Introduction
Epithelial ovarian cancer (EOC) is the most lethal
gynecologic malignancy worldwide, with a 5-year survival rate of
46.5% between 2005 to 2011 (1), as
well as 230,000 new cases and 150,000 deaths annually in 2012
(2). Due to the increasing research
on traditional and novel targeted drugs, the lives of patients with
EOC are being prolonged. Researchers have revealed that after
treatment with surgery and platinum-based chemotherapy, 50% of
patients with EOC with stage III disease could live for >5 years
(3). At present, the median survival
of recurrent patients sensitive to platinum is 3 years; however,
for patients with EOC who are resistant to platinum it is only 1
year (4). The remission rate of
patients with EOC has increased markedly with the combination of
platinum-based traditional chemotherapy and targeting therapies,
including VEGF inhibitors and poly(ADP-ribose) polymerase 1
inhibitors (3,5). However, these two classes of drug are
only used for patients who are sensitive to platinum (6). Therefore, exploring novel efficient
targeted drugs that can enhance the sensitivity of patients with
EOC to platinum is of significance to improve patient
prognosis.
DNA double-strand breaks (DSBs) are a key mechanism
of platinum-based chemotherapy in EOC. When entering EOC cells,
platinum binds to DNA spontaneously to induce platinum-DNA
cross-links that damage the normal structure and function of DNA
(7). As a result, cell cycle
progression is blocked and cell death is induced (8,9). DNA
damage repair could be induced by excessive DNA DSB aggregation in
EOC cells, which causes cells to be resistant to platinum (9). The sensitivity of EOC cells to platinum
depends on their ability to induce DNA damage repair spontaneously
(5). DNA DSBs are mainly repaired by
homologous recombination signaling pathways (10). Previous research has demonstrated
that homologous recombination defects are obvious in patients with
ovarian cancer with BRCA1 gene mutations, resulting in higher
sensitivity to chemotherapy than in patients without BRCA1 gene
mutations (11). It is estimated
that ~50% of high-grade serous adenocarcinomas feature homologous
recombination (3,12). Therefore, identifying molecules that
can inhibit homologous recombination in EOC cells is key to
enhancing the sensitivity of patients with EOC to platinum.
Kin17 DNA and RNA binding protein (KIN17), a DNA-
and RNA-binding protein that is extremely well conserved across
biological evolution, is considered critical to the proliferation
and survival of mammalian cells, including normal and cancerous
cells (13). Importantly, the KIN17
protein has been demonstrated to be involved in the DNA damage
repair process by regulating the homologous recombination signaling
pathway (14,15). When the KIN17 gene is silenced, DNA
replication, DNA damage repair, cell cycle progression and
proliferation are inhibited in breast cancer cells; however,
sensitivity to chemotherapy is increased (16). Proliferation, invasion and metastasis
are inhibited by downregulation of the KIN17 protein, thereby
inducing apoptosis and cell cycle arrest in cervical cancer cells
(17). The KIN17 protein can induce
the proliferation of liver cancer cells (18), and its upregulation is associated
with the invasion and metastasis of non-small cell lung cancer
(19). However, to the best of our
knowledge, the effect of KIN17 in ovarian cancer remains unclear
and requires further exploration.
The aim of the present study was to investigate the
possible association between KIN17 expression and the
clinical/pathological features, as well as the overall survival
(OS) of patients with EOC, and to verify the effects of KIN17 on
EOC in vitro in order to provide a theoretical basis for
future research on novel targeted drugs.
Materials and methods
Bioinformatics analysis
The Oncomine database (https://www.oncomine.org/resource/login.html) was used
to analyze the difference in KIN17 gene expression between EOC and
normal ovarian tissues [datasets from The Cancer Genome Atlas
(TCGA) Ovarian (Affymetrix ID: 205664_at), Lu Ovarian (Affymetrix
ID: 37778_at) (20) and TCGA Ovarian
2 (Affymetrix ID: 0-007853661)], as well as between borderline
ovarian surface epithelial-stromal tumor and ovarian cancer tissues
[dataset from Anglesio Ovarian (GSE12172, Affymetrix ID:
205664_at)] (21). Furthermore, the
association between KIN17 mRNA expression and the sensitivity of
EOC cells to platinum was examined using the Oncomine database
[dataset derived from Gyorffy CellLine (GSE11812, Affymetrix ID:
205664_at)] (22). The Kaplan-Meier
database (http://kmplot.com/analysis/index.php?p=service&cancer=ovar)
was used to examine the association between the transcriptional
levels of KIN17 and the prognoses of patients with ovarian cancer.
All the data in this part were analyzed from 15 datasets (23). The follow-up time was between 2 and
240 months. The expression range of the probe in 205664_at was
between 4 and 1,845; 533 and 377 were selected as the cut-off
values for OS and progression-free survival (PFS) analysis,
respectively. The expression range of the probe in 236887_at was
between 6 and 558; 127 was selected as the cut-off value for OS
analysis. All cut-off values between the lower and upper quartiles
were computed, and the best performing threshold was used as the
cut-off.
Specimens
The present study was approved by the Research
Ethics Committee of The Second Hospital of Jilin University
(Changchun, China). All patients were informed and agreed to
participate in the present study. Paraffin-embedded specimens were
collected at The Second Hospital of Jilin University (Changchun,
China) from 56 patients with EOC and five patients with mucous
cystadenoma who were diagnosed between January 2012 and January
2018. The median age of the patients was 52 years (range, 32–76
years). All patients were female. The inclusion criteria were as
follows: i) Initially diagnosed with EOC and treated with early
comprehensive staging laparotomy or ideal cytoreductive surgery;
ii) the EOC diagnosis was confirmed by the Pathology Department of
The Second Hospital of Jilin University; iii) postoperative
pathology results were measured strictly by Federation
International of Gynecology and Obstetrics (FIGO) staging criteria
(24); iv) standardized
platinum-based chemotherapy was administered postoperatively; and
v) complete follow-up data were available. The exclusion criteria
were as follows: i) Individual history of other malignant tumors;
ii preoperative chemo/radiotherapy; and iii) secondary ovarian
cancer. All patient blood samples were collected before surgery and
immediately analyzed for the levels of CA125 and human epididymis
protein 4 (HE4). Fresh frozen (−80°C) specimens, including cancer
tissues and adjacent tissues, were collected from 20 patients with
EOC who were diagnosed at the same hospital between February 2018
and August 2019. All patients with EOC received standard
platinum-based chemotherapy following cytoreductive surgeries. As
shown in Table I, available
clinical/pathological data were extracted from the Medical Record
Database of The Second Hospital of Jilin University. All 56
patients with EOC were followed up, and the OS of these patients
was calculated. The median follow-up for patients with EOC was 31
months, ranging between 7 and 77 months.
 | Table I.Association between KIN17 protein
expression and clinical features of patients with epithelial
ovarian cancer. |
Table I.
Association between KIN17 protein
expression and clinical features of patients with epithelial
ovarian cancer.
|
|
| KIN17
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical
features | No. of cases
(%) | High expression,
n | Low expression,
n | χ2 | P-value |
|---|
| Age, years |
|
|
|
|
|
|
<52 | 27 (48.21) | 16 | 11 |
|
|
|
≥52 | 29 (51.79) | 16 | 13 | 0.095 | 0.757 |
| FIGO stage |
|
|
|
|
|
|
I+II | 23 (41.07) | 10 | 13 |
|
|
|
III+IV | 33 (58.93) | 22 | 11 | 2.976 | 0.085 |
| Tumor size, cm |
|
|
|
|
|
| ≤8 | 28 (50.00) | 14 | 14 |
|
|
|
>8 | 28 (50.00) | 18 | 10 | 1.167 | 0.280 |
| EOC
typea |
|
|
|
|
|
| Type
I | 26 (46.43) | 14 | 12 |
|
|
| Type
II | 30 (53.57) | 18 | 12 | 0.215 | 0.643 |
| Pathological
grade |
|
|
|
|
|
| Low
level | 23 (41.07) | 13 | 10 |
|
|
| High
level | 33 (58.93) | 19 | 14 | 0.006 | 0.938 |
| Ascites volume,
ml |
|
|
|
|
|
|
<500 | 31 (55.36) | 16 | 15 |
|
|
|
≥500 | 25 (44.64) | 16 | 9 | 0.867 | 0.352 |
| Lymphatic
metastasisc |
|
|
|
|
|
|
Positive | 17 (37.78) | 13 | 4 |
|
|
|
Negative | 28 (62.22) | 12 | 16 | 4.840 | 0.028b |
| CA125,
U/mld |
|
|
|
|
|
|
<500 | 25 (47.17) | 17 | 8 |
|
|
|
≥500 | 28 (52.83) | 15 | 13 | 1.149 | 0.284 |
| HE4,
pmol/ld |
|
|
|
|
|
|
<500 | 26 (68.42) | 17 | 9 |
|
|
|
≥500 | 12 (31.58) | 6 | 6 | 0.813 | 0.367 |
Immunohistochemistry
Immunohistochemical (IHC) staining was performed as
described previously (25). Briefly,
after being fixed in 10% formalin at room temperature for 24 h, EOC
tissues were embedded in paraffin and cut into 3-µm thick sections.
After being deparaffinized by xylol and rehydrated by descending
alcohol series (100, 85 and 70% alcohol, 10 min for each
concentration) at room temperature, the sections were soaked in
EDTA retrieval buffers (cat. no. AR0023; Wuhan Boster Biological
Technology, Ltd.) and heated in a microwave oven at 98°C for 15
min. Then the sections were cooled naturally at room temperature.
Non-specific binding was blocked using 5% bovine serum albumin
(BSA; cat. no. AR1006; Wuhan Boster Biological Technology, Ltd.) at
room temperature for 20 min. Subsequently, the histological
sections were stained with rabbit anti-KIN17 antibody (dilution,
1:100; cat. no. PB0639; Wuhan Boster Biological Technology, Ltd.)
at 4°C overnight. Goat anti-rabbit IgG conjugated with horseradish
peroxidase (dilution, 1:200; cat. no. S0001; Affinity Biosciences)
was used as the secondary antibody, and this staining procedure was
carried out at 37°C for 30 min. Reactive products were visualized
with 3,3′-diaminobenzidene (cat. no. AR1022; Wuhan Boster
Biological Technology, Ltd.) as the chromogen, and the sections
were counterstained with hematoxylin (0.1%; cat. no. AR0005; Wuhan
Boster Biological Technology, Ltd.) at room temperature for 2 min.
Histological images were captured under a light microscope (BX51;
Olympus Corporation) with an objective magnification of ×200 or
×400. The intensity was scored as follows: 0, no color; 1, light
yellow; 2, yellow brown; and 3, brown. The proportion of positive
tumor cells was scored as: 0, 0–4; 1, 5–24; 2, 25–49; 3, 50–74; and
4, 75–100%. The overall score of each sample was obtained by
multiplying the two scores. A score of >4 was defined as high
KIN17 expression, and a score of ≤4 was defined as low KIN17
expression. The IHC staining was double-blinded and scored
independently by two experienced pathologists working at The
Pathology Department of the Second Hospital of Jilin
University.
Western blot analysis
Protein samples from fresh frozen tissues or stably
transfected cells were extracted using cold RIPA lysis buffer (cat.
no. P0013K; Beyotime Institute of Biotechnology). Subsequently, the
protein samples were quantified according to the Bradford method
using an Easy Protein Quantitative Kit (cat. no. DQ101-01; TransGen
Biotech Co., Ltd.). A total of 30 µg protein sample was added to
each lane, resolved by 12% SDS-PAGE, separated electrophoretically
and transferred onto polyvinylidene fluoride membranes (EMD
Millipore). The non-specific binding sites of the membrane were
blocked with 5% dried skim milk at 37°C for 1 h. Subsequently, the
membrane was incubated with primary antibodies, including rabbit
anti-KIN17 (dilution, 1:2,000; cat. no. PB0639; Wuhan Boster
Biological Technology, Ltd.) and mouse anti-β-actin (dilution,
1:1,500; cat. no. HC201-01; TransGen Biotech Co., Ltd.) at 4°C
overnight, followed by incubation with horseradish
peroxidase-tagged goat anti rabbit IgG (dilution, 1:4,000; cat. no.
S0001) and goat anti mouse IgG (dilution, 1:4,000; cat. no. S0002)
(both Affinity Biosciences) at room temperature for 2 h,
respectively. Finally, the protein expression levels were detected
with an electrochemiluminescence plus kit (cat. no. KF001; Affinity
Biosciences), and the densities of the specific bands were
semi-quantified using an imaging densitometer (Clinx Science
Instruments Co., Ltd.) and the software Gel Analysis (Clinx Science
Instruments Co., Ltd.) that is provided with the densitometer.
Cell culture and stably transfected
cell line development
The human EOC cell line SKOV3 was originally
obtained from The Basic Medical College of Jilin University
(Changchun, China). The cells were cultured in Iscove's modified
Dulbecco's medium (IMDM; HyClone; Cytiva) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere with 5%
CO2.
The construction of short hairpin RNA (shRNA)-KIN17
(NM_012311.3) was performed by GeneCopoeia, Inc. The stably
transfected SKOV3 cell line was transfected with shRNA-KIN17 (cat.
no. CS-RSH086442-CU6-01; GeneCopoeia, Inc.) based with the plasmid
psi-U6 (GeneCopoeia, Inc.) using Lipofectamine 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. SKOV3 cells were seeded into a six-well
plate at a density of 8×105 cells/well. Then, 2.5 µg
shRNA-KIN17 or shRNA-Scramble (treatment groups) and only
Lipofectamine 3000 reagent (negative control) were added into the
wells of different groups. Following incubation at 37°C for 96 h,
the transfected cells were selected with 5 µg/ml puromycin (cat.
no. P1033; Biotopped Life Sciences) for 2 weeks before
experimentation, and the surviving cells were continuously cultured
as a stably transfected cell line. The transfection efficiency was
detected by fluorescence microscopy, reverse
transcription-quantitative PCR (RT-qPCR) and western blotting.
RT-qPCR
RNA was extracted from the stably transfected cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
followed by quantification with a NanoDrop 2000 instrument
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.). Following
reverse transcription with a cDNA synthesis kit (Beijing Transgen
Biotech Co., Ltd.), cDNA was used for PCR amplification using a
SYBR Green real-time PCR kit (Beijing Transgen Biotech Co., Ltd.)
using an ABI-Q3 system (Thermo Fisher Scientific, Inc.). The
primers used were as follows: KIN17 Gene forward,
5′-GGACCCAGAAACTATCCGCC-3′ and reverse, 5′-TTCCCTTCCAGGCCTCTTCT-3′;
and GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse,
5′-GAAGATGGTGATGGGATTTC-3′. GAPDH was used as an internal control
gene. Reverse transcription was performed with Anchored Oligo(dT)
at 42°C for 15 min, followed by incubating with Random Primer (N9)
at 25°C for 10 min, 42°C for 15 min and 85°C for 5 sec using a
96-well Thermal Cycler (Thermo Fisher Scientific, Inc.). PCR was
performed at 94°C for 30 sec, followed by 45 cycles of
amplification at 94°C for 5 sec, 51°C for 15 sec and 72°C for 10
sec using an ABI-Q3 system (Thermo Fisher Scientific, Inc.).
Relative mRNA expression levels were calculated using the
2−ΔΔCq method (26).
Cell counting kit-8 (CCK-8) assay
After being transfected with shRNA-KIN17 or shRNA
Scramble, these SKOV3 cells and untreated SKOV3 cells were seeded
into 96-well plates (1,000 cells/well). After 24, 48, 72, 96 and
120 h, CCK-8 reagent (10 µl/well; BIOSS) was added to each well.
Following culture at 37°C for 2 h, the absorbance of each well was
measured at 450 nm using a microplate reader (Cytation 5; BioTek
Instruments, Inc.). The stably transfected SKOV3 cells were seeded
into 96-well plates at a density of 5×103 cells/well and
incubated at 37°C overnight. After exposure to cisplatin at 0, 0.5,
1, 2, 4, 8 or 16 µg/ml at 37°C for 48 h, CCK-8 reagent (10 µl/well;
BIOSS) was used to detect the proliferation rates of SKOV3 cells
and sensitivity of SKOV3 cells to cisplatin at 37°C for 2 h.
Transwell assay
For the migration assay, following 12 h of serum
starvation treatment, untreated SKOV3, shRNA-KIN17-transfected
SKOV3 and shRNA-Scramble-transfected SKOV3 cells were adjusted to a
density of 3×105 cells/ml using serum-free Iscove's
modified Dulbecco's medium (IMDM; containing or not containing
cisplatin at 1 µg/ml). Subsequently, 100 µl single cell suspension
was added to the apical chamber (8-µm polycarbonate membrane;
Corning, Inc.), and 600 µl IMDM with 10% FBS (containing or not
containing cisplatin at 1 µg/ml) was added to the bottom chamber.
The cells were allowed to migrate through the polycarbonate
membrane for 40 h at 37°C with 5% CO2. Subsequently, the
cells remaining on the upper surface were wiped off with a cotton
ball. The migration cells were fixed with 4% paraformaldehyde at
room temperature for 30 min and stained with 0.1% crystal violet
(cat. no. C6470; Biotopped Life Sciences) at room temperature for
10 min, and then observed using an optical light microscope
(magnification, ×200). Subsequently, the cells were observed and
quantified in five random fields of view (magnification, ×200).
Mean integrated optical density (IOD) was measured using Image-Pro
Plus 6.0 software (Media Cybernetics, Inc). The IOD fold changes of
the treatment groups were also analyzed. All experiments were
performed in triplicate.
Statistical analysis
Continuous data are presented as mean and ± standard
deviation. The association between the expression levels of KIN17
and the clinical/pathological features of patients with EOC was
analyzed using the χ2 test. Student's two-tailed t-test
was used to verify significant differences in KIN17 expression
between EOC and EOC-adjacent tissues. In addition, one-way ANOVA
and Tukey's test were used for multiple comparisons. OS curves were
generated using the Kaplan-Meier method and compared using a
log-rank test. Univariate analyses were performed using
Kaplan-Meier. Multivariate analyses were performed using Cox
regression models. All experiments were repeated three times
independently. All data were analyzed using SPSS software (version
23.0; IBM Corp.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Association between KIN17 DNA/mRNA
levels and EOC
Using the Oncomine database, the association between
the gene expression of KIN17 and EOC was analyzed. As shown in
Fig. 1A, compared with that in
normal ovary tissues, the transcription of KIN17 was upregulated in
ovarian serous adenocarcinoma tissues (P<0.05), which was also
demonstrated in Fig. 1B (P<0.05).
Nevertheless, there was no significant difference in KIN17 mRNA
expression between normal ovary tissues and other EOC tissues,
including ovarian mucinous, endometrioid and clear cell
adenocarcinoma tissues (Fig. 1B).
Fig. 1C shows that the KIN17 DNA
levels were upregulated in ovarian serous adenocarcinoma tissues
compared with ovarian tissues (P<0.05). Furthermore, the
transcription of KIN17 was higher in ovarian cancer tissues than
borderline ovarian surface epithelial-stromal tumor tissues
(P<0.05; Fig. 1D). Gyorffy Cell
Line Statistics obtained and analyzed from the Oncomine database
revealed that cisplatin-resistant cancer cell lines exhibited high
mRNA expression levels of KIN17 compared with cisplatin-sensitive
and intermediate sensitive cancer cell lines (Fig. 1E). Additionally, KIN17 mRNA
expression was identified to be increased in the ES-2 cell line, a
cisplatin-resistant EOC cell line with a KIN17 expression value of
0.08, compared with the OVCAR-3 cells with an expression value of
−1.24, FU-OV-1 cells with an expression value of −1.07 and OAW42
cells with an expression value of −0.66 (cisplatin sensitivity), as
well as the SKOV3 cell line with an expression value of −0.56
(cisplatin intermediate sensitivity; Fig. 1F).
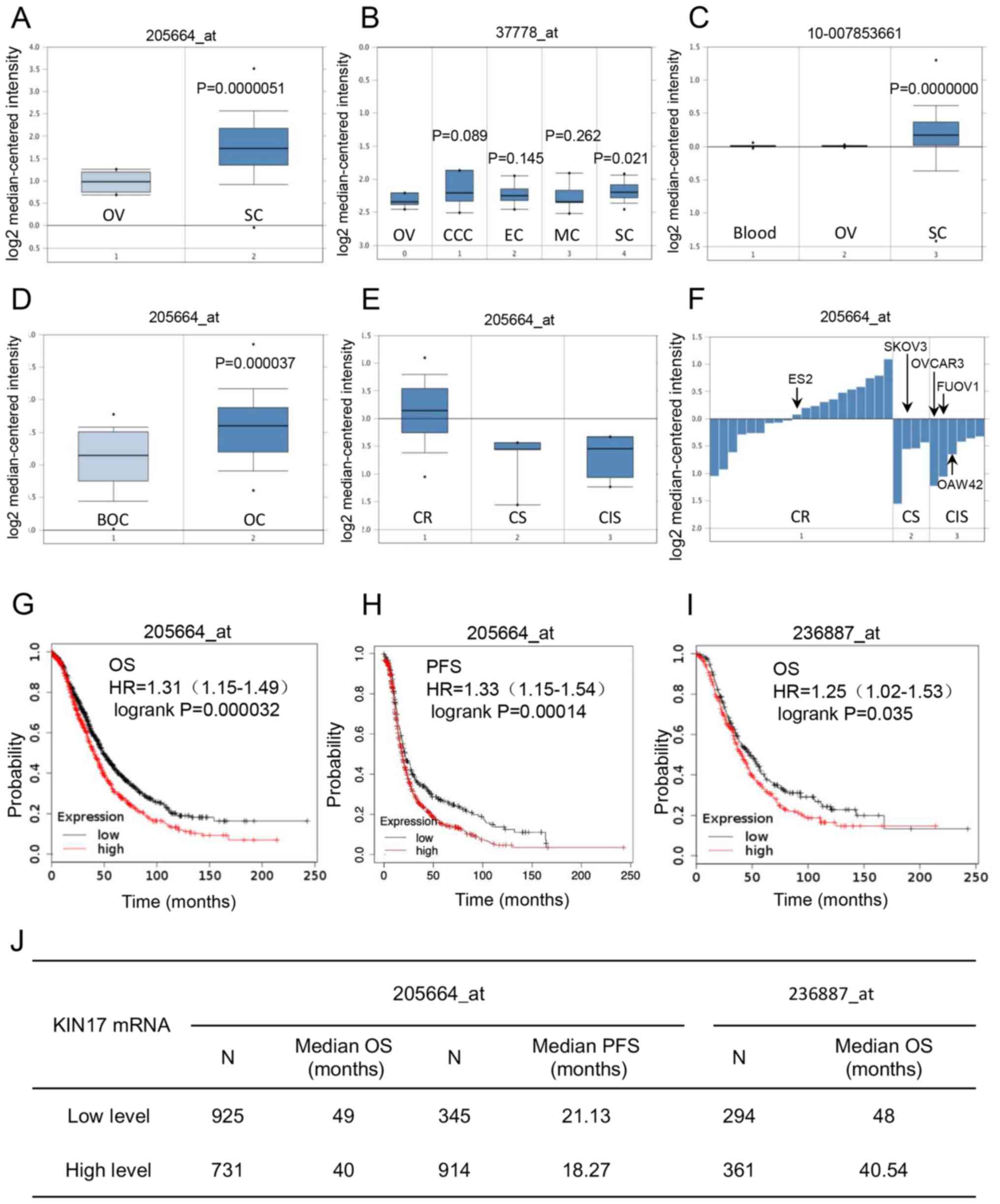 | Figure 1.Bioinformatics analysis of the role
of KIN17 gene expression in the development of OC and prognosis of
patients with OC. (A) mRNA expression levels of KIN17 in OV and SC
tissues (Affymetrix ID: 205664_at; 1. OV, n=8; 2. SC, n=586). (B)
mRNA expression levels of KIN17 in OV and EOC tissues (Affymetrix
ID: 37778_at; 0. OV, n=5; 1. CCC, n=7; 2. EC, n=9; 3. MC, n=9; 4.
SC, n=20). (C) DNA expression levels of KIN17 in blood, OV and SC
tissues (Affymetrix ID: 10-007853661; 1. Blood, n=431; 2. OV,
n=130; 3. SC, n=607). (D) mRNA expression levels of KIN17 in BOC
and OC tissues (Affymetrix ID: 205664_at; 1. BOC, n=30; 2. OC,
n=44). (E) mRNA expression levels of KIN17 in Gyorffy cell lines
(Affymetrix ID: 205664_at; 1. CR, n=20; 2. CS, n=6; 3. CIS, n=4).
(F) mRNA expression levels of KIN17 in different EOC cell lines
(Affymetrix ID: 205664_at; 1. CR; 2. CS; 3. CIS). (G) Association
between KIN17 mRNA expression and the OS of patients with OC
(Affymetrix ID: 205664_at). (H) Association between KIN17 mRNA
expression and the PFS of patients with OC (Affymetrix ID:
205664_at). (I) Association between KIN17 mRNA expression and the
OS of patients with OC (Affymetrix ID: 236887_at). (J) Sample
numbers and median survival (OS or PFS) of patients with OC in
different datasets. BOC, borderline ovarian surface
epithelial-stromal tumor; CCC, clear cell adenocarcinoma; CIS,
cisplatin intermediate sensitivity; CR, cisplatin resistant; CS,
cisplatin sensitivity; EC, endometrioid adenocarcinoma; EOC,
epithelial ovarian cancer; HR, hazard ratio; KIN17, Kin17 DNA and
RNA binding protein; MC, mucinous adenocarcinoma; OC, ovarian
cancer; OS, overall survival; OV, normal ovary; PFS,
progression-free survival; SC, ovarian serous adenocarcinoma. |
Association between KIN17 mRNA
expression and the prognosis of patients with EOC
Using the Kaplan-Meier database, the association
between the transcription of KIN17 and EOC prognosis was
investigated. As shown in Fig. 1G, I and
J, compared with patients with EOC with low mRNA expression
levels of KIN17, patients with EOC with high expression levels of
KIN17 had a poorer OS (median, 49 months vs. 40 months in
205664_at, P<0.01; median, 48 months vs. 40.54 months in
236887_at, P<0.05). Furthermore, compared with patients with EOC
with low mRNA expression levels of KIN17, patients with EOC with
high expression levels of KIN17 had a poorer PFS (median, 21.13
months vs. 18.27 months in 205664_at; P<0.01; Fig. 1H and J).
Difference in KIN17 expression between
EOC and EOC-adjacent tissues
The expression levels of KIN17 were detected by
western blotting in EOC and adjacent tissues. As shown in Fig. 2A and B, compared with EOC-adjacent
tissues, EOC tissues exhibited higher KIN17 protein expression
(P<0.05).
 | Figure 2.Analysis of the roles of KIN17
protein in the development and predictive prognosis of patients
with EOC using IHC staining and western blotting. (A) Western blot
of KIN17 in EOC and adjacent tissues. (B) Analysis of KIN17
expression level in EOC and adjacent tissues. (C) Typical IHC
staining images for HGSC, HGMC, HGEC, MD, LGSC, LGMC, LGEC and CCC
(magnification, ×200 or ×400). MD was used as a negative control.
KIN17 protein expression in the cytoplasm was indicated using red
arrows and KIN17 protein expression in the nucleus was indicated
using black arrows. (D) Overall survival of patients with EOC
according to Federation International of Gynecology and Obstetrics
stages, EOC types, pathological grades, lymphatic metastasis and
KIN17 expression. **P<0.01 vs. adjacent tissue group. CCC, clear
cell carcinoma; EOC, epithelial ovarian cancer; HGEC, high-grade
endometrioid carcinoma; HGMC, high-grade mucous carcinoma; HGSC,
high-grade serous carcinoma; IHC, immunohistochemical; KIN17, Kin17
DNA and RNA binding protein; LGEC, low-grade endometrioid
carcinoma; LGMC, low-grade mucous carcinoma; LGSC, low-grade serous
carcinoma; MD, mucous cystadenoma. |
Analysis of the association between
KIN17 expression and the clinical/pathological features of patients
with EOC
Using IHC staining and scoring, the expression
levels of KIN17 in EOC tissues were used to divide cases into low
expression (score ≤4; 24 cases) and high expression (score >4;
32 cases) groups. Cellular yellowish or brownish staining was
scored as positive. IHC staining of KIN17 in various pathologic
categories of low grade or high grade is shown in Fig. 2C. The nuclear and cytoplasmic
expression of KIN17 is also indicated in Fig. 2C. In the present study, the median
age of the 56 patients with EOC was 52 years. All features,
including age, preoperative CA125 and HE4, tumor size, lymphatic
metastasis, FIGO stage, EOC type, pathological grade, and ascites
volume, are summarized in Table I.
Following further analysis, only lymphatic metastasis was
identified to be associated with the expression levels of KIN17
(P<0.05). More specifically, the protein expression levels of
KIN17 were higher in patients with EOC who underwent lymphatic
metastasis compared with in patients with EOC without lymphatic
metastasis (76.47 vs. 42.85%; P<0.05).
Analysis of risk factors for the OS of
patients with EOC
After following up the 56 patients with EOC, 3- and
5-year OS rates were calculated, and all features, as well as the
expression levels and location of KIN17, were evaluated (Table II). As shown in Table II and Fig. 2D, compared with those of patients
with EOC with high protein expression levels of KIN17, both the 3-
and 5-year OS rates were higher in patients with EOC with low
protein expression levels of KIN17 (3-year OS, 82.0% in the EOC
patients with low protein expression levels of KIN17 vs. 56.8% in
the EOC patients with high protein expression levels of KIN1;
5-year OS, 60.7 vs. 35.4%; P<0.05). This data suggested that
high KIN17 expression was one of the risk factors for OS in
patients with EOC. However, no association was observed between the
location of KIN17 expression and OS (P>0.05). Furthermore, it
was identified that advanced stage (stage III+IV), type II, high
grade and positive lymphatic metastasis were risk factors for the
OS of patients with EOC (Table II;
Fig. 2D; P<0.05).
 | Table II.Kaplan-Meier univariate analysis of
OS in patients with EOC. |
Table II.
Kaplan-Meier univariate analysis of
OS in patients with EOC.
| Clinical
features | No. of cases | 3-year OS, % | 5-year OS, % | P-value |
|---|
| Age, years |
|
|
|
|
|
<52 | 27 | 72.6 | 25.8 |
|
|
≥52 | 29 | 64.2 | 52.3 | 0.851 |
| FIGO stage |
|
|
|
|
|
I+II | 23 | 91.1 | 72.9 |
|
|
III+IV | 33 | 51.6 | 24.1 | 0.001a |
| Tumor size, cm |
|
|
|
|
| ≤8 | 28 | 70.4 | 55.4 |
|
|
>8 | 28 | 65.5 | 36.8 | 0.636 |
| EOC type |
|
|
|
|
| I | 26 | 84.3 | 54.5 |
|
| II | 30 | 52.4 | 36.9 | 0.036a |
| Pathological
grade |
|
|
|
|
|
Low | 23 | 86.5 | 76.9 |
|
|
High | 33 | 54.5 | 23.3 | 0.006a |
| Ascites volume,
ml |
|
|
|
|
|
<500 | 31 | 73.6 | 57.2 |
|
|
≥500 | 25 | 61.7 | 36.0 | 0.488 |
| Lymph node
metastasisb |
|
|
|
|
|
Positive | 17 | 59.6 | – |
|
|
Negative | 28 | 81.6 | 58.7 | 0.028a |
| CA125, U/ml |
|
|
|
|
|
<500 | 25 | 67.0 | 48.0 |
|
|
≥500 | 28 | 66.2 | 52.9 | 0.794 |
| HE4, pmol/l |
|
|
|
|
|
<500 | 26 | 56.1 | 48.1 |
|
|
≥500 | 12 | 91.7 | 67.9 | 0.150 |
| KIN17
expression |
|
|
|
|
| High
level | 32 | 56.8 | 35.4 |
|
| Low
level | 24 | 82.0 | 60.7 | 0.024a |
| KIN17
localization |
|
|
|
|
|
Cytoplasm | 18 | 62.9 | 52.4 |
|
|
Nucleus | 26 | 63.2 | 45.1 | 0.585 |
In addition, to exclude the impact of confounding
factors, Cox regression analysis was utilized to verify the
independent risk factors for OS in patients with EOC. It was
concluded that both advanced stage and high expression levels of
KIN17 were independent risk factors for OS in patients with EOC
(Table III). More precisely, the
risk of death in patients with advanced-stage EOC was 6.510 times
higher than that in patients who were at an early stage (stage
I+II; P<0.05). The risk of death in patients with EOC with high
expression levels of KIN17 was 2.828 times greater than that in
patients with low expression (P<0.05).
 | Table III.Multivariate Cox regression analysis
of overall survival in patients with epithelial ovarian cancer. |
Table III.
Multivariate Cox regression analysis
of overall survival in patients with epithelial ovarian cancer.
| Prognostic
factors | Estimation of
regression coefficient (B) | HR (95% CI) | P-value |
|---|
| FIGO stage | 1.873 | 6.510 (1.877,
22.575) | 0.006a |
| KIN17
expression | 1.040 | 2.828 (1.035,
7.732) | 0.043a |
Association between KIN17 expression
and the proliferation, as well as cisplatin sensitivity, of ovarian
cancer cells
To further investigate the effects of KIN17 on EOC,
shRNA-KIN17 was used to silence KIN17 expression, and a stably
transfected SKOV3 cell line was generated. The transfection
efficiency was examined by fluorescence microscopy (Fig. 3A), RT-qPCR (P<0.05; Fig. 3B) and western blotting (P<0.05;
Fig. 3C and D). As shown in Fig. 3E, from day 4, compared with the
shRNA-Scramble group, the cell proliferation of SKOV3 cells in the
shRNA-KIN17 group was markedly lower (P<0.05), which implied
that KIN17 knockdown could inhibit the proliferation of SKOV3
cells. Fig. 3F shows that between
the concentrations of cisplatin 0.5 to 4 µg/ml, compared with the
shRNA-Scramble group, the cell proliferation of SKOV3 cells in the
shRNA-KIN17 group was markedly lower (P<0.05), which suggested
KIN17 knockdown sensitized SKOV3 cells to cisplatin.
Migration ability of EOC cells is
inhibited by downregulation of KIN17
A trans-well assay was used to detect the migration
ability of EOC cells. As shown in Fig.
3G, cisplatin at 1 µg/ml inhibited the migration ability of
SKOV3 cells. Following treatment with shRNA-KIN17, the number of
cells migrating through the polycarbonate membrane was
significantly decreased in the combine group (Fig. 3G), indicating that KIN17 knockdown
could inhibit the migration ability of EOC cells. Therefore, the
experimental data implied a notable role of KIN17 in the migration
ability of EOC cells.
Discussion
Although the KIN17 protein serves an important role
in human cells, it is not a highly expressed protein in all human
tissues. Normally, KIN17 expression in the majority of normal
tissues is low, while KIN17 upregulation has been detected in
various cancer tissues (27), such
as breast cancer (16), cervical
cancer (17), liver cancer (18) and lung cancer (19). Compared with the normal breast cell
line HS578Bst, the expression levels of KIN17 are upregulated in
MDA-MB-231, SKBr-3, BT474, and MCF-7 cell lines (breast carcinoma)
(16). Moreover, compared with
immortalized epidermal keratinocyte line HaCaT, the expression
levels of KIN17 are upregulated in H1299, K562 (colorectal
carcinoma), PC3 (prostatic carcinoma) and HeLa cell lines (cervical
carcinoma) (28). These results
suggest that KIN17 might be closely associated with the development
of malignancies.
In the present study, the expression levels of KIN17
were demonstrated to be markedly upregulated in EOC tissues
compared with EOC-adjacent tissues, borderline EOC tissues and
normal ovary tissues. The present in vitro experimental
results demonstrated that KIN17 silencing inhibited the
proliferation of EOC cells. These results implied that the
expression levels of KIN17 were associated with the oncogenesis and
development of EOC. Furthermore, advanced stages (stage III+IV),
type II, high grade and high KIN17 expression were identified as
risk factors for OS in patients with EOC based on Kaplan-Meier
univariate analysis. In addition, the study revealed that both
advanced disease stage and high expression levels of KIN17 were
independent risk factors based on Cox multivariate analysis. These
results provided strong clinical evidence that KIN17 may have
prognostic value in patients with EOC. The clinical data
demonstrated that the KIN17 protein was closely associated with the
OS of patients with EOC.
The combination of cytoreductive surgery and
platinum-based chemotherapy is the standard treatment for patients
with EOC (29). Homologous
recombination is one of the important mechanisms of DNA damage
repair and subsequent chemotherapeutic resistance in tumor cells
(30). Therefore, exploring
molecules that could regulate homologous recombination and thus
prevent platinum resistance in EOC is an important line of
research. Previous studies have suggested that KIN17 could induce
chemotherapy resistance in cancer cells by initiating the
homologous recombination signaling pathway, resulting in the poor
prognosis of patients with cancer (14,31–33). The
results of the bioinformatics and clinical analyses demonstrated
that patients with EOC with high KIN17 expression had a poorer
prognosis as compared with the patients with low KIN17 expression,
which suggested that KIN17 could be a prognostic biomarker for EOC.
However, to the best of our knowledge, the mechanisms are still
unclear. Bioinformatics data suggested that high mRNA expression
levels of KIN17 might be associated with cisplatin resistance in
EOC cell lines. Furthermore, the present in vitro analysis
revealed that KIN17 knockdown could increase the sensitivity of EOC
cells to cisplatin, which might be associated with the failure of
the homologous recombination signaling pathway in the cells. These
results imply that patients with EOC with low expression levels of
KIN17 might be more sensitive to cisplatin and have an improved
prognosis.
Migration of EOC cells is markedly related to the
prognosis of patients (34). A
previous study suggested that KIN17 might be a prognostic biomarker
of non-small cell lung cancer (NSCLC), since it serves a key role
in the invasion and metastasis of NSCLC cells (19). The effect of KIN17 on cell invasion
has also been observed in cervical cancer (17). In the present study, the clinical
data suggested that patients with EOC with high KIN17 expression
were more likely to suffer from lymph node metastasis. Subsequent
experiments indicated that KIN17 silencing resulted in inhibition
of EOC cell migration in vitro. Furthermore, the
anti-migration effect of cisplatin might be enhanced by KIN17
knockdown in vitro. Therefore, it was hypothesized that EOC
cells in the tissues of patients with low expression levels of
KIN17 may be less likely to migration, leading to an improved
prognosis.
In conclusion, the present study revealed that the
expression levels of KIN17 were increased in EOC. High KIN17
expression may be associated with the oncogenesis and development
of EOC. Additionally, high expression levels of KIN17 were
associated with lymph node metastasis, which might lead to a poor
prognosis. Furthermore, KIN17 knockdown inhibited the proliferation
and migration of EOC cells and sensitized EOC cells to cisplatin
in vitro. Therefore, KIN17 may be an ideal candidate for
therapy and a prognostic biomarker of EOC, which warrants further
exploration.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Jilin Province Development and Reform Commission (grant no.
2014G073), the Jilin Province of Department Finance (grant no.
2019SCZT040) and the Jilin Province Science and Technology
Department (grant no. 20200201589JC).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC, SZ and LZ conceived and designed the
experiments. YX, YP, SW, WL and HZ performed the experiments. TW,
ZY and YP collected and analyzed the data. JC and SW interpreted
the results and wrote the manuscript. JC, YX and SZ assessed and
confirmed the authenticity of all the raw data. All authors agree
to be accountable for all aspects of the research and to guarantee
the accuracy and integrity of any part of the work. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
standard guidelines and was approved by the Ethics Committee of the
Second Hospital of Jilin University (approval no. 2018218;
Changchun, China). All patients provided written informed consent
prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lheureux S, Gourley C, Vergote I and Oza
AM: Epithelial ovarian cancer. Lancet. 393:1240–1253. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Griffiths RW, Zee YK, Evans S, Mitchell
CL, Kumaran GC, Welch RS, Jayson GC, Clamp AR and Hasan J: Outcomes
after multiple lines of chemotherapy for platinum-resistant
epithelial cancers of the ovary, peritoneum, and fallopian tube.
Int J Gynecol Cancer. 21:58–65. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boussios S, Karihtala P, Moschetta M,
Karathanasi A, Sadauskaite A, Rassy E and Pavlidis N: Combined
strategies with poly (ADP-Ribose) polymerase (PARP) inhibitors for
the treatment of ovarian cancer: A literature review. Diagnostics
(Basel). 9:872019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luvero D, Milani A and Ledermann JA:
Treatment options in recurrent ovarian cancer: Latest evidence and
clinical potential. Ther Adv Med Oncol. 6:229–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang QE, Milum K, Han C, Huang YW, Wani G,
Thomale J and Wani AA: Differential contributory roles of
nucleotide excision and homologous recombination repair for
enhancing cisplatin sensitivity in human ovarian cancer cells. Mol
Cancer. 10:242011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Borst P, Rottenberg S and Jonkers J: How
do real tumors become resistant to cisplatin? Cell Cycle.
7:1353–1359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Norouzi-Barough L, Sarookhani MR, Sharifi
M, Moghbelinejad S, Jangjoo S and Salehi R: Molecular mechanisms of
drug resistance in ovarian cancer. J Cell Physiol. 233:4546–4562.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burdova K, Storchova R, Palek M and
Macurek L: WIP1 promotes homologous recombination and modulates
sensitivity to PARP inhibitors. Cells. 8:12582019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lord CJ and Ashworth A: The DNA damage
response and cancer therapy. Nature. 481:287–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang QG, Xiong CF and Lv YX: Kin17
facilitates thyroid cancer cell proliferation, migration, and
invasion by activating p38 MAPK signaling pathway. Mol Cell
Biochem. 476:727–739. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
The Cancer Genome Atlas Research Network,
. Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Denis SFB, Miccoli L, Despras E, Frobert
Y, Creminon C and Angulo JF: Ionizing radiation triggers
chromatin-bound kin17 complex formation in human cells. J Biol
Chem. 277:19156–19165. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng T, Gao H, Yu P, He H, Ouyang X, Deng
L and Zhang Y: Up-regulation of kin17 is essential for
proliferation of breast cancer. PLoS One. 6:e253432011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Gao H, Gao X, Huang S, Wu K, Yu
X, Yuan K and Zeng T: Elevated expression of Kin17 in cervical
cancer and its association with cancer cell proliferation and
invasion. Int J Gynecol Cancer. 27:628–633. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kou WZ, Xu SL, Wang Y, Wang LW, Wang L,
Chai XY and Hua QL: Expression of Kin17 promotes the proliferation
of hepatocellular carcinoma cells in vitro and in
vivo. Oncol Lett. 8:1190–1194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Huang S, Gao H, Wu K, Ouyang X,
Zhu Z, Yu X and Zeng T: Upregulation of KIN17 is associated with
non-small cell lung cancer invasiveness. Oncol Lett. 13:2274–2280.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu KH, Patterson AP, Wang L, Marquez RT,
Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I,
et al: Selection of potential markers for epithelial ovarian cancer
with gene expression arrays and recursive descent partition
analysis. Clin Cancer Res. 10:3291–3300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Anglesio MS, Arnold JM, George J, Tinker
AV, Tothill R, Waddell N, Simms L, Locandro B, Fereday S,
Traficante N, et al: Mutation of ERBB2 provides a novel alternative
mechanism for the ubiquitous activation of RAS-MAPK in ovarian
serous low malignant potential tumors. Mol Cancer Res. 6:1678–1690.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Györffy B, Surowiak P, Kiesslich O,
Denkert C, Schäfer R, Dietel M and Lage H: Gene expression
profiling of 30 cancer cell lines predicts resistance towards 11
anticancer drugs at clinically achieved concentrations. Int J
Cancer. 118:1699–1712. 2006. View Article : Google Scholar
|
|
23
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Heintz AP, Odicino F, Maisonneuve P, Quinn
MA, Benedet JL, Creasman WT, Ngan HY, Pecorelli S and Beller U:
Carcinoma of the ovary. FIGO 26th annual report on the results of
treatment in gynecological cancer. Int J Gynaecol Obstet. 95 (Suppl
1):S161–S192. 2006. View Article : Google Scholar
|
|
25
|
Kong D, Chen J, Sun X, Lin Y, Du Y, Huang
D, Cheng H, He P, Yang L, Wu S, et al: GRIM-19 over-expression
represses the proliferation and invasion of orthotopically
implanted hepatocarcinoma tumors associated with downregulation of
Stat3 signaling. Biosci Trends. 13:342–350. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kannouche P, Mauffrey P, Pinon-Lataillade
G, Mattei MG, Sarasin A, Daya-Grosjean L and Angulo JF: Molecular
cloning and characterization of the human KIN17 cDNA encoding a
component of the UVC response that is conserved among metazoans.
Carcinogenesis. 21:1701–1710. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Despras E, Miccoli L, Créminon C,
Rouillard D, Angulo JF and Biard DS: Depletion of KIN17, a human
DNA replication protein, increases the radiosensitivity of RKO
cells. Radiat Res. 159:748–758. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ovarian cancer. Nat Rev Dis Primers.
2:160622016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Haynes B, Saadat N, Myung B and Shekhar
MP: Crosstalk between translesion synthesis, Fanconi anemia
network, and homologous recombination repair pathways in
interstrand DNA crosslink repair and development of
chemoresistance. Mutat Res Rev Mutat Res. 763:258–266. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Prat J: New insights into ovarian cancer
pathology. Ann Oncol. 23 (Suppl 10):x111–x117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Doufekas K and Olaitan A: Clinical
epidemiology of epithelial ovarian cancer in the UK. Int J Womens
Health. 6:537–545. 2014.PubMed/NCBI
|
|
33
|
Chen H, Lisby M and Symington LS: RPA
coordinates DNA end resection and prevents formation of DNA
hairpins. Mol Cell. 50:589–600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xue Z, Zhu X and Teng Y: Long non-coding
RNA CASC2 inhibits progression and predicts favorable prognosis in
epithelial ovarian cancer. Mol Med Rep. 18:5173–5181.
2018.PubMed/NCBI
|

















