Introduction
Hepatocellular carcinoma (HCC), a malignant tumor
arising from the liver, accounts for 80% of primary liver cancer
(1), and is the third leading cause
of cancer-related deaths globally (2) with a mortality of 0.598 million
according to the report released by WHO International Agency in
2018 (3). The 5-year survival rate
of patients with HCC is as low as 30% (4). It is imperative to improve
understanding of hepatocarcinogenesis and to develop novel
strategies for the treatment of HCC.
Long non-coding RNAs (lncRNAs), are a class of
non-coding transcripts with lengths exceeding 200 nucleotides,
which participate in a variety of physiological and pathological
processes, including embryogenesis, tumorigenesis and some
cardiovascular diseases (5). lncRNA
maternally expressed gene 3 (MEG3) has been reported to be
abnormally expressed in HCC tissues (6,7),
however, its role in HCC and the related mechanisms are not well
understood. MicroRNAs (miRs) are small (~22 nucleotide) non-coding
RNAs which also participate in various biological processes, such
as development, growth, organogenesis and angiogenesis (8,9). lncRNAs
and miRNAs can interact with each other to serve a wide spectrum of
biological functions, such as neovascularization, angiogenesis and
vasculogenic mimicry (10,11). miR-9-5p (GenBank: LM379059.1) is a
well-known microRNA which serves important roles in the growth,
angiogenesis, radio-resistance and metastasis of various types of
cancers, such as colorectal cancer, acute myeloid leukemia, and
gastric cancer (12–14). MEG3 has been reported to serve as a
competing endogenous RNA (ceRNA) for miR-9-5p in prostate cancer
(15) and esophageal cancer
(16) to mediate tumor
progression.
Midkine (MDK) was initially found as a growth factor
in retinoic acid-induced embryonic tumor cells (17). Currently, MDK is viewed as a
multifaceted factor promoting cell growth, survival, metastasis,
migration, and angiogenesis, in particular during tumorigenesis in
cancer (18).
Phosphoinositide-dependent kinase 1 (PDK1) is a serine/threonine
protein kinase which serves central roles in signal transduction
(19). Several studies have
demonstrated that PDK1 can promote cell proliferation by activating
the Akt/mTOR signaling pathway (20,21). A
recent study by Lu et al (22) reported that miR-9-5p can inhibit
angiogenesis by targeting MDK and regulating the PDK/AKT pathway in
nasopharyngeal carcinoma.
The present study aimed to investigate the effect of
MEG3 on HCC cell viability, apoptosis and migration. In addition,
the interaction between MEG3, miR-9-5p and MDK, and the activation
of PDK/AKT pathway in HCC cells was also assessed. The present
study provides new insights into the molecular mechanisms of MEG3
in HCC and suggests a novel therapeutic target for HCC.
Materials and methods
Cell culture
MHCC-97L cell line (cat. no. BNCC337741) was
purchased from Beijing Beina Chuanglian Institute of Biotechnology.
The cells were maintained in RPMI-1640 medium (Nanjing KeyGen
Biotech. Co. Ltd.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific Inc.) and 100 U/ml penicillin G
sodium and 0.1 mol/ml streptomycin sulfate, at 37°C for 48 h in a
humidified atmosphere of 5% CO2.
Transfection
miR-9-5p mimic (3.75 µl, 25 nM), miR-9-5p negative
control (NC; 3.75 µl, 25 nM), lncRNA MEG3 overexpression vector
(2.5 µg, 2.5 µg/ml) and lncRNA MEG3 NC (2.5 µg, 2.5 µg/ml) were
purchased from General Biosystems, Inc. The sequences of miR-9-5p
mimic and the corresponding NC were: miR-9-5p mimic sense,
5′-UCUUUGGUUAUCUAGCUGUAUGA-3′, antisense,
5′-UCAUACAGCUAGAUAACCAAAGA-3′; NC: Sense,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′, antisense,
5′-UCUACUCUUUCUAGGAGGUUGUGA-3′. Cell transfection was conducted
using Lipofectamine 3000® transfection reagent (Thermo
Fisher Scientific Inc.) according to the manufacturer's
instructions. Cells were incubated with the complexes at 37°C for 6
h, then the transfection media was replaced with culture medium. 48
h later, the cells were subjected to subsequent experimentation.
Cells in the control group were untransfected cells.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from MHCC-97L cells using
the Ultrapure RNA kit (CoWin Biosciences), and then
reverse-transcribed into cDNA using a HiFiScriptcDNA synthesis kit
(CoWin Biosciences). Reverse transcription was performed at 50°C
for 15 min, and then at 85°C for 5 min. Primers used for
amplification were: MEG3 forward, 5′-CATACAAAGCAGCCACTCAC-3 and
reverse, 5′-GGGATCCTTCCATTCAGGAC-3′; GAPDH forward,
5′-CAATGACCCCTTCATTGACC-3 and reverse, 5′-GAGAAGCTTCCCGTTCTCAG-3′;
miR-9-5p forward, 5′-TCTTTGGTTATCTAGCTGTATGA and reverse,
5′-CCAGCTATGCGCCATTAGCAA-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3 and reverse,
CGCTTCACGAATTTGCGTGTCAT-3′ (CoWin Biosciences). RT-qPCR assays were
performed using the UltraSYBR Mixture (CoWin Biosciences). Data
were analyzed using the 2−ΔΔCq method (23). Expression of MEG3 was normalized to
GAPDH, and expression of miR-9-5p was normalzed to U6.
Western blotting
Total proteins were extracted from MHCC-97L cells
using RIPA Cell Lysis Buffer (Applygen Technologies Inc.) and then
protein concentrations were quantified using the bicinchoninic acid
(BCA) method (Thermo Fisher Scientific Inc.). A total of 50 µg of
protein was separated using 10% SDS-PAGE gels and transferred to
PVDF membranes (EMD Millipore). After blocking overnight at 4°C in
5% BSA, the PVDF membranes were incubated with the primary
antibodies, including Rabbit Anti-caspase-3 (1:5,000; cat. no.
ab32351), rabbit anti-caspase-9 (1:2,000; cat. no. ab202068),
rabbit `nti-MDK (1:1,000; cat. no. ab52637), rabbit anti-PDK1
(1:2,000; cat. no. ab207450) (all Abcam), rabbit anti-AKT (1:500;
cat. no. bs-2720R; BIOSS), rabbit anti-matrix metalloproteinase-1
(MMP-1) (1:1,000; cat. no. bs-4597R; BIOSS), Rabbit
Anti-phosphorylated (p)-AKT (cat. no. bs-2720R; 1:1,000; BIOSS),
Rabbit Anti p-PDK1 (cat. no. AF3018; 1:1,000; Affinity Biosciences)
and Mouse Monoclonal Anti-GAPDH (cat. no. TA-08; 1:2,000;
OrigeneTechnologies, Inc.) at 4°C overnight. Horseradish
peroxidase-labeled secondary antibodies, including Goat Anti-Mouse
IgG (H+L) (cat. no. ZB-2305) and Goat Anti-Rabbit IgG (H+L) (cat.
no. ZB-2301) (both OrigeneTechnologies, Inc.), were used at 1:2,000
dilution and the membranes were incubated at room temperature for 2
h. GAPDH was used as the internal loading control. Signals were
detected with the SuperSignal® west pico
chemiluminescent substrate (Thermo Fisher Scientific Inc.), and
band density was determined by ImageLab software v.5.2 125104
(Bio-Rad Laboratories Inc.).
Luciferase reporter assay
Wild-type lncRNA MEG3 3′-UTR and MDK 3′-UTR
containing the putative binding sites of miR-9-5p (15,22) were
amplified and inserted into the firefly luciferase reporter vector
pmirGLO (Shanghai Enzyme Research Biotechnology Co., Ltd.).
miR-9-5p mimic and miR-NC were synthesized by Universal biological
systems (Anhui) Co., Ltd. The sequences of miR-9-5p mimic and the
corresponding NC were: miR-9-5p mimic sense,
5′-UCUUUGGUUAUCUAGCUGUAUGA-3′, antisense,
5′-UCAUACAGCUAGAUAACCAAAGA-3′; NC: Sense,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′, antisense,
5′-UCUACUCUUUCUAGGAGGUUGUGA-3′. Cells were co-transfected with the
miR-NC/miR-9-5p mimic and the wild-type 3′-UTR of MDK or lncRNA
MEG3 using Lipofectamine 3000® (Invitrogen; Thermo
Fisher Scientific Inc.). Firefly and Renilla luciferase
activities were detected 48 h later by the Dual-Luciferase Reporter
Assay System (Beyotime Institute of Biotechnology) using the Dual
Luciferase Reporter Gene Assay kit (Beyotime Institute of
Biotechnology). The firefly luciferase activity was normalized to
the Renilla luciferase activity for each individual
analysis.
CCK8 assay
Cells were seeded at 1×104 cells/well in
96-well plates and maintained in RPMI-1640 medium at 37°C in an
atmosphere of 5% CO2 for 48 h. Cell viability was
measured using a CCK8 Kit (Nanjing KeyGen Biotech. Co. Ltd.)
according to the manufacturer's instructions. Cells were incubated
with the CCK8 reagent (10 µl) at 37°C for 1 h. Absorbance at 450 nm
was measured using a Tecan Safire II Microplate Reader (Tecan Group
Ltd.).
Cell cycle analysis using flow
cytometry (FCM)
FCM analysis of the cell cycle of MHCC-97L cells was
performed using a Cell Cycle Staining kit [MultiSciences (Lianke)
Biotech Co., Ltd.] according to the manufacturer's instructions.
The cell suspension was centrifuged at 978 × g for 3 min, and the
cells were subsequently fixed in ethanol at 4°C for 2 h. After
washing with PBS for 3 times, 1 ml of DNA staining solution (PI
containing RNase A) was added to the tubes. The cells were
incubated at room temperature in the dark for 30 min, and the cell
cycle was analyzed using the NovoCyte 2060R flow cytometer (ACEA
Biosciences, Inc.) with NovoExpress software v.1.2.5 (ACEA
Biosciences, Inc.).
FCM analysis of cell apoptosis
Apoptosis rate of MHCC-97L cells was determined by
FCM using an Annexin V-FITC/propidium iodide PI Apoptosis kit
[MultiSciences (Lianke) Biotech Co., Ltd.]. Briefly,
1×106 cells were washed with cold PBS twice, and
re-suspended in 300 µl binding buffer. After incubation with 3 µl
Annexin V-FITC and 3 µl PI-PE at room temperature for 10 min in the
dark, the cells were mixed with 200 µl binding buffer, and cell
apoptosis rate was analyzed using a NovoCyte 2060R flow cytometer
(ACEA Biosciences, Inc.) with NovoExpress software v.1.2.5 (ACEA
Biosciences, Inc.). Early and late apoptosis were both
analyzed.
Wound healing assay
MHCC-97L cells were grown in FBS free medium to
~100% confluence in 6-well plates following transfection. The
confluent monolayers were scratched by using a 10 µl pipette tip,
and then washed with PBS 3 times. Images were taken by an inverted
fluorescence microscope (MF53; Guangzhou Mingmei Photoelectric Co.,
Ltd.) immediately or 24 h after wounding and migration was
quantified. Migration rate was calculated as the migration distance
divided by the migration time.
Statistical analyses
Statistical analyses were performed using SPSS
v.19.0 (IBM, Corp.). Al experiments were performed at least 3
times. Experimental data were presented as the mean ± SD, and the
statistical significance was assessed by one-way analysis of
variance followed by the post hoc least significant difference
(LSD) test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Effect of lncRNA MEG3 on HCC cell
viability, apoptosis and migration
In preliminary experiments, the expression of lncRNA
MEG3 was examined in 5 HCC cell lines and it was found that
MHCC-97L had the lowest lncRNA MEG3 expression (data not shown).
Therefore, the MHCC-97L cell line was chosen for subsequent
gain-off-function experiments. lncRNA MEG3 overexpression vector
was transfected into MHCC-97L cells to investigate the effect of
lncRNA MEG3 on HCC cell viability, apoptosis and migration. As
demonstrated in Fig. 1A, lncRNA MEG3
NC did not affect lncRNA MEG3 expression. Compared with the lncRNA
MEG3 NC group, lncRNA MEG3 was significantly increased in the
lncRNA MEG3 group (P<0.01; Fig.
1A). The effect of lncRNA MEG3 on cell viability was examined
by the CCK8 assay and FCM. The results from CCK8 assay demonstrated
that cell viability in the lncRNA MEG3 group was significantly
decreased when compared with that in the lncRNA MEG3 NC group
(P<0.01; Fig. 1B). FCM analysis
of cell cycle demonstrated that the fraction of S-phase cells was
significantly higher and the fraction of G2-phase cells
was significantly lower in the lncRNA MEG3 group compared with the
lncRNA MEG3 NC group (P<0.01; Fig. 1C
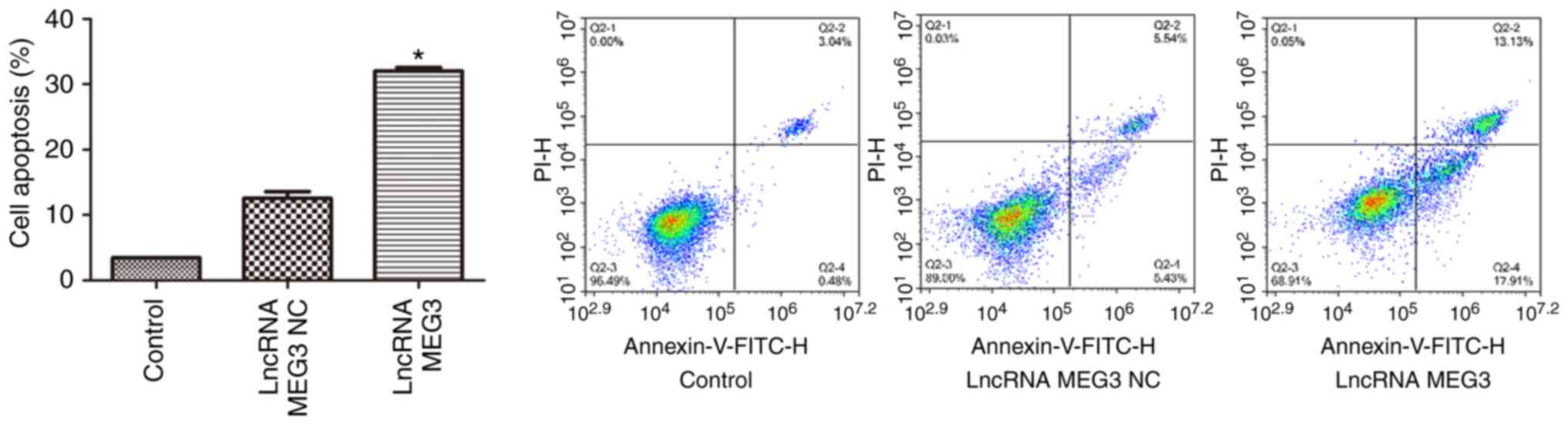
and D). FCM analysis of cell apoptosis revealed that compared
with the cells transfected with lncRNA MEG3 NC, cell apoptosis rate
was significantly increased in the cells transfected with lncRNA
MEG3 overexpression vector (P<0.01; Fig. 2). Wound healing assays were performed
to examine the effect of lncRNA MEG3 on cell migration. The results
demonstrated that compared with the lncRNA MEG3 NC group, cell
migration was inhibited in the lncRNA MEG3 group (P<0.01;
Fig. 3). Compared with the control
group, lncRNA MEG3 NC did not show any effects on cell viability,
apoptosis or migration (Figs.
1–3).
lncRNA MEG3 targets miR-9-5p and
miR-9-5p targets MDK
Luciferase reporter assays were used to investigate
whether lncRNA MEG3 targets miR-9-5p. As shown in Fig. 4A, compared with the lncRNA
MEG3+miR-9-5p mimic NC group, the luciferase activity was
significantly decreased in the lncRNA MEG3+miR-9-5p mimic group
(P<0.01). In addition, whether MDK was a direct target of
miR-9-5p was investigated. It was revealed that luciferase activity
was significantly decreased in the MDK+miR-9-5p mimic group when
compared with that in the MDK + miR-9-5p mimic NC group (P<0.01;
Fig. 4B).
Effect of lncRNA MEG3 on the
expression of miR-9-5p, MDK, apoptosis and migration-related
proteins and PDK/AKT signaling
lncRNA MEG3 effect on miR-9-5p expression was
determined by RT-qPCR analysis. As shown in Fig. 5A, compared with the lncRNA MEG3 NC
group, miR-9-5p expression was significantly decreased in the
lncRNA MEG3 group (P<0.01). The effect of lncRNA MEG3 on MDK
expression was determined by western blotting and it was revealed
that the relative protein level of MDK was significantly increased
in the lncRNA MEG3 group when compared with that in the lncRNA MEG3
NC group (P<0.01; Fig. 5B and C).
Western blotting also showed that the protein levels of caspase-3
and 9 were significantly upregulated in lncRNA MEG3-transfected
cells compared with lncRNA MEG3 NC-transfected cells (P<0.01;
Fig. 5B and C). Western blotting
indicated that the expression level of MMP1 was significantly lower
in the lncRNA MEG3-transfected cells compared with the lncRNA MEG3
NC-transfected cells (P<0.01; Fig. 5B
and C). In order to verify the effect of lncRNA MEG3 on PDK/AKT
signaling, western blotting was performed to examine p-PDK1 and
p-AKT expression. Compared with the lncRNA MEG3 NC group, the
protein expression of p-PDK1 and p-AKT was upregulated in the
lncRNA MEG3 group (both P<0.01; Fig.
5B and D).
 | Figure 5.Effect of lncRNA MEG3 on the
expression of miR-9-5p, MDK, apoptosis and migration-related
proteins, and PDK/AKT pathway activation. (A) Effect of lncRNA MEG3
on miR-9-5p expression. (B) Western blot images. (C) Relative
protein expression of MDK, caspase-3, caspase-9 and MMP1 determined
by western blotting. (D) Effect of lncRNA MEG3 on PDK/AKT
signaling. Cells in the control group were untransfected cells
(n=3). *P<0.01 compared with the lncRNA MEG3 NC group. lncRNA,
long non-coding RNA; MEG3, maternally expressed gene 3 (MEG3); HCC,
hepatocellular carcinoma; NC, negative control; miR, microRNA; MDK,
Midkine; PDK1, phosphoinositide-dependent kinase 1, p,
phosphorylated; MMP-1, matrix metalloproteinase 1. |
Discussion
The pathogenesis of HCC is complex and numerous
studies have demonstrated that lncRNAs are involved in the
initiation, development and metastasis of HCC (24,25).
Some studies have shown that MEG3 is significantly downregulated in
HCC tissues compared with carcinoma-adjacent tissues (6,7).
Restoring MEG3 expression in cancer cells can suppress cell
proliferation, invasion as well as angiogenesis, and induce cell
apoptosis (26,27). These studies indicated that MEG3 is
one of the lncRNAs with tumor suppressor activity (28). In the present study, the MEG3
overexpression vector was transfected into MHCC-97L cells to
investigate the effect of MEG3 on cell viability, apoptosis and
migration in HCC. The findings of the present study demonstrated
that MEG3 inhibited HCC cell viability and a large number of cells
accumulated in the S phase. Similarly, He et al (29) confirmed that MEG3 overexpression
suppressed clone formation of hepatoma cells. Caspase proteins
serve important roles in the process of apoptosis (30). Caspase-9 is an initiator of
apoptosis, and caspase-3 is the final executor of apoptosis
(30). In the present study,
compared with lncRNA MEG3 negative control, MEG3 overexpression led
to increased caspase-3 and caspase-9 expression, as well as an
increased cell apoptosis rate indicating that MEG3 promotes cell
apoptosis. In addition, in the present study MMP1 expression was
inhibited by MEG3 in MHCC-97L cells. MMP1 is involved in invasion
and metastasis during the development of cancer, and high
expression of MMP1 is associated with poor prognosis of patients
with HCC (31). The wound healing
assay results of the present study further verified the inhibitory
effect of MEG3 on cell migration. These results of the present
study were consistent with the previous studies (6,24), and
suggested that MEG3 may be a tumor suppressor in HCC.
lncRNAs can serve as a kind of ceRNA to regulate
target genes through interacting with miRNAs (32,33).
Recent studies have clarified that lncRNA-miRNA-mRNA form a novel
regulatory network in various pathophysiological processes, such as
differention, growth, necrosis and autophagy (34,35). The
role of MEG3 in cancers has been reported in several studies, in
which the interaction between lncRNAs, miRNA and mRNA has been
highlighted (36,37). In glioma, MEG3 inhibited the
proliferation, migration and invasion of glioma cells by regulating
miR-96-5p/MTSS I-BAR Domain Containing 1 (38). In multiple myeloma, MEG3 inhibited
tumor progression through regulating the miR-181a/Homeobox A11 axis
(39). Particularly, previous
studies have suggested that MEG3/miR-9-5p is involved in regulating
the progression of prostate cancer and esophageal cancer by
targeting Quaking 5 and Forkhead Box O1 (15,16).
Recently, a study by Liu et al (40) reported that MEG3 can serve as a ceRNA
for miR-9-5p to mediate the development of HCC through upregulating
SOX11. To further explore the molecular mechanisms underlying the
role of MEG3 in HCC, the present study investigated whether MEG3
exerts its protective effects on HCC via regulating miR-9-5p/MDK
axis. In the present study, to verify the targeting relationship
between MEG3, miR-9-5p and MDK, a dual luciferase reporter assay
was performed. Consistent with previous studies (15,40),
MEG3 directly targeted miR-9-5p in the present study. Subsequently,
the present study demonstrated that MDK is a direct target of
miR-9-5p. This finding was in agreement with a previous study which
demonstrated that exosomal miR-9 targets MDK in nasopharyngeal
carcinoma (22). In addition, the
present study revealed that the expression of miR-9-5p was
downregulated, and the expression of MDK was upregulated by MEG3,
indicating that MEG3 can regulate the miR-9-5p/MDK axis in HCC. The
PDK/AKT signaling pathway has been suggested to be linked with MDK
in angiogenesis, and MDK knockdown led to the inhibition of PDK1
and AKT in HUVECs (22). In the
present study, the results revealed that the PDK/AKT pathway was
activated by MEG3 in HCC. Altogether, the findings of the present
study demonstrated that MEG3 targets the miR-9-5p/MDK axis and
modulates PDK/AKT pathway in HCC.
In the present study, the MHCC-97L cell line was
chosen for gain-off-function experiments. Indeed, using at least
one other HCC cell line would make the present study more
convincing. It is a limitation of the present study that only 1
cell line was used. lncRNA and miRNA may also have negative
feedback regulation. Another limitation of the present study is
that it only demonstrated the effect of lncRNA MEG3 overexpression
on miR-9-5p, without reverse validation. The MHCC-97L cell line is
representative of HCC cell lines in previous studies (41,42). Li
et al (41) only selected the
MHCC-97L cell line to investigate the effects of miR-34a-5p on
proliferation and chemical resistance of HCC cells (41). To examine the role of TIMP
Metallopeptidase Inhibitor 2 (TIMP2) in HCC metastasis, Kai et
al (42) chose the MHCC-97L cell
line for research and found that the ectopic expression of the
TIMP2 open reading frame in highly metastatic HCC cell line
MHCC-97L significantly reduced the progress of HCC. Future studies
should continue investigating the effect of lncRNA MEG3 on HCC, and
to further explore the related mechanisms in different HCC line
cells.
In conclusion, the present study demonstrated that
MEG3 affects HCC cell viability, apoptosis and migration through
its targeting of miR-9-5p/MDK and regulation of the PDK/AKT
pathway. The present study provided new insights into the molecular
mechanisms of MEG3 in HCC, and suggested that the MEG3/miR-9-5p/MDK
axis is a potential therapeutic target in HCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the First-Class
Discipline Construction Project in Guizhou Province [Chinese
Pharmacy; grant no. (2017)008], Science and Technology Plan Project
of Guizhou Province [grant no. (2017)1073], and Department of
Science and Technology Qian Ke He Platform Talent of Guizhou [grant
no. (2017) 5655].
Availability of data and materials
The data sets generated and analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
DW and QL designed the study, analyzed the data and
prepared the manuscript. DW, ZM and DM conducted the experiments.
All authors were substantially involved in the research,
acquisition of data, analysis and manuscript preparation. DW and QL
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McGlynn KA, Petrick JL and London WT:
Global epidemiology of hepatocellular carcinoma: An emphasis on
demographic and regional variability. Clin Liver Dis. 19:223–238.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Korean Liver Cancer Association, National
Cancer Center, . 2018 Korean Liver Cancer Association-National
Cancer Center Korea Practice Guidelines for the Management of
Hepatocellular Carcinoma. Gut Liver. 13:227–299. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Liu J, Lv Y, Zhang C and Guo S:
lncRNA meg3 suppresses hepatocellular carcinoma in vitro and vivo
studies. Am J Transl Res. 11:4089–4099. 2019.PubMed/NCBI
|
|
7
|
Zhuo H, Tang J, Lin Z, Jiang R, Zhang X,
Ji J, Wang P and Sun B: The aberrant expression of MEG3 regulated
by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol
Carcinog. 55:209–219. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu F, Yang Z and Li G: Role of specific
microRNAs for endothelial function and angiogenesis. Biochem
Biophys Res Commun. 386:549–553. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lopez-Urrutia E, Bustamante Montes LP,
Ladron de Guevara Cervantes D, Perez-Plasencia C and Campos-Parra
AD: Crosstalk between long non-coding RNAs, Micro-RNAs and mRNAs:
Deciphering molecular mechanisms of master regulators in cancer.
Front Oncol. 9:6692019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tam C, Wong JH, Tsui SKW, Zuo T, Chan TF
and Ng TB: lncRNAs with miRNAs in regulation of gastric, liver, and
colorectal cancers: Updates in recent years. Appl Microbiol
Biotechnol. 103:4649–4677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Snezhkina AV, Krasnov GS, Zhikrivetskaya
SO, Karpova IY, Fedorova MS, Nyushko KM, Belyakov MM, Gnuchev NV,
Sidorov DV, Alekseev BY, et al: Overexpression of microRNAs miR-9,
−98, and −199 correlates with the downregulation of HK2 expression
in colorectal cancer. Mol Biol (Mosk). 52:220–230. 2018.(In
Russian). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou L, Fu L, Lv N, Chen XS, Liu J, Li Y,
Xu QY, Huang S, Zhang XD, Dou LP, et al: A minicircuitry comprised
of microRNA-9 and SIRT1 contributes to leukemogenesis in t(8;21)
acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 21:786–794.
2017.PubMed/NCBI
|
|
14
|
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC,
Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, et al: Aberrant
hypermethylation of miR-9 genes in gastric cancer. Epigenetics.
6:1189–1197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu M, Huang Y, Chen T, Wang W, Yang S, Ye
Z and Xi X: lncRNA MEG3 inhibits the progression of prostate cancer
by modulating miR-9-5p/QKI-5 axis. J Cell Mol Med. 23:29–38. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong Z, Zhang A, Liu S, Lu F, Guo Y, Zhang
G, Xu F, Shi Y, Shen S, Liang J and Guo W: Aberrant
methylation-mediated silencing of lncRNA MEG3 functions as a ceRNA
in esophageal cancer. Mol Cancer Res. 15:800–810. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kretschmer PJ, Fairhurst JL, Decker MM,
Chan CP, Gluzman Y, Böhlen P and Kovesdi I: Cloning,
characterization and developmental regulation of two members of a
novel human gene family of neurite outgrowth-promoting proteins.
Growth Factors. 5:99–114. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Filippou PS, Karagiannis GS and
Constantinidou A: Midkine (MDK) growth factor: A key player in
cancer progression and a promising therapeutic target. Oncogene.
39:2040–2054. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang F, Shan S, Huo Y, Xie Z, Fang Y, Qi
Z, Chen F, Li Y and Sun B: miR-155-5p inhibits PDK1 and promotes
autophagy via the mTOR pathway in cervical cancer. Int J Biochem
Cell Biol. 99:91–99. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng F, Zhang Y, Li X, Wang J and Wang Z:
Clinical significance of miR-138 in patients with malignant
melanoma through targeting of PDK1 in the PI3K/AKT autophagy
signaling pathway. Oncol Rep. 38:1655–1662. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chuang CK, Lin HCA, Tasi HY, Lee KH, Kao
Y, Chuang FL, Chang YH, Lin PH, Liu CY and Pang ST: Clinical
presentations and molecular studies of invasive renal epithelioid
angiomyolipoma. Int Urol Nephrol. 49:1527–1536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu J, Liu QH, Wang F, Tan JJ, Deng YQ,
Peng XH, Liu X, Zhang B, Xu X and Li XP: Exosomal miR-9 inhibits
angiogenesis by targeting MDK and regulating PDK/AKT pathway in
nasopharyngeal carcinoma. J Exp Clin Cancer Res. 37:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Abbastabar M, Sarfi M, Golestani A and
Khalili E: lncRNA involvement in hepatocellular carcinoma
metastasis and prognosis. EXCLI J. 17:900–913. 2018.PubMed/NCBI
|
|
25
|
Huang Z, Zhou JK, Peng Y, He W and Huang
C: The role of long noncoding RNAs in hepatocellular carcinoma. Mol
Cancer. 19:772020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
27
|
Zhang CY, Yu MS, Li X, Zhang Z, Han CR and
Yan B: Overexpression of long non-coding RNA MEG3 suppresses breast
cancer cell proliferation, invasion, and angiogenesis through AKT
pathway. Tumour Biol. 39:10104283177013112017.PubMed/NCBI
|
|
28
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv
YB, Li YG and Cao MR: Overexpression of long non-coding RNA meg3
inhibits proliferation of hepatocellular carcinoma Huh7 Cells via
negative modulation of miRNA-664. J Cell Biochem. 118:3713–3721.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu H, Zhou Y and Tang L: Caffeine induces
sustained apoptosis of human gastric cancer cells by activating the
caspase-9/caspase-3 signalling pathway. Mol Med Rep. 16:2445–2454.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Altadill A, Rodríguez M, González LO,
Junquera S, Corte MD, González-Dieguez ML, Linares A, Barbón E,
Fresno-Forcelledo M, Rodrigo L and Vizoso FJ: Liver expression of
matrix metalloproteases and their inhibitors in hepatocellular
carcinoma. Dig Liver Dis. 41:740–748. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Veneziano D, Marceca GP, Di Bella S,
Nigita G, Distefano R and Croce CM: Investigating miRNA-lncRNA
interactions: Computational tools and resources. Methods Mol Biol.
1970:251–277. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang X, Su R, Guo Q, Liu J, Ruan B and
Wang G: Competing endogenous RNA (ceRNA) hypothetic model based on
comprehensive analysis of long non-coding RNA expression in lung
adenocarcinoma. PeerJ. 7:e80242019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang Y: The novel regulatory role of
lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med.
22:5768–5775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen K, Ma Y, Wu S, Zhuang Y, Liu X, Lv L
and Zhang G: Construction and analysis of a lncRNA-miRNA-mRNA
network based on competitive endogenous RNA reveals functional
lncRNAs in diabetic cardiomyopathy. Mol Med Rep. 20:1393–1403.
2019.PubMed/NCBI
|
|
36
|
Zhu D, Xiao Z, Wang Z, Hu B, Duan C, Zhu
Z, Gao N, Zhu Y and Wang H: MEG3/MIR-376B-3P/HMGA2 axis is involved
in pituitary tumor invasiveness. J Neurosurg. 2020.(Ahead of
print).
|
|
37
|
Lu WX: Long non-coding RNA MEG3 represses
cholangiocarcinoma by regulating miR-361-5p/TRAF3 axis. Eur Rev Med
Pharmacol Sci. 23:7356–7368. 2019.PubMed/NCBI
|
|
38
|
Zhang S and Guo W: Long non-coding RNA
MEG3 suppresses the growth of glioma cells by regulating the
miR-96-5p/MTSS1 signaling pathway. Mol Med Rep. 20:4215–4225.
2019.PubMed/NCBI
|
|
39
|
Shen X, Bai H, Zhu H, Yan Q, Yang Y, Yu W,
Shi Q, Wang J, Li J and Chen L: Long non-coding RNA MEG3 functions
as a competing endogenous RNA to regulate HOXA11 expression by
sponging miR-181a in multiple myeloma. Cell Physiol Biochem.
49:87–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Z, Chen JY, Zhong Y, Xie L and Li JS:
lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells
by sponging miR-9-5p to upregulate SOX11. Braz J Med Biol Res.
52:e86312019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li XY, Wen JY, Jia CC, Wang TT, Li X, Dong
M, Lin QU, Chen ZH, Ma XK, Wei LI, et al: MicroRNA-34a-5p enhances
sensitivity to chemotherapy by targeting AXL in hepatocellular
carcinoma MHCC-97L cells. Oncol Lett. 10:2691–2698. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kai AK, Chan LK, Lo RC, Lee JM, Wong CC,
Wong JC and Ng IO: Down-regulation of TIMP2 by
HIF-1α/miR-210/HIF-3α regulatory feedback circuit enhances cancer
metastasis in hepatocellular carcinoma. Hepatology. 64:473–487.
2016. View Article : Google Scholar : PubMed/NCBI
|