Introduction
Acute myeloid leukemia (AML) is a highly
heterogeneous disease that originates from myeloid progenitor cells
(1). AML has a very high mortality
rate, particularly in patients over 65 years of age (2). At present, although approaches such as
chemotherapy and stem cell transplantation have achieved
significant success in the treatment of AML, >70% of 65-year-old
or older patients will die within 1 year of diagnosis (3,4). Even
patients who achieve complete remission may eventually die from the
emergence of drug resistance, which is a major obstacle for AML
treatment (5). Therefore, it is
necessary to investigate the molecular mechanisms of AML
pathogenesis to develop novel therapeutic strategies.
A total of 90% of the human genome is comprised of
non-coding RNAs (ncRNAs), which are divided into numerous different
subtypes (6). As subtypes of ncRNAs,
long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have
attracted increasing attention in recent years in the study of AML
pathogenesis (7–9). LncRNAs exceed 200 nucleotides in
length. Studies have revealed that lncRNAs serve a critical role in
numerous life processes, including gene regulation, cell cycle
regulation and cell differentiation regulation (10,11).
LncRNAs that are abnormally expressed may act as oncogenes or tumor
suppressor genes in numerous types of malignancy (12). For example, lncRNA HOTAIR has been
demonstrated to be involved in metastasis and poor prognosis of
liver, colorectal and pancreatic cancer types (13–15). The
expression level of the lncRNA MALAT1 in lung tumors was three-fold
higher than that in healthy controls (16). Furthermore, lncRNA LINC01018 was
correlated with the molecular pathways of tumors (17). Another study demonstrated that the
expression levels of liver metabolic genes were regulated by
LINC01018 (18). At present,
research on LINC01018 is scarce, and the role of LINC01018 in AML
progression and drug resistance is not completely clear.
MiRNAs, short ncRNAs with a length of 19–25 nt
(19), have been proven to be
associated with the initiation and progression of AML in previous
studies. For example, a study undertaken by Song et al
(20) determined that miR-22 was a
proto-oncogene and that abnormalities in the miR-22-TET2 axis were
common in hematopoietic malignancies. In addition, Bousquet et
al (21) indicated that miR-125b
interferes with the differentiation of primary human
CD34+ cells and inhibits terminal differentiation in
leukemic cells. At present, studies have demonstrated that
miR-499a-5p is associated with the occurrence and development of
lung adenocarcinoma, pancreatic cancer and oral squamous cell
carcinoma (22–24). However, whether miR-499a-5p serves a
role in the pathogenesis of AML remains undetermined.
Programmed cell death 4 (PDCD4) is a tumor repressor
that is usually decreased in various cancer types (25–27). It
has been shown to effectively inhibit cancer cell promotion,
progression and proliferation (28).
A recent study has indicated that PDCD4 may suppress mTORC2
activation, which also serves a significant role in the regulation
of cell invasion, migration and metastasis (29). Furthermore, several studies have
reported that PDCD4 may inhibit the translation of a series of
genes, including p53, Bcl-xl and XIAP, thereby suppressing the
occurrence of tumors (30–32). However, the role of PDCD4 in AML
pathogenesis has not been extensively investigated.
In the present study, the expression levels of
LINC01018, miR-499a-5p and PDCD4 were detected in AML tissues and
cell lines, and the interaction between PDCD4 and LINC01018 or
miR-499a-5p was analyzed, providing a novel available therapeutic
target for the treatment of AML.
Materials and methods
AML tissues and cell lines
A total of 40 bone marrow specimens were collected
from patients with AML (mean age, 62.3 years; age range, 32 to 71
years) at the Affiliated Hangzhou First People's Hospital from May
2016 to Feb 2020. Bone marrow specimens were obtained from 40
healthy volunteers (mean age, 60.3 years; age range, 36 to 72
years) at the Affiliated Hangzhou First People's Hospital from May
2018 to Feb 2020. The present study was approved by the Ethics
Committee of Affiliated Hangzhou First People's Hospital (Hangzhou,
China; approval no. 146-01). Written informed consent was provided
by all patients prior to the study start. The clinical
characteristics of the patients are presented in Tables I and II. AML HL-60 and THP-1 cell lines and the
normal BM HS cell line were supplied by the American Type Culture
Collection. All cell lines were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.). All cells were
cultured in a humidified incubator with 5% CO2 at
37°C.
 | Table I.Clinical characteristics of patients
with AML (n=40). |
Table I.
Clinical characteristics of patients
with AML (n=40).
| Parameter | AML patients,
n |
|---|
| Age, years |
|
|
>60 | 32 |
|
≤60 | 8 |
| Sex |
|
|
Male | 22 |
|
Female | 18 |
| Diabetes |
|
| No | 34 |
|
Yes | 6 |
| Length of hospital
stay, days |
|
|
>20 | 11 |
|
≤20 | 29 |
| Glucocorticoid |
|
| No | 40 |
|
Yes | 0 |
| Chemotherapy |
|
| No | 40 |
|
Yes | 0 |
| Radiotherapy |
|
| No | 40 |
|
Yes | 0 |
| FAB
classification |
|
|
M1-M3 | 17 |
|
M4-M6 | 23 |
| Cytogenetic
risk |
|
|
Favorable | 5 |
|
Intermediate | 31 |
|
Unfavorable | 4 |
| NCCN risk
stratification |
|
|
Favorable | 9 |
|
Intermediate | 17 |
|
Unfavorable | 14 |
 | Table II.Comparison between patients with AML
and controls in age, sex, diabetes and length of stay. |
Table II.
Comparison between patients with AML
and controls in age, sex, diabetes and length of stay.
| Parameter | All cases | Health control | AML | P-value |
|---|
| Age, years |
|
|
|
|
|
>60 | 32 | 15 | 17 | 0.695 |
|
≤60 | 8 | 5 | 3 |
|
| Sex |
|
|
|
|
|
Male | 22 | 9 | 13 | 0.525 |
|
Female | 18 | 10 | 8 |
|
| Diabetes |
|
|
|
|
| No | 34 | 19 | 15 | 0.398 |
|
Yes | 6 | 2 | 4 |
|
| Length of hospital
stay, days |
|
|
|
|
|
>20 | 11 | 5 | 6 | 0.477 |
|
≤20 | 29 | 18 | 11 | – |
Transfection of oligonucleotides
pcDNA3.1/LINC01018, a miR-499a-5p mimic
(5′-UUAAGACUUGCAGUGAUGUUU-3′), miR-499a-5p inhibitor
(5′-AAACATCTCTGCAAGTCTTAA-3′), miR mimic control
(5′-ACUACUGAGUGACAGUAGA-3′), miR inhibitor control
(5′-CAGUACUUUUGUGUAGUACAA-3′), si-PDCD4 (sense,
5′-GGAGCGGUUUGUAGAAGAAdTdT-3′ and antisense,
3′-dTdTCCUCGCCAAACAUCUUCUU-5′), si-control (sense,
5′-UUCCCUUUGUCAUCCUUUGCCUdTdT-3′ and antisense,
3′-dTdTAAGGGAAACAGUAGGAAACGGA-5′) and a pcDNA3.1 empty vector were
all purchased from Shanghai GenePharma Co., Ltd. HL-60 and THP-1
cells were transfected with pcDNA3.1 (50 ng), pcDNA3.1/LINC01018
(50 ng), miR-499a-5p mimic (100 nM), mimic control (100 nM),
miR-499a-5p inhibitor (100 nM), inhibitor control (100 nM),
si-PDCD4 (50 nM), and si-control (50 nM) at 37°C for 48 h using
Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Cells were collected and used 48 h later for subsequent
experimentation.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
Total RNA from AML tissues and cells was extracted
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
and its quality was detected via NanoDrop 2000c (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Next,
the RNA samples were converted into cDNA using M-MLV reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.). The
reverse transcription protocol was 70°C for 10 min followed by 2
min on ice. RT-qPCR was conducted on an ABI 7900 system with SYBR
Green Real-Time PCR master mixes (Thermo Fisher Scientific, Inc.).
The thermocycling conditions were as follows: Denaturation at 95°C
for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1
min. GAPDH was used as an internal control for LINC01018 and PDCD4,
and U6 small nuclear RNA served as a control for miR-499a-5p. The
relative expression was analyzed using the 2−ΔΔCq method
(33). The sequences of all primers
are presented in Table III.
 | Table III.Primer sequences used for
quantitative PCR. |
Table III.
Primer sequences used for
quantitative PCR.
| ID | Sequence
(5′-3′) |
|---|
| GAPDH | Forward:
TGTTCGTCATGGGTGTGAAC |
| GAPDH | Reverse:
ATGGCATGGACTGTGGTCAT |
| LINC01018 | Forward:
TGGATTCACATCTGCTGGGT |
| LINC01018 | Reverse:
TGGCCAACATTTGTCAAGGG |
| PDCD4 | Forward:
CCTGAATTAGCACTGGATACTCCT |
| PDCD4 | Reverse:
CTAGCCTGCACACAATCTACAGTT |
| miR-499a-5p | Forward:
ATGTAGCGTGCGACCG |
| miR-499a-5p | Reverse:
CAGGCTGACGCACTCTGTGCT |
| U6 | Forward:
CCATCGGAAGCTCGTATACGAAATT |
| U6 | Reverse:
GGCCTCTCGAACTTGCGTGTCAG |
Cell viability assessment
The effects of LINC01018 and miR-499a-5p on AML cell
viability were evaluated using the Cell Counting kit-8 (CCK-8;
Sigma-Aldrich; Merck KGaA) assay. AML cells were seeded onto
96-well plates (4×103) and cultured overnight at 37°C
followed by transfection of the indicated oligonucleotides. After
12, 24, 48 and 72 h of transfection, the absorbance of each well
was detected at 450 nm by a spectrophotometer. The results
represent the mean of three replicates under the same
conditions.
Cell proliferation analysis
AML cell proliferation was analyzed by an EdU assay
kit (Guangzhou RiboBio Co., Ltd.), according to the manufacturer's
protocols. Cell nuclei were stained with Hoechst 3344 (4 µg/ml,
Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for
30 min. EdU-positive cells were red.
Cell apoptosis analysis
Apoptosis of AML cells was detected by TUNEL
staining using a cell death detection reagent, fluorescein (Roche
Diagnostics). Briefly, transfected AML cells (1×106)
were added into the slides with polylysine. Cells were fixed with
2% formaldehyde at 4°C for 30 min and permeabilized using 0.2%
Triton-X. After washing three times with PBS, cells were processed
with 20 µg/ml Proteinase K (Sigma-Aldrich; Merck KGaA; cat. no.
P2308) at room temperature for 20 min. Next, cells were washed with
PBS three times followed by a 1 h incubation of the TUNEL reaction
mixture at 37°C according to the manufacturer's instructions.
Nucleic acids were stained using DAPI (1:500, Sigma-Aldrich, cat.
no. D9564) for 15 min at room temperature. Subsequently, cells were
incubated with 50 µl DAB solution at room temperature for 20 min,
and the nuclei were stained using hematoxylin at room temperature
for 3 min. After dehydration and transparency, the signals were
scanned from 6 random fields using a confocal microscope (Olympus
BX51TRF; Olympus Corporation) (magnification, ×100) and the number
of TUNEL-positive cells was counted. The apoptosis index was used
to measure the degree of cell apoptosis.
RNA immunoprecipitation (RIP)
RIP was performed to analyze the interplay between
LINC01018 and miR-499a-5p using an EZ-Magna RIP kit (Merck KGaA).
Following cell lysis, the cell extract was incubated with RIP
buffer containing an AGO2 antibody coated on magnetic beads, and an
IgG antibody was used as a control. The precipitated RNAs were
quantified and purified and then reverse transcribed into cDNA, and
RT-qPCR was performed to analyze LINC01018 and miR-499a-5p
levels.
Western blotting
Total protein was extracted from HL-60 and THP-1
cells using cell lysis buffer (cat. no. AR0105; Wuhan Boster
Biological Technology, Ltd.). Following the protein concentration
being determined using a BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.), 50 µg samples were isolated by 10% SDS-PAGE
gels, transferred onto PVDF membranes (Merck KGaA) and blocked with
5% skimmed milk for 1 h at room temperature. Subsequently, the
membranes were incubated with Bax antibody (dilution, 1:1,000; cat.
no., ab182733; Abcam) or Bcl-2 antibody (dilution, 1:1,000; cat.
no., ab32124; Abcam) at 4°C overnight, followed by incubation with
the corresponding HRP-conjugated secondary antibodies (1:2,000;
cat. nos. ab205719 or ab6721; Abcam) at room temperature for 1 h.
Finally, the protein bands were visualized using an enhanced
chemiluminescence reagent (Merck KGaA) and GAPDH was used as an
internal control.
Bioinformatics analysis
miR-499a-5p information was obtained through miR
Base (http://www.mirbase.org/index.shtml). The prediction of
LINC01018 and miR-499a-5p was conducted using starbase v2.0
(http://starbase.sysu.edu.cn). The target
genes including PDCD4 of miR-499a-5p were predicted using starbase
v2.0, TargetScan (http://www.targetscan.org/) and MicroRanda (http://www.microrna.org/).
Dual-luciferase reporter assay
The interaction between miR-499a-5p and LINC01018 or
PDCD4 was verified by dual-luciferase assays. In brief, the
wild-type (WT) and mutant (MUT) sequences of LINC01018 or PDCD4,
which contained miR-499a-5p binding sites, were amplified and
inserted into pmirGLO (Promega Corporation). The recombinant
luciferase plasmids were named LINC01018-WT, PDCD4-WT1, PDCD4-WT2,
PDCD4-WT3, LINC01018-MUT, PDCD4-MUT1, PDCD4-MUT2, PDCD4-MUT3. AML
cells were co-transfected with miR-499a-5p mimics or negative
control and the indicated recombinant luciferase plasmids (0.8 µg)
and 0.08 µg renilla plasmid (RL-SV40; Promega Corporation) using
Lipofectamine 3000® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h of transfection, the luciferase
activity was analyzed by a dual-luciferase reporter assay system
(Promega Corporation) according to the manufacturer's instructions.
The results were normalized using Renilla luciferase
activity.
Statistical analysis
All results are presented as the mean ± standard
deviation. The two groups were compared using the unpaired
Student's t-test, and multiple comparisons were conducted using
one-way analysis of variance, followed by Tukey's post hoc
test in GraphPad Prism 7 (GraphPad Software, Inc.). Correlation
analysis was conducted using the Pearson correlation coefficient.
P<0.05 was considered to indicate a statistically significant
difference.
Results
LINC01018 is downregulated while
miR-499a-5p is upregulated in AML tissues and cell lines
RNAalifold Web Server was used to obtain the
predicted secondary structures of LINC01018: Centroid and minimum
free energy predictions. A mountain plot of the secondary
structures is shown in Fig. 1A. The
expression of LINC01018 and miR-499a-5p in AML tissues and cell
lines was measured using RT-qPCR. Compared with that in healthy
controls (n=40), the expression level of LINC01018 was decreased,
while miR-499a-5p was increased, in AML tissues (n=40; P<0.05;
Fig. 1B and C). The results in
Fig. 1D show a negative correlation
between LINC01018 and miR-499a-5p expression in patients with AML
(r=−0.3881; P=0.0134). In addition, RT-qPCR analysis indicated that
LINC01018 levels were markedly decreased in HL-60 and THP-1 cell
lines compared with that in HS cells (P<0.05; Fig. 1E). The expression of miR-499a-5p was
significantly higher in HL-60 and THP-1 cells than in HS cells
(P<0.05, Fig. 1F). These data
demonstrated that LINC01018 was expressed at low levels, while
miR-499a-5p was highly expressed, in AML tissues and cell
lines.
MiR-499a-5p reverses the suppressive
effects of LINC01018-overexpression on AML cell growth
Following investigating the levels of LINC01018 and
miR-499a-5p and verifying their correlation, the biological
functions of miR-499a-5p and LINC01018 in AML were analyzed. To
begin with, RT-qPCR was applied to investigate the overexpression
efficiency of LINC01018 in AML cells. The results indicated that
the LINC01018 level was significantly higher in the
pcDNA3.1/LINC01018 group than in the blank and negative control
groups (P<0.05; Fig. 2A). Next,
CCK-8 and EdU staining assays were performed to detect the
proliferation of HL-60 and THP-1 cells following transfection of
LINC01018 alone or LINC01018 plus miR-499a-5p. The results revealed
that LINC01018 transfection significantly suppressed cell
proliferation; however, this effect was reversed by the
overexpression of miR-499a-5p (P<0.05; Fig. 2B-F). Furthermore, the TUNEL assay
verified that LINC01018-overexpression increased the apoptosis rate
of HL-60 and THP-1 cells, while miR-499a-5p overexpression reversed
this effect (P<0.05; Fig. 2G-I).
In addition, western blot analysis demonstrated that the protein
expression levels of Bax and Bcl-2 were increased in HL-60 and
THP-1 cells following transfection with pcDNA3.1/LINC01018, while
miR-499a-5p overexpression abolished this upregulation of Bax and
Bcl-2 (P<0.05; Fig. 2J-K). In
conclusion, LINC01018-overexpression inhibited the growth of HL-60
and THP-1 cells; however, miR-499a-5p blocked these effects.
LINC01018 acts as a sponge of
miR-499a-5p, and PDCD4 is targeted by miR-499a-5p
To further study the mechanism of LINC01018 and
miR-499a-5p in AML, the association between LINC01018 and
miR-499a-5p and the potential targeted genes of miR-499a-5p was
investigated by bioinformatics analysis. To begin with, the
transfections were successful with miR-499a-5p or miR-499a-5p
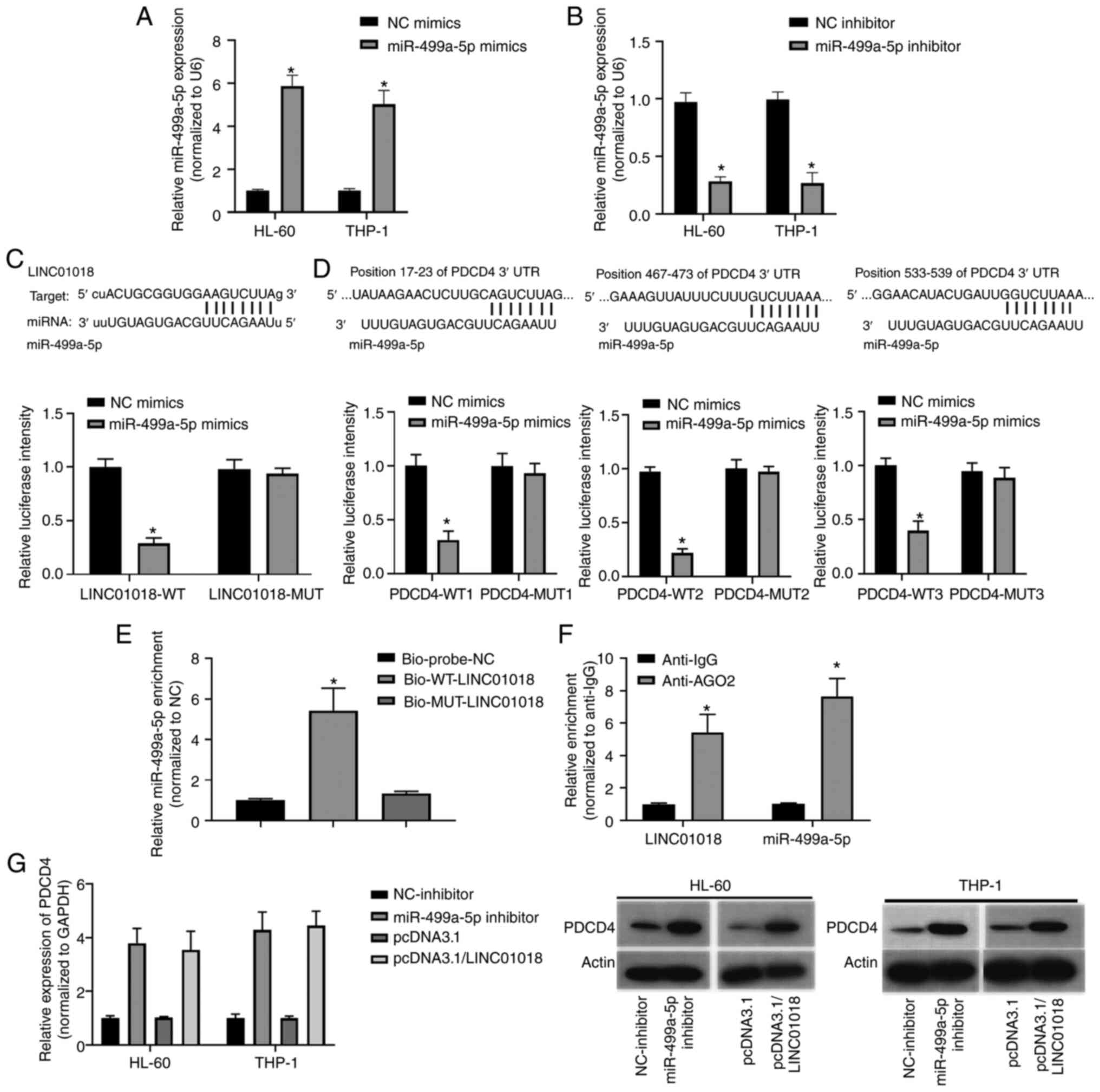
inhibitors in all cell types validated by qPCR (Fig. 3A and B). One complementary binding
site was identified between LINC01018 and miR-499a-5p (Fig. 3C; upper panel). The luciferase
reporter assay results demonstrated that the transfection of
miR-499a-5p mimics strongly decreased the luciferase activity of
cells following transfection with LINC01018-WT (P<0.05; Fig. 3C; lower panel). Furthermore, three
binding sites between miR-499a-5p and PDCD4 were predicted using a
bioinformatics tool (Fig. 3D; upper
panel). The luciferase reporter results demonstrated that the
luciferase activities were markedly decreased following
co-transfection with miR-499a-5p and PDCD4-WT1, PDCD4-WT2 or
PDCD4-WT3 relative to NC mimics, while co-transfection with
miR-499a-5p and PDCD4-MUT1, PDCD4-MUT2 or PDCD4-MUT3 caused no
notable change relative to NC mimics (P<0.05; Fig. 3D; lower panel). Next, an RNA
pull-down assay was performed to investigate the interaction
between LINC01018 and miR-499a-5p. Compared with that in the
Bio-probe-NC group, miR-499a-5p was significantly enriched in the
Bio-WT-LINC01018 group but not in the Bio-MUT-LINC01018 group
(P<0.05; Fig. 3E). Furthermore, a
RIP assay was used to further confirm the interplay between
LINC01018 and miR-499a-5p. The results indicated that LINC01018 and
miR-499a-5p were significantly enriched in the anti-AGO2 group
compared with the anti-IgG group (P<0.05; Fig. 3F). In addition, western blotting was
performed to measure PDCD4 protein expression. The expression of
PDCD4 was markedly increased in HL-60 and THP-1 cells following
knockdown of miR-499a-5p or overexpression of LINC01018 relative to
the negative control (Fig. 3G).
Taken together, these results indicated that PDCD4 is a target gene
of miR-499a-5p and that its expression level is regulated by
LINC01018 through miR-499a-5p.
Knockdown of PDCD4 abrogates the
effects of the miR-499a-5p inhibitor on AML cell lines
Following confirming the result that PDCD4 was a
target gene of miR-499a-5p, the effects of PDCD4 on miR-499a-5p
were investigated. To begin with, a PDCD4-knockdown siRNA was
designed and its efficiency was validated (Fig. 4A). Next, western blot analysis of
PDCD4 protein expression demonstrated that PDCD4 was significantly
increased by transfection of HL-60 and THP-1 cells with a
miR-499a-5p inhibitor, while transfection of si-PDCD4 reversed this
effect (P<0.05; Fig. 4B). The EdU
proliferation assay was performed to determine the effects of
co-transfection of miR-499a-5p inhibitor and si-PDCD4 on cell
proliferation. The results demonstrated that knockdown of
miR-499a-5p inhibited the proliferation of HL-60 and THP-1 cells
compared with the blank and negative controls, while si-PDCD4
transfection abolished the inhibitory effects of miR-499a-5p on
cell proliferation (P<0.05; Fig. 4C
and D). TUNEL assay results demonstrated that the apoptosis
rate was upregulated by knockdown of miR-499a-5p in HL-60 and THP-1
cells, while transfection of si-PDCD4 reversed the promotive
effects of miR-499a-5p-knockdown on cell apoptosis (P<0.05;
Fig. 4E and F). Furthermore, the
expression levels of Bax and Bcl-2 were increased by miR-499a-5p
inhibitor transfection, while co-transfection of miR-499a-5p
inhibitor and si-PDCD4 abrogated this effect (P<0.05; Fig. 4G and H). These data demonstrated that
the miR-499a-5p inhibitor suppressed the proliferation of HL-60 and
THP-1 cells but promoted cell apoptosis, which was accomplished
through regulation of PDCD4.
PDCD4 is downregulated in AML
To investigate the correlation between PDCD4 and
LINC01018 or miR-499a-5p, the expression level of PDCD4 was
measured by RT-qPCR. The results demonstrated that PDCD4 expression
was downregulated in HL-60 and THP-1 cells (P<0.05; Fig. 5A) and AML tissues (P<0.05;
Fig. 5B), compared with HS cells and
healthy controls. In addition, the results in Fig. 5C demonstrated a positive correlation
between PDCD4 and LINC01018 expression levels in patients with AML
(r=0.5098; P=0.0008). By contrast, PDCD4 and miR-499a-5p were
negatively correlated in patients with AML (r=−0.3342; P=0.0296;
Fig. 5D). In summary, these results
suggested that LINC01018 contributed toward AML cell growth by
modulating PDCD4 through suppression of miR-499a-5p (Fig. 5E).
Discussion
Increasing evidence has suggested that several
lncRNAs are involved in multiple cell processes in AML. Wang et
al (34) reported that the
lncRNA, CRNDE, which inhibits the apoptosis and promotes the
proliferation of U937 cells, acted as a molecular marker for the
treatment of AML. Linc-223 was reported to be decreased in AML
cells and regulated their proliferation and differentiation
(34). A novel axis of
LINC01018/miR-182-5p/FOXO1 was identified in hepatocellular
carcinoma, and the antitumor effect of LINC01018 was verified in
vivo (35). However, no previous
studies have identified the mechanism of LINC01018 in AML. The
present study confirmed the decreased expression of LINC01018 in
AML tissues and cell lines, and that LINC01018-overexpression
inhibited cell proliferation and promoted apoptosis in AML cells.
Therefore, the antitumor effect of LINC01018 in AML has also been
substantiated.
At present, numerous studies have confirmed that
miRNAs serve key roles in the initiation and progression of AML.
For example, miR-34a exhibited lower expression and was identified
as a tumor suppressor promoting apoptosis in AML cell lines
(36). A study proved that miR-125b
induces myeloid and B-cell leukemia by suppressing IRF4 through
different mechanisms (37). MiR-135a
was revealed to be downregulated in AML cells, and its
overexpression inhibited proliferation and the cell cycle and
promoted cellular apoptosis via HOXA10 (38). MiR-182-5p promotes cell proliferation
in AML cell lines and patient blood samples and reverses cisplatin
resistance (39). The molecular
mechanism of miR-499a-5p has previously been demonstrated in
certain cancer studies (24,40–42).
MiR-499a-5p exhibits high expression and carcinogenic effects
through the TOR pathway in highly metastatic lung cancer exosomes
(22). MiR-499a-5p acts as a
non-invasive biomarker and may increase the diagnostic sensitivity
of pancreatic cancer when combined with CA199 (24). Nevertheless, the mechanisms of
miR-499a-5p in AML pathogenesis remain largely unknown. The results
of the present study suggested that miR-499a-5p was upregulated in
AML tissues and cell lines, knockdown of miR-499a-5p suppressed
cell proliferation and induced cell apoptosis, and miR-499a-5p was
sponged by LINC01018. A negative correlation was identified between
LINC01018 and miR-499a-5p.
PDCD4 may interfere with the translation process by
directly binding with target mRNA. Multiple targets of PDCD4 are
involved in cell survival, proliferation and invasion. Identifying
more translational targets of PDCD4 will provide insight into how
PDCD4 inhibits tumorigenesis. For example, in CML primary
CD34+ cells and AML cell models, including MOLM13 and
MV4.11, the phospho-STAT5-miR21-PDCD4 pathway is active (43). Several studies have demonstrated that
PDCD4, as a target gene of miRNAs, serves an important role in AML
(44–48). These findings are consistent with the
results of the present study. In the present study the expression
levels of PDCD4 in AML tissues and cell lines were measured, and
the results revealed high expression. Next, bioinformatics analysis
and the dual-luciferase reporter assay were used to determine that
PDCD4 was the target gene of miR-499a-5p, and knockdown of PDCD4
could reverse the effects of the miR-499a-5p inhibitor on AML
progression.
In summary, the results of the present study
suggested that the low level of LINC01018 is associated with AML
pathogenesis by inhibiting AML cell proliferation and promoting
apoptosis, which is mediated via knockdown of miR-499a-5p and
regulation of PDCD4 expression. These observations provide a
feasible theoretical basis for the treatment of AML.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81600129) and the
Science and Technology Project of Hangzhou (grant no. 2016Z01).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
HZ and QX designed the experiments. HZ, QX, PS and
XJ performed the experiments. HZ collected the data. QX analyzed
the data. HZ and QX drafted the initial manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Affiliated Hangzhou First People's Hospital (Hangzhou,
China; approval no. 146-01). Written informed consent was provided
by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
De Kouchkovsky I and Abdul-Hay M: ‘Acute
myeloid leukemia: A comprehensive review and 2016 update’. Blood
Cancer J. 6:e4412016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shah A, Andersson TM, Rachet B, Björkholm
M and Lambert PC: Survival and cure of acute myeloid leukaemia in
England, 1971–2006: A population-based study. Br J Haematol.
162:509–516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Davila J, Slotkin E and Renaud T: Relapsed
and refractory pediatric acute myeloid leukemia: Current and
emerging treatments. Paediatr Drugs. 16:151–168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meyers J, Yu Y, Kaye JA and Davis KL:
Medicare fee-for-service enrollees with primary acute myeloid
leukemia: An analysis of treatment patterns, survival, and
healthcare resource utilization and costs. Appl Health Econ Health
Policy. 11:275–286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi Z, Tiwari AK, Patel AS, Fu LW and Chen
ZS: Roles of sildenafil in enhancing drug sensitivity in cancer.
Cancer Res. 71:3735–3738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang JX, Rastetter RH and Wilhelm D:
Non-coding RNAs: An introduction. Adv Exp Med Biol. 886:13–32.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dong X, Chen K, Cuevas-Diaz Duran R, You
Y, Sloan SA, Zhang Y, Zong S, Cao Q, Barres BA and Wu JQ:
Comprehensive identification of long non-coding RNAs in purified
cell types from the brain reveals functional LncRNA in OPC fate
determination. PLoS Genet. 11:e10056692015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wrang X, Chen H, Bai J and He A: MicroRNA:
An important regulator in acute myeloid leukemia. Cell Biol Int.
41:936–945. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lei L, Xia S, Liu D, Li X, Feng J, Zhu Y,
Hu J, Xia L, Guo L, Chen F, et al: Genome-wide characterization of
lncRNAs in acute myeloid leukemia. Brief Bioinform. 19:627–635.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Puvvula PK: LncRNAs regulatory networks in
cellular senescence. Int J Mol Sci. 20:26152019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miao Y, Sui J, Xu SY, Liang GY, Pu YP and
Yin LH: Comprehensive analysis of a novel four-lncRNA signature as
a prognostic biomarker for human gastric cancer. Oncotarget.
8:75007–75024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ruan X, Li P, Chen Y, Shi Y, Pirooznia M,
Seifuddin F, Suemizu H, Ohnishi Y, Yoneda N, Nishiwaki M, et al: In
vivo functional analysis of non-conserved human lncRNAs associated
with cardiometabolic traits. Nat Commun. 11:452020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shang H, Sun L, Braun T, Si Q and Tong J:
Association between miR-124 rs531564 and miR-100 rs1834306
polymorphisms and cervical cancer: A meta-analysis. Eur J Gynaecol
Oncol. 40:925–931. 2019.
|
|
20
|
Song SJ, Ito K, Ala U, Kats L, Webster K,
Sun SM, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, Delwel R
and Pandolfi PP: The oncogenic microRNA miR-22 targets the TET2
tumor suppressor to promote hematopoietic stem cell self-renewal
and transformation. Cell Stem Cell. 13:87–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bousquet M, Quelen C, Rosati R, Mansat-De
Mas V, La Starza R, Bastard C, Lippert E, Talmant P,
Lafage-Pochitaloff M, Leroux D, et al: Myeloid cell differentiation
arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid
leukemia with the t(2;11)(p21;q23) translocation. J Exp Med.
205:2499–2506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He S, Li Z, Yu Y, Zeng Q, Cheng Y, Ji W,
Xia W and Lu S: Exosomal miR-499a-5p promotes cell proliferation,
migration and EMT via mTOR signaling pathway in lung
adenocarcinoma. Exp Cell Res. 379:203–213. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou YY, Lee JH, Chen HC, Yang CM, Huang
SJ, Liou HH, Chi CC, Tsai KW and Ger LP: The association between
miR-499a polymorphism and oral squamous cell carcinoma progression.
Oral Dis. 21:195–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi Q, Feng K, Xia L, Wang C and Zhu J:
Combined use of Serum miR-499a-5p and CA199 increases the
diagnostic sensitivity of pancreatic cancer. Clin Lab. 65:2019.
View Article : Google Scholar
|
|
25
|
Elmore S: Apoptosis: A review of
programmed cell death. Toxicol Pathol. 35:495–516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Manabe A, Coustan-Smith E, Kumagai M, Behm
FG, Raimondi SC, Pui CH and Campana D: Interleukin-4 induces
programmed cell death (apoptosis) in cases of high-risk acute
lymphoblastic leukemia. Blood. 83:1731–1737. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Long J, Yin Y, Guo H, Li S, Sun Y, Zeng C
and Zhu W: The mechanisms and clinical significance of PDCD4 in
colorectal cancer. Gene. 680:59–64. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Q and Yang HS: The role of Pdcd4 in
tumour suppression and protein translation. Biol Cell. May
28–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou H and Huang S: Role of mTOR signaling
in tumor cell motility, invasion and metastasis. Curr Protein Pept
Sci. 12:30–42. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fehler O, Singh P, Haas A, Ulrich D,
Müller JP, Ohnheiser J and Klempnauer KH: An evolutionarily
conserved interaction of tumor suppressor protein Pdcd4 with the
poly(A)-binding protein contributes to translation suppression by
Pdcd4. Nucleic Acids Res. 42:11107–11118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liwak U, Thakor N, Jordan LE, Roy R, Lewis
SM, Pardo OE, Seckl M and Holcik M: Tumor suppressor PDCD4
represses internal ribosome entry site-mediated translation of
antiapoptotic proteins and is regulated by S6 kinase 2. Mol Cell
Biol. 32:1818–1829. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wedeken L, Singh P and Klempnauer KH:
Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J
Biol Chem. 286:42855–42862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Zhou Q and Ma JJ: High expression
of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and
promotes the malignant progression in acute myeloid leukemia cell
line U937. Eur Rev Med Pharmacol Sci. 22:763–770. 2018.PubMed/NCBI
|
|
35
|
Wang S, Xu M, Sun Z, Yu X, Deng Y and
Chang H: LINC01018 confers a novel tumor suppressor role in
hepatocellular carcinoma through sponging microRNA-182-5p. Am J
Physiol Gastrointest Liver Physiol. 317:G116–G126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Ren W and Chen K: MiR-34a promotes
apoptosis and inhibits autophagy by targeting HMGB1 in acute
myeloid leukemia cells. Cell Physiol Biochem. 41:1981–1992. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
So AY, Sookram R, Chaudhuri AA,
Minisandram A, Cheng D, Xie C, Lim EL, Flores YG, Jiang S, Kim JT,
et al: Dual mechanisms by which miR-125b represses IRF4 to induce
myeloid and B-cell leukemias. Blood. 124:1502–1512. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu H and Wen Q: Downregulation of miR135a
predicts poor prognosis in acute myeloid leukemia and regulates
leukemia progression via modulating HOXA10 expression. Mol Med Rep.
18:1134–1140. 2018.PubMed/NCBI
|
|
39
|
Zhang S, Zhang Q, Shi G and Yin J:
MiR-182-5p regulates BCL2L12 and BCL2 expression in acute myeloid
leukemia as a potential therapeutic target. Biomed Pharmacother.
97:1189–1194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gu X, Dong M, Liu Z, Yang J and Shi Y:
MiR-499a-5p inhibits proliferation, invasion, migration, and
epithelial-mesenchymal transition, and enhances radiosensitivity of
cervical cancer cells via targeting eIF4E. Onco Targets Ther.
13:2913–2924. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Z, Li C, Fan XY and Liu LJ: Circular
RNA circ_0079593 promotes glioma development through regulating
KPNA2 expression by sponging miR-499a-5p. Eur Rev Med Pharmacol
Sci. 24:1288–1301. 2020.PubMed/NCBI
|
|
42
|
Zhao L, Jiang P, Zheng H, Chen P and Yang
M: Downregulation of miR-499a-5p predicts a poor prognosis of
patients with non-small cell lung cancer and restrains the
tumorigenesis by targeting fibroblast growth factor 9. Technol
Cancer Res Treat. 19:15330338209570012020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Espadinha AS, Prouzet-Mauléon V, Claverol
S, Lagarde V, Bonneu M, Mahon FX and Cardinaud B: A tyrosine
kinase-STAT5-miR21-PDCD4 regulatory axis in chronic and acute
myeloid leukemia cells. Oncotarget. 8:76174–76188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gu J, Zhu X, Li Y, Dong D, Yao J, Lin C,
Huang K, Hu H and Fei J: miRNA-21 regulates arsenic-induced
anti-leukemia activity in myelogenous cell lines. Med Oncol.
28:211–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ozpolat B, Akar U, Steiner M,
Zorrilla-Calancha I, Tirado-Gomez M, Colburn N, Danilenko M,
Kornblau S and Berestein GL: Programmed cell death-4 tumor
suppressor protein contributes to retinoic acid-induced terminal
granulocytic differentiation of human myeloid leukemia cells. Mol
Cancer Res. 5:95–108. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Riccioni R, Lulli V, Castelli G, Biffoni
M, Tiberio R, Pelosi E, Lo-Coco F and Testa U: miR-21 is
overexpressed in NPM1-mutant acute myeloid leukemias. Leuk Res.
39:221–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Simmons HM, Ruis BL, Kapoor M, Hudacek AW
and Conklin KF: Identification of NOM1, a nucleolar, eIF4A binding
protein encoded within the chromosome 7q36 breakpoint region
targeted in cases of pediatric acute myeloid leukemia. Gene.
347:137–145. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang JJ, Wang ZY, Chen R, Xiong J, Yao YL,
Wu JH and Li GX: Macrophage-secreted exosomes delivering miRNA-21
inhibitor can regulate BGC-823 cell proliferation. Asian Pac J
Cancer Prev. 16:4203–4209. 2015. View Article : Google Scholar : PubMed/NCBI
|