
support and made substantial contributions to the conception. XZ
collected the data. ZD and TT performed data analysis. YC
contributed to manuscript revisions and participated in data
collection and sorting. YW was involved in revising the manuscript
critically for important intellectual content, and analysis and
interpretation of data. XLZ, WP, YC and YW confirm the authenticity
of all raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by The Ethics Committee of
the Zhejiang Provincial People's Hospital (Hangzhou, China;
approval no. 2020QT103).
Patient consent for publication
The study received an informed consent exemption
approved by The Ethics Committee of the Zhejiang Provincial
People's Hospital.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Baffy G, Brunt EM and Caldwell SH:
Hepatocellular carcinoma in non-alcoholic fatty liver disease: an
emerging menace. J Hepatol. 56:1384–1391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy KR: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roayaie S, Obeidat K, Sposito C, Mariani
L, Bhoori S, Pellegrinelli A, Labow D, Llovet JM, Schwartz M and
Mazzaferro V: Resection of hepatocellular cancer ≤2 cm: Results
from two Western centers. Hepatology. 57:1426–1435. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yousefnia S, Momenzadeh S, Seyed Forootan
F, Ghaedi K and Esfahani MH: The influence of peroxisome
proliferator-activated receptor γ (PPARγ) ligands on cancer cell
tumorigenicity. Gene. 649:14–22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li S, Yang B, Du Y, Lin Y, Liu J, Huang S,
Zhang A, Jia Z and Zhang Y: Targeting PPARα for the treatment and
understanding of cardiovascular diseases. Cell Physiol Biochem.
51:2760–2775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Magadum A and Engel FB: PPARβ/δ: Linking
metabolism to regeneration. Int J Mol Sci. 19:20132018. View Article : Google Scholar
|
|
9
|
Lehrke M and Lazar MA: The many faces of
PPARgamma. Cell. 123:993–999. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Skat-Rørdam J, Højland Ipsen D,
Lykkesfeldt J and Tveden-Nyborg P: A role of peroxisome
proliferator-activated receptor γ in non-alcoholic fatty liver
disease. Basic Clin Pharmacol Toxicol. 124:528–537. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsu HT and Chi CW: Emerging role of the
peroxisome proliferator-activated receptor-gamma in hepatocellular
carcinoma. J Hepatocell Carcinoma. 1:127–135. 2014.PubMed/NCBI
|
|
12
|
Lee YH, Seo D, Choi KJ, Andersen JB, Won
MA, Kitade M, Gómez-Quiroz LE, Judge AD, Marquardt JU, Raggi C, et
al: Antitumor effects in hepatocarcinoma of isoform-selective
inhibition of HDAC2. Cancer Res. 74:4752–4761. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giannini EG, Aglitti A, Borzio M, Gambato
M, Guarino M, Iavarone M, Lai Q, Levi Sandri GB, Melandro F,
Morisco F, et al: Overview of immune checkpoint inhibitors therapy
for hepatocellular carcinoma, and The ITA.LI.CA cohort derived
estimate of amenability rate to immune checkpoint inhibitors in
clinical practice. Cancers (Basel). 11:16892019. View Article : Google Scholar
|
|
14
|
Anstee QM, Reeves HL, Kotsiliti E, Govaere
O and Heikenwalder M: From NASH to HCC: Current concepts and future
challenges. Nat Rev Gastroenterol Hepatol. 16:411–428. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choudhary NS, Kumar N and Duseja A:
Peroxisome proliferator-activated receptors and their agonists in
nonalcoholic fatty liver disease. J Clin Exp Hepatol. 9:731–739.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Westerbacka J, Kolak M, Kiviluoto T,
Arkkila P, Sirén J, Hamsten A, Fisher RM and Yki-Järvinen H: Genes
involved in fatty acid partitioning and binding, lipolysis,
monocyte/macrophage recruitment, and inflammation are overexpressed
in the human fatty liver of insulin-resistant subjects. Diabetes.
56:2759–2765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanaka N, Aoyama T, Kimura S and Gonzalez
FJ: Targeting nuclear receptors for the treatment of fatty liver
disease. Pharmacol Ther. 179:142–157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng X, Yu W, Li X, Zhou F, Zhang W, Shen
Q, Li J, Zhang C and Shen P: Apigenin, a modulator of PPARγ,
attenuates HFD-induced NAFLD by regulating hepatocyte lipid
metabolism and oxidative stress via Nrf2 activation. Biochem
Pharmacol. 136:136–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Samuel VT and Shulman GI: Nonalcoholic
fatty liver disease as a nexus of metabolic and hepatic diseases.
Cell Metab. 27:22–41. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Silva AKS and Peixoto CA: Role of
peroxisome proliferator- activated receptors in non-alcoholic fatty
liver disease inflammation. Cell Mol Life Sci. 75:2951–2961. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao M, Ma Y, Alsaggar M and Liu D: Dual
outcomes of rosiglitazone treatment on fatty liver. AAPS J.
18:1023–1031. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang SJ, Choi JM, Chae SW, Kim WJ, Park
SE, Rhee EJ, Lee WY, Oh KW, Park SW, Kim SW and Park CY: Activation
of peroxisome proliferator-activated receptor gamma by
rosiglitazone increases sirt6 expression and ameliorates hepatic
steatosis in rats. PLoS One. 6:e170572011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu S, Matsusue K, Kashireddy P, Cao WQ,
Yeldandi V, Yeldandi AV, Rao MS, Gonzalez FJ and Reddy JK:
Adipocyte-specific gene expression and adipogenic steatosis in the
mouse liver due to peroxisome proliferator-activated receptor
gamma1 (PPARgamma1) overexpression. J Biol Chem. 278:498–505. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng J, Dai W, Mao Y, Wu L, Li J, Chen K,
Yu Q, Kong R, Li S, Zhang J, et al: Simvastatin re-sensitizes
hepatocellular carcinoma cells to sorafenib by inhibiting
HIF-1α/PPAR-γ/PKM2-mediated glycolysis. J Exp Clin Cancer Res.
39:242020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Piccinin E, Villani G and Moschetta A:
Metabolic aspects in NAFLD, NASH and hepatocellular carcinoma: The
role of PGC1 coactivators. Nat Rev Gastroenterol Hepatol.
16:160–174. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shu Y, Lu Y, Pang X, Zheng W, Huang Y, Li
J, Ji J, Zhang C and Shen P: Phosphorylation of PPARγ at Ser84
promotes glycolysis and cell proliferation in hepatocellular
carcinoma by targeting PFKFB4. Oncotarget. 7:76984–76994. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou GX, Liu P, Yang J and Wen S: Mining
expression and prognosis of topoisomerase isoforms in
non-small-cell lung cancer by using Oncomine and Kaplan-Meier
plotter. PLoS One. 12:e01745152017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: ISBN 3-900051-07-0, 2012. http://www.R-project.org/
|
|
33
|
Bierley JD, Gospodarowicz MK and Wittekind
C: The TNM classification of malignant tumours. 8th edition.
Oxford: Wiley, Blackwell; 2017
|
|
34
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen S, Dong Z, Yang P, Wang X, Jin G, Yu
H, Chen L, Li L, Tang L, Bai S, et al: Hepatitis B virus X protein
stimulates high mobility group box 1 secretion and enhances
hepatocellular carcinoma metastasis. Cancer Lett. 394:22–32. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nojima H, Kuboki S, Shinoda K, Shimizu H,
Ohtsuka M, Kato A, Yoshitomi H, Furukawa K, Takayashiki T and
Miyazaki M: Activation of peroxisome proliferator-activated
receptor-gamma inhibits tumor growth by negatively regulating
nuclear factor-κB activation in patients with hepatocellular
carcinoma. J Hepatobiliary Pancreat Sci. 23:574–584. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cervello M, Emma MR, Augello G, Cusimano
A, Giannitrapani L, Soresi M, Akula SM, Abrams SL, Steelman LS,
Gulino A, et al: New landscapes and horizons in hepatocellular
carcinoma therapy. Aging (Albany NY). 12:3053–3094. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu Z, Meng SH, Bai JG, Sun C, Zhao LL,
Tang RF, Yin ZL, Ji JW, Yang W and Ma GJ: C/EBPα regulates FOXC1 to
modulate tumor growth by interacting with PPARγ in hepatocellular
carcinoma. Curr Cancer Drug Targets. 20:59–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tan J, Shen W, Shi W, Chen X, Sun D, Xu C,
Yan Q, Cheng H, Lai Y and Ji H: ONTD induces growth arrest and
apoptosis of human hepatoma Bel-7402 cells though a peroxisome
proliferator-activated receptor γ-dependent pathway. Toxicol In
Vitro. 45:44–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schaefer KL, Wada K, Takahashi H,
Matsuhashi N, Ohnishi S, Wolfe MM, Turner JR, Nakajima A, Borkan SC
and Saubermann LJ: Peroxisome proliferator-activated receptor gamma
inhibition prevents adhesion to the extracellular matrix and
induces anoikis in hepatocellular carcinoma cells. Cancer Res.
65:2251–2259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu J, Shen B, Chu ES, Teoh N, Cheung KF,
Wu CW, Wang S, Lam CN, Feng H, Zhao J, et al: Inhibitory role of
peroxisome proliferator-activated receptor gamma in
hepatocarcinogenesis in mice and in vitro. Hepatology.
51:2008–2019. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koga H, Sakisaka S, Harada M, Takagi T,
Hanada S, Taniguchi E, Kawaguchi T, Sasatomi K, Kimura R, Hashimoto
O, et al: Involvement of p21(WAF1/Cip1), p27(Kip1), and p18(INK4c)
in troglitazone-induced cell-cycle arrest in human hepatoma cell
lines. Hepatology. 33:1087–1097. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu J, Qiao L, Zimmermann L, Ebert MP,
Zhang H, Lin W, Röcken C, Malfertheiner P and Farrell GC:
Troglitazone inhibits tumor growth in hepatocellular carcinoma in
vitro and in vivo. Hepatology. 43:134–143. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu CW, Farrell GC and Yu J: Functional
role of peroxisome-proliferator-activated receptor gamma in
hepatocellular carcinoma. J Gastroenterol Hepatol. 27:1665–1669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cao LQ, Shao ZL, Liang HH, Zhang DW, Yang
XW, Jiang XF and Xue P: Activation of peroxisome
proliferator-activated receptor-γ (PPARγ) inhibits hepatoma cell
growth via downregulation of SEPT2 expression. Cancer Lett.
359:127–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee HJ, Su Y, Yin PH, Lee HC and Chi CW:
PPAR(gamma)/PGC-1(alpha) pathway in E-cadherin expression and
motility of HepG2 cells. Anticancer Res. 29:5057–5063.
2009.PubMed/NCBI
|
|
47
|
Hsu HT, Sung MT, Lee CC, Kuo YJ, Chi CW,
Lee HC and Hsia CY: Peroxisome proliferator-activated receptor γ
expression is inversely associated with macroscopic vascular
invasion in human hepatocellular carcinoma. Int J Mol Sci.
17:12262016. View Article : Google Scholar
|
|
48
|
Han M, Gao H, Ju P, Gao MQ, Yuan YP, Chen
XH, Liu KL, Han YT and Han ZW: Hispidulin inhibits hepatocellular
carcinoma growth and metastasis through AMPK and ERK signaling
mediated activation of PPARγ. Biomed Pharmacother. 103:272–283.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lin L, Yan L, Liu Y, Qu C, Ni J and Li H:
The burden and trends of primary liver cancer caused by specific
etiologies from 1990 to 2017 at the global, regional, national,
age, and sex level results from the global burden of disease study
2017. Liver Cancer. 9:563–582. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Singal AG, Lampertico P and Nahon P:
Epidemiology and surveillance for hepatocellular carcinoma: New
trends. J Hepatol. 72:250–261. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Oliveros E, Somers VK, Sochor O, Goel K
and Lopez-Jimenez F: The concept of normal weight obesity. Prog
Cardiovasc Dis. 56:426–433. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tu Q, Hu C, Zhang H, Peng C, Kong M, Song
M, Zhao C, Wang Y, Li J, Zhou C, et al: Establishment and
validation of novel clinical prognosis nomograms for luminal a
breast cancer patients with bone metastasis. Biomed Res Int.
2020:19720642020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang X, Yuan K, Wang H, Gong P, Jiang T,
Xie Y, Sheng L, Liu D, Liu X and Xu G: Nomogram to predict
mortality of endovascular thrombectomy for ischemic stroke despite
successful recanalization. J Am Heart Assoc. 9:e0148992020.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bookman MA: Can we predict who lives long
with ovarian cancer? Cancer. 125 (Suppl 24):S4578–S4581. 2019.
View Article : Google Scholar
|