Introduction
Epithelial ovarian cancer (EOC) is consistently one
of the leading causes of cancer-associated mortality with a
mortality rate of ~9% in Taiwan and various other areas of the
world (1,2). EOC has several subtypes with different
origins, multiple molecular characteristics and a range of outcomes
(3). EOC consists of five
histological subtypes, namely serous, mucinous, clear cell,
endometrioid and transitional cell/Brenner tumor subtypes (4). Ovarian clear cell carcinoma (OCCC) is a
distinct type of ovarian cancer, and is associated with both a poor
survival and resistance to platinum-based chemotherapy (3). OCCC is the second most common EOC
subtype in Taiwan and Japan (2),
whereas it ranks fourth in North America (5). Despite significant efforts to develop
new targeted diagnostic and therapeutic approaches aimed at
decreasing mortality, these have been largely unsuccessful, as
metastasis remains the main cause of mortality and accounts for
~90% of all OCCC-associated deaths (6,7).
Metastasis is a multistep process that is a
significant cause of cancer-associated mortality in humans
(8). Therefore, identifying genes
and molecular pathways in OCCC that are associated with metastasis
may lead to advances in therapeutics. Our previous studies have
revealed that low expression levels of slit guidance ligand 2
(SLIT2) are associated with a poor survival and promote esophageal
cancer metastasis (9,10). SLIT2 is a secreted glycoprotein of
the SLIT family and is the human orthologue of the
Drosophila Slit2 protein (11). SLIT2 is the ligand of the receptor
roundabout guidance receptor 1 (ROBO1), which is known to
participate in intercellular signal transduction via GTPase
activation protein (12). Moreover,
this signaling serves an important role in cell migration (9,12). In
addition, SLIT/ROBO signaling has been revealed to be involved in
the development of a number of organs, including the heart and
organs of the reproductive tract and nervous system (13).
Our previous study has indicated that SLIT2 may be a
candidate tumor suppressor that may be silenced in epithelial
tumors of the aerodigestive tract via genetic deletion and
epigenetic promoter hypermethylation (10). Furthermore, the epigenetic silencing
of SLIT2 has been observed in serous ovarian cancer (14–16). A
previous study has indicated low SLIT2 expression in EOC samples
compared with in the normal human ovarian surface epithelium
(17). Additionally, SLIT2
expression can significantly decrease the invasion and migration of
endometrial carcinoma cells (18).
Moreover, injecting exogenous ROBO1-expressing cells into nude mice
decreases the size of breast tumors (19). However, to the best of our knowledge,
there are no published studies that have investigated changes in
SLIT2/ROBO1 signaling in OCCC. Therefore, the present study
performed a range of molecular analyses on human normal and
malignant OCCC samples, as well as on SKOV3 cells that are able to
form OCCC. The current findings revealed that SLIT2 may be a
potential molecular target for the treatment of human OCCC.
Materials and methods
Cell culture
The SKOV3 low-mobility (SKOV3-L) and high-mobility
(SKOV3-H) cell lines were kindly provided by Dr Lu-Hai Wang
(Institute of Molecular and Genomic Medicine, National Health
Research Institute, Miaoli, Taiwan) (20). SKOV3 cells were maintained in
RPMI-1640 medium with 10% FBS (both Thermo Fisher Scientific,
Inc.). All cells were incubated at 37°C in a humidified atmosphere
with 5% CO2.
Western blot analysis
Cells were lysed in RIPA buffer (0.05 M Tris-HCl, pH
7.4, 0.15 M NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA, 0.5
mM DTT, 1 mM phenylmethylsulfonyl fluoride, 5 µg/ml leupeptin and
10 µg/ml aprotinin) on ice, and then centrifuged at 15,000 × g at
4°C for 5 min. The protein concentration was estimated by a BSA
standard curve. Subsequently, SDS gel loading buffer (60 mM Tris
base, 2% SDS, 10% glycerol and 5% β-mercaptoethanol) was added to
the samples, and 50 µg protein/lane was separated by 8% SDS-PAGE.
The proteins were electro-blotted onto Immobilon-P membranes (EMD
Millipore) using transfer buffer. The membranes were blocked with
5% skim milk in phosphate-buffered saline with 0.1% Tween-20 (PBST)
for 1 h at room temperature. Immunoblotting was performed using
primary anantibodies against SLIT2 (cat. no. AB5701; 1:800;
MilliporeSigma), β-catenin (cat. no. 9582; 1:1,000; Cell Signaling
Technology, Inc.), phosphorylated (p)AKT (cat. no. sc-7985-R;
1:500; Santa Cruz Biotechnology, Inc.), AKT (cat. no. 4691; 1:800;
Cell Signaling Technology, Inc.) and snail family transcriptional
repressor 1 (SNAI1; cat. no. 3895; 1:500; Cell Signaling
Technology, Inc.). β-actin (cat. no. GTX109639; 1:1,000; GeneTex,
Inc.) was used as an internal control to confirm that equal amounts
of proteins had been loaded onto the gel. The membranes were
subsequently probed with a horseradish peroxidase-conjugated
secondary antibody (cat. no. #12-371, 1:5,000; MilliporeSigma) for
1 h at room temperature. The bands were visualized using a western
blot chemiluminescence reagent (MilliporeSigma).
Reverse transcription-quantiative PCR
(RT-qPCR)
Total RNA was prepared from tumor cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). cDNA was synthesized using Oligo(dT)18 primer (Genedirex,
Inc.), dNTPs (Protech, Inc.), RT buffer (Bioline, Inc.) and
SuperScript™ Reverse Transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
mRNA expression levels of SLIT2 were measured using the Applied
Biosystems StepOne™ Real-Time PCR System (Thermo Fisher Scientific,
Inc.) and SYBR®-Green (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions with β-actin as an
internal control. The mRNA expression levels were evaluated using
the following primers: SLIT2 forward, 5′-GGTGTCCTCTGTGATGAAGAG-3′
and reverse, 5′-GTGTTTAGGACACACACCTCG-3′; and β-actin forward,
5′-GGCGGCACCACCATGTACCCT-3′ and reverse
5′-AGGGGCCGGACTCGTCATACT-3′. Reactions were performed in a volume
of 25 µl with 1 µl cDNA and 0.25 pmol primers. The PCR protocol
involved 35 cycles of three sequential steps: 95°C for 30 sec, 58°C
for 30 sec and 72°C for 30 sec. The mRNA expression levels were
calculated using the 2−ΔΔCq method
(ΔCq=CqSLIT2-Cq-β-actin) (21).
TissueScan array
The present study was performed after approval from
the research ethics committee review board of the Tzu-Chi Hospital
(approval no: 101-04; Hualien, Taiwan). TissueScan Ovarian Cancer
Panels (cat. nos. HORT502 and HORT504; OriGene Technologies, Inc.)
were purchased on 4 April, 2018, to examine the mRNA expression
levels of SLIT2 in cDNA samples that had been prepared from normal
ovarian and ovarian tumor tissue samples. Briefly, the TissueScan
plate was removed from −20°C storage and allowed to warm at room
temperature. A PCR pre-mix was prepared containing SYBR®
Green master mix, the aforementioned primers and double-distilled
H2O. A total of 25 µl of the PCR pre-mix was added to
each well in the 96-well TissueScan plate. The PCR was performed as
aforementioned.
Methylation-specific PCR (MSP)
assay
The methylation status of the promoter region of the
SLIT2 gene was assessed by treating cells with sodium bisulfite,
followed by an MSP assay. A total of 500 ng genomic DNA was
denatured at 95°C for 5 min and incubated with NaOH (final
concentration, 0.2 M) at 37°C for 15 min. Hydroquinone (10 nM;
Sigma-Aldrich; Merck KGaA) and sodium bisulfite (3 M;
Sigma-Aldrich; Merck KGaA) were added and incubated at 50°C for 18
h. Subsequently, modified DNA was purified using a Microcon YM-50
DNA purification column (EMD Millipore). Treatment of genomic DNA
with sodium bisulfite converts unmethylated but not methylated
cytosine residues to uracil residues, which are then converted to
thymidine residues during the subsequent PCR step. The primers and
PCR conditions were as described previously (9). A hypermethylated gene was defined as a
gene in tumor cells from which the methylated PCR products were
amplified more than the unmethylated PCR products (10).
5-aza-2′-Deoxycytidine (5-aza-dC)
treatment
In total, 1×105 cancer cells were seeded
in a 100-mm culture dish before 5-aza-dC treatment. Cancer cells
were allowed to double three times (mean duration, 144 h) in the
presence of 5 µmol/l 5-aza-dC (Sigma-Aldrich; Merck KGaA) at 37°C.
Subsequently, the cells were collected and subjected to MSP,
RT-qPCR and western blot analysis, as aforementioned.
Analysis of SLIT2 knockdown,
overexpression and treatment with SLIT2 protein
Short hairpin RNA (shRNA) plasmids, consisting of
shRNA-control (non-targeting) and shRNA-SLIT2, were purchased from
the RNAi Core Lab of Academia Sinica. The empty pCMV6 and
pCMV6-SLIT2 overexpression vectors were obtained from OriGene
Technologies, Inc. The SLIT2 knockdown was performed in SKOV3-L
cells, whereas the SLIT2 overexpression was performed in SKOV3-H
cells. A total of 1×105 cancer cells were transfected
with 4 µg shRNA-control, shRNA-SLIT2, pCMV6 or pCMV6-SLIT2 using
Lipofectamine® 2000 transfection reagent (Thermo Fisher
Scientific, Inc.) for 4 h according to the manufacturer's
instructions. Following a 48-h incubation, the cells were subjected
to RT-qPCR and western blot analysis as aforementioned.
Human SLIT2 protein was purchased from Abcam. A
total of 1×105 cells were treated with 5 ng/ml SLIT2
protein at 37°C for 48 h, and the results were analyzed by
migration assay.
Gap closure assay
The gap closure assay was conducted using a culture
insert (Ibidi GmbH), and SKOV3-L and SKOV3-H cells were used in
this assay. The cells used for the gap closure assays were
transfected with the empty vector, SLIT2 overexpression plasmid,
shRNA-control or shRNA-SLIT2, as aforementioned. The cells were
serum-starved for 16 h before conducting the migration experiment.
A cell-free gap of 500-µm was created by removing the culture
insert, and serum-free medium was added after the wounds were
created. The cell-free gaps that remained after 12 h of culture
were photographed under a light microscope and measured using
ImageJ software (Version 1.51, National Institutes of Health). The
area of the open wound was calculated. A total of three independent
experiments were performed for each cell type.
Transwell migration assay
Transwell assays were performed to determine the
migratory ability of the various types of SKOV3 cells transfected
with SLIT2 overexpression or knockdown plasmids, as aforementioned.
The Transwell system (Falcon; Corning Life Sciences) consisted of
upper and lower chambers. The cells seeded in the upper chamber
were able to migrate through the membrane to the lower chamber. In
total, 3×105 cells were seeded into the upper chamber in
serum-free RPMI-1640 medium and the lower chamber was filled with
RPMI-1640 medium with 10% FBS. After incubation for 24 h at 37°C,
the cells that were attached to the reverse side of the Transwell
membrane were stained using crystal violet for 30 min, and total
cells were counted under a light microscope at ×100 magnification.
In total, three independent experiments were performed for each
cell type.
Adhesion assay
An appropriate number of 6-well microplates were
pre-coated with extracellular matrix (ECM) proteins (Sigma-Aldrich;
Merck KGaA) for 16 h at 4°C and were then blocked with 1% BSA
(Sigma-Aldrich; Merck KGaA) at 37°C for 1 h. Subsequently,
6×104 cells/well were seeded in the pre-coated wells in
RPMI-1640 medium containing 10% FBS and were then incubated in a
rotating incubator at 37°C for 2 h to recover. Next, the cells were
incubated for 30 min at 37°C in 5% CO2, followed by
three washes with PBS to remove any unattached cells. Subsequently,
the remaining attached cells were fixed and stained with 1% crystal
violet/MeOH for 10 min at room temperature. Finally, the cells were
lysed using DMSO and the absorbance was measured at 550 nm. The
absorbance at 550 nm was proportional to the number of cells
attached to the pre-coated ECM wells. In total, three independent
experiments were performed for each cell type.
Data collection
The gene expression profiling and methylation
profiling datasets were selected from the National Center of
Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database based
on the keywords ‘ovarian cancer’ and ‘Homo sapiens’, and with the
following inclusion criteria: i) Ovarian clear cell
carcinomas[title]; ii) ovarian cancer[title], clear; iii) the
dataset contained >50 samples; and iv) the dataset contained
survival data. Datasets using serum and cell line samples were
excluded. A total of two gene expression profiling (GSE8841 and
GSE65986) and one methylation profiling (GSE51820) datasets were
selected (22–24). The 83 ovarian tumor samples of the
GSE8841 dataset contained 16 clear cell samples. The GSE65986
dataset contained 55 epithelial ovarian cancer samples, including
25 clear cell samples. The GSE51820 dataset contained 96 ovarian
cancer samples, including 13 clear cell and 4 normal ovarian
surface epithelium samples. In GSE65986 and GSE8841, the low
expression group was defined as the expression value of SLIT2 <
mean-1SD, and the other samples were defined as the high expression
group (25). In the GSE51820
dataset, hypermethylation of the SLIT2 gene was defined as a probes
value > mean + 1SD of the OCCC samples (26).
Statistical analysis
The results of the RT-qPCR, gap closure and
Transwell assays were analyzed using an unpaired Student's t-test.
The Kaplan-Meier method was used to construct progression-free
survival curves, and then a comparison was performed using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference. SPSS version 19.0 (IBM Corp.) was used for
all statistical analyses.
Results
Aggressive phenotype of OCCC cells and
promoter methylation of SLIT2
OCCC cell models, which consisted of SKOV3-L and
SKOV3-H cells, were used in the various experiments. A number of
different cancer cell properties, including the cell migration rate
and cell adhesion, were compared between SKOV3-L and SKOV3-H cells
(Fig. 1). The gap closure rates of
SKOV3-L and SKOV3-H cells were measured in order to assess their
migratory ability. The SKOV3-H cells had a significantly higher
migratory ability compared with the SKOV3-L cells (Fig. 1A). The adhesion of the SKOV3-L and
SKOV3-H cells was also evaluated. The results demonstrated that
SKOV3-H cells had a significantly decreased adhesive capacity
compared with SKOV3-L cells, as determined via a cell adhesion
assay (Fig. 1B).
Whether the expression levels of SLIT2 were altered
in OCCC cells was further examined by measuring the protein
expression levels of SLIT2 in SKOV3-L and SKOV3-H cells. The
results indicated that there was lower SLIT2 expression in the
SKOV3-H cells compared with that in the SKOV3-L cells (Fig. 1C). Subsequently, whether the low
SLIT2 protein expression was due to downregulated SLIT2 mRNA
expression was investigated. Consistent with the SLIT2 protein
results, significantly lower SLIT2 mRNA expression was detected in
the SKOV3-H cells compared with in the SKOV3-L cells (Fig. 1D). To determine if there was an
epigenetic effect associated with the expression of the SLIT2 gene,
DNA methylation assays of the SLIT2 gene were conducted on the two
types of SKOV3 cells. DNA hypermethylation of the SLIT2 gene was
observed in SKOV3-H cells, whereas a normal degree of methylation
was observed in SKOV3-L cells (Fig.
1E).
It was then evaluated whether the low expression
levels of SLIT2 were caused by the DNA methylation present in the
SKOV3-H cells. The demethylating agent 5-aza-dC was used to treat
SKOV3-H cells. As shown in Fig. 1F,
demethylation of the SLIT2 gene by 5-aza-dC treatment was able to
increase SLIT2 mRNA and protein expression in SKOV3-H cells.
To confirm whether the hypermethylation of SLIT2 was
associated with patients with OCCC, the publicly available GSE51820
dataset was analyzed. This methylation profile of 96 primary
ovarian cancer samples included 13 clear cell ovarian cancer
samples and 4 normal ovarian surface epithelium samples. A SLIT2
methylation probes value of OCCC higher than the mean of the 4
normal ovarian surface epithelium samples + 1SD indicated
hypermethylation. Based on this criterion, the frequency of SLIT2
methylation was between 31 and 85% in patients with OCCC (Table SI).
Migration of OCCC cells is suppressed
by overexpression of SLIT2 or by the addition of purified SLIT2
protein
Our previous studies have revealed that low SLIT2
expression is able to promote the migration of lung and esophageal
cancer cells (9,10). Therefore, it was hypothesized that
SLIT2 may act as a cell migration suppressor in OCCC. To verify the
role of SLIT2 reactivation in ovarian cancer cell migration, gap
closure and Transwell assays were performed using SKOV3-H cells in
the presence and absence of the ectopic expression of SLIT2. The
overexpression of SLIT2 resulted in high SLIT2 protein expression
in SKOV3-H cells (Fig. 2A).
Moreover, a significant decrease in the migration of SKOV3-H cells
was observed with SLIT2 overexpression compared with the control
cells (Fig. 2B and C).
Since SLIT2 is a secreted glycoprotein, a gap
closure assay was used to examine whether purified SLIT2 protein
was able to affect the mobility of SKOV3-H cells. The results
demonstrated that addition of purified SLIT2 protein (5 ng/ml) was
able to significantly decrease the migratory capacity of SKOV3-H
cells compared with the control cells (Fig. 2D).
Knockdown of SLIT2 increases the
motility of OCCC cells
To further confirm the presence of a reciprocal
association between SLIT2 expression and cell motility in OCCC,
shRNA technology was used to knock down the SLIT2 gene in the
SKOV3-L cells, which expressed SLIT2 at a high level and exhibited
a low level of migration in vitro prior to transfection.
Using western blot analysis, it was found that SLIT2-knockdown in
SKOV3-L cells markedly inhibited the protein expression levels of
SLIT2 compared with those in cells transfected with the
shRNA-control (Fig. 3A). Next, the
motility of these knockdown cells was determined using gap closure
and Transwell assays. As shown in Fig.
3B and C, SKOV3-L cells transfected with shSLIT2-2 exhibited a
significantly greater migratory capacity compared with
control-treated SKOV3-L cells.
β-catenin/pAKT/SNAI1 involvement in
the SLIT2-mediated mobility of OCCC cells
The epithelial-mesenchymal transition (EMT) of
cancer cells is a fundamental feature of tumor invasion and
metastasis (27). The transcription
factors β-catenin and SNAI1, together with hyperactivation of the
AKT signaling pathway, have been reported to be involved in the EMT
process (28). To determine whether
the β-catenin/pAKT/AKT/SNAI1 axis was involved in the mechanism
underlying SLIT2-mediated migration of OCCC cells, the protein
expression levels of β-catenin, pAKT/AKT and SNAI1 were measured in
the SLIT2-knockdown SKOV3-L cells and in SKOV3-H cells
overexpressing SLIT2. SLIT2-knockdown SKOV3-L cells exhibited
higher protein expression levels of β-catenin, pAKT and SNAI1
compared with the control cells (Fig.
4). Conversely, overexpression of SLIT2 in SKOV3-H cells
resulted in decreased protein expression levels of β-catenin, pAKT
and SNAI1 compared with the empty vector cells (Fig. 4).
Low SLIT2 expression occurs in human
OCCC and is associated with a poor prognosis
The aforementioned results indicated that SLIT2
expression was associated with the migration of OCCC cells in
vitro. However, to the best of our knowledge, no studies have
examined the mRNA expression levels of SLIT2 in OCCC tissue samples
and their association with the clinical prognosis of patients.
Therefore, RT-qPCR was performed to measure SLIT2 mRNA expression
in two commercial array panels (HORT102 and HORT104), which
contained 96 cDNA samples from 14 normal ovarian tissues and 82
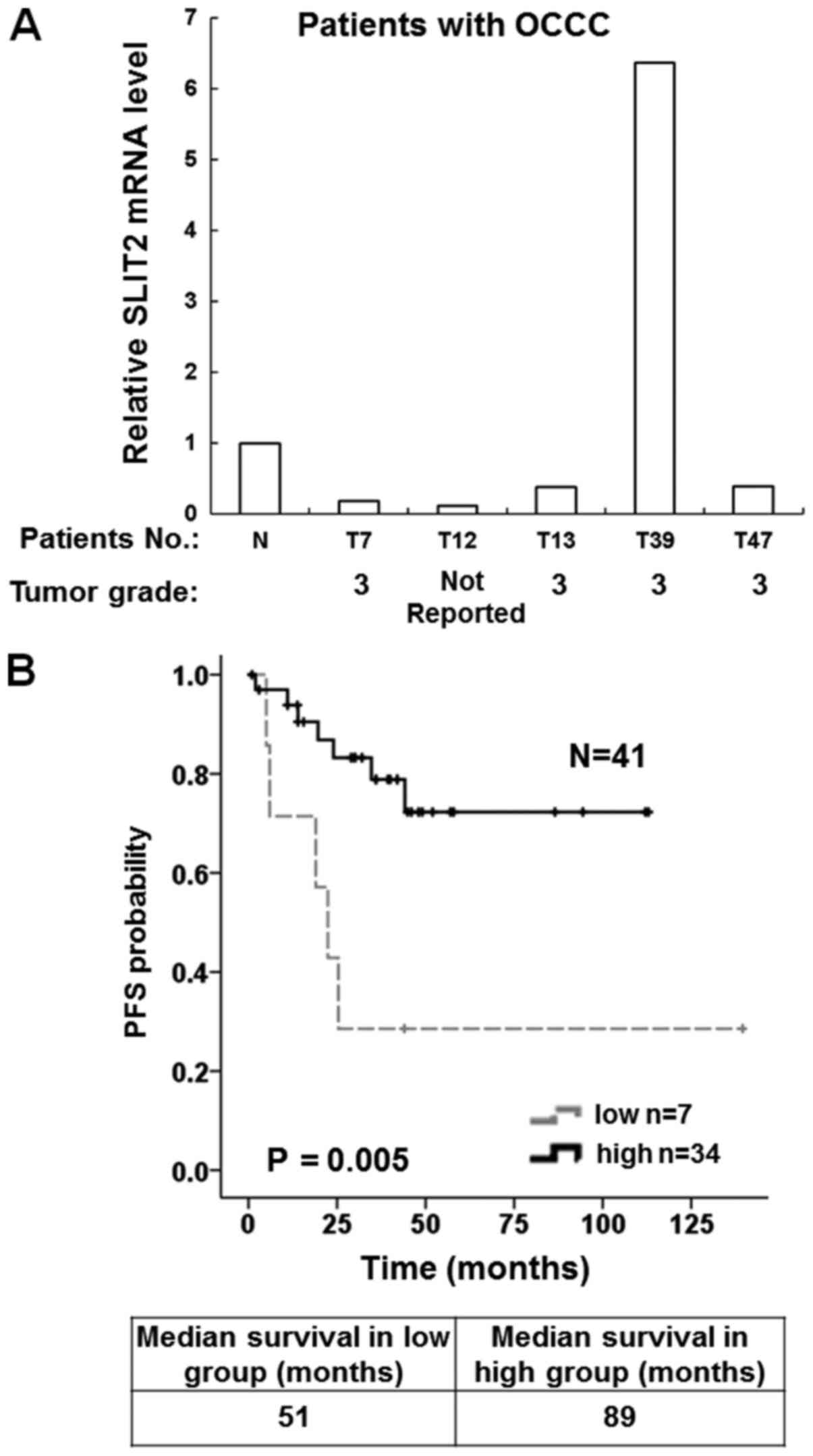
ovarian tumor tissues including five OCCC samples (Table SII). The results demonstrated that
four of the OCCC tissue samples had lower SLIT2 mRNA expression
compared with the normal ovarian tissue samples (Fig. 5A).
To confirm whether low SLIT2 expression was
associated with prognosis in patients with OCCC, survival curves
were drawn using the Kaplan-Meier method. The SLIT2 mRNA expression
of OCCC samples present in the publicly available GSE8841 and
GSE65986 microarray datasets (22,23) were
evaluated. There were 16 OCCC samples collected by the GSE8841
project and 25 OCCC samples collected by the GSE65986 project
(Table SIII). The results
demonstrated that 17% (7/41) of tumors exhibited low expression
levels of SLIT2. It was found that the median survival time was 51
months in the low SLIT2 expression group and 89 months in the high
SLIT2 expression group. Moreover, high SLIT2 expression was
associated with a longer progression-free survival in patients with
OCCC (Fig. 5B).
Discussion
To further understand the effects of changes in the
expression levels of SLIT2 in OCCC, the present study performed a
molecular analysis of OCCC cells, which included measuring the
SLIT2 mRNA and protein expression levels and assessing the DNA
methylation status of the SLIT2 gene. The current study identified
an epigenetic mechanism that appeared to be associated with OCCC
and involved SLIT2 gene silencing. Furthermore, the present study
provided evidence that higher expression levels of SLIT2 may
inhibit the progression of OCCC.
It is known that tumor suppressor genes can be
inactivated by both genetic and epigenetic changes (29). In the present study, epigenetic
silencing of SLIT2 by hypermethylation was observed, as determined
by treating OCCC cells with 5-aza-dC. Consistent with these
results, it was also revealed that the frequency of SLIT2
methylation ranged between 31–85% in 13 patients with OCCC from the
GSE51820 project (Table SI). In
addition, a study by Dong et al (16) reported that hypermethylation of the
SLIT2 gene was frequently observed in patients with serous ovarian
cancer. However, the possibility that somatic mutations of SLIT2
may also be implicated in carcinogenesis remains to be determined.
Thus, detailed mutational analysis on the SLIT2 gene is required in
order to obtain an improved overall understanding of OCCC
tumorigenesis. Qiu et al (14) found that decreased SLIT2 expression
was significantly associated with SLIT2 promoter hypermethylation
in 66 ovarian cancer samples. Conversely, Dai et al
(15) found that SLIT2 was widely
expressed in human normal and malignant ovarian tissue microarrays
(4 adult granular cell tumor, 5 dysgerminoma, 8 adenocarcinoma, 5
teratoma malignant change, 6 yolk sac tumor, 20 mucinous
adenocarcinoma and 136 serous adenocarcinoma), although the
malignant ovarian tissue microarrays did not include OCCC samples.
The present results provided evidence that lower SLIT2 expression
was associated with epigenetic silencing of SLIT2. However, the
lack of immunohistochemical analysis of SLIT2 in other subtypes,
such as high-grade serous carcinoma or endometrial carcinoma, was a
limitation of the present study. The role of SLIT2 in other
histological subtypes, such as high-grade serous carcinoma, should
be further investigated in future studies.
Previous studies have reported that SLIT2 may be a
tumor suppressor gene (10,14,16);
however, to the best of our knowledge, the anti-migration
properties of this gene have not yet been reported for human OCCC.
The present findings demonstrated that the motility of OCCC cells
was decreased when SLIT2 was overexpressed, while SLIT2-knockdown
resulted in an increase in cell motility. Consistent with the
current findings, Dickinson et al (30) revealed that inhibiting SLIT/ROBO
activity induced an increase in the migration of primary luteal
cell cultures. Moreover, another study by Dickinson et al
(31) found that blocking SLIT/ROBO
activity resulted in decreased apoptosis of SKOV3 cells. Jeon et
al (32) observed that SLIT2
overexpression inhibited the proliferation, migration and invasion
of thyroid cancer cells. In addition, a previous study reported
that disrupting SLIT2-ROBO signaling in pancreatic ductal
adenocarcinoma cells enhanced cancer metastasis and predisposed to
neural invasion (33). Collectively,
the aforementioned findings support the hypothesis that SLIT2 may
serve a role as an inhibitor of cell migration.
In the present study, the expression levels of SLIT2
in OCCC were found to be associated with the expression levels of
β-catenin and pAKT, which is in line with the results of studies
targeting breast and lung cancer (19,34).
Additionally, the current findings demonstrated that the protein
expression levels of β-catenin, pAKT and SNAI1 were increased in
OCCC cells treated with shRNA-SLIT2. Conversely, low protein
expression levels of β-catenin, pAKT and SNAI1 were identified in
OCCC cells when SLIT2 was overexpressed. The EMT in ovarian cancer
is induced by multiple factors, including hepatocyte growth factor
(HGF) signaling (35). A previous
study demonstrated that shRNA-SLIT2 was able to enhance HGF-induced
migration, while overexpression of SLIT2 was able to inhibit
HGF-triggered motile responses in SKOV3 cells (36). Thus, it is worth investigating
whether there are other signaling pathways that are associated with
SLIT2 signaling in order to further understand the role of SLIT2.
SLIT2 is a secreted glycoprotein and its expression appears to be
frequently downregulated in various types of human cancer, such as
esophageal, thyroid and lung cancer (10,31,32,34).
Thus, the SLIT2 peptides may represent a new target when designing
novel therapeutic molecules that may help to attenuate human OCCC
metastasis.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Tang-Yuan Chus
(Department of Obstetrics and Gynecology, Tzu Chi General Hospital,
Hualien, Taiwan) for his helpful discussions regarding this
research.
Funding
The present study was supported in part by the
Ministry of Science and Technology (grant no. MOST
NSC101-2320-B-320-008) and the Buddhist Tzu Chi Medical Foundation
(grant no. TCMMP108-01-03).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Authors' contributions
CJL performed the experiments and participated in
the analysis of the data. WRH participated in the acquisition and
analysis of the data. CZW performed the clinical experiments. RCT
designed this study, analyzed the data and drafted the manuscript.
CJL and RCT confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Website of Health Promotion
Administration, Ministry of Health and Welfare, Taiwan, .
2019.https://www.hpa.gov.tw/Pages/ashx/File.ashx?FilePath=~/File/Attach/10227/File_11644.pdf
|
|
3
|
Tan DS, Miller RE and Kaye SB: New
perspectives on molecular targeted therapy in ovarian clear cell
carcinoma. Br J Cancer. 108:1553–1559. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaw TJ, Senterman MK, Dawson K, Crane CA
and Vanderhyden BC: Characterization of intraperitoneal,
orthotopic, and metastatic xenograft models of human ovarian
cancer. Mol Ther. 10:1032–1042. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seyfried TN and Huysentruyt LC: On the
origin of cancer metastasis. Crit Rev Oncog. 18:43–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okamoto A: Management and new strategy of
ovarian clear cell carcinoma. Int J Clin Oncol. 25:4182020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang L, Wang Y, Rong Y, Xu L, Chu Y,
Zhang Y and Yao Y: MiR-1179 promotes cell invasion through
SLIT2/ROBO1 axis in esophageal squamous cell carcinoma. Int J Clin
Exp Pathol. 8:319–327. 2015.PubMed/NCBI
|
|
10
|
Tseng RC, Chang JM, Chen JH, Huang WR,
Tang YA, Kuo IY, Yan JJ, Lai WW and Wang YC: Deregulation of
SLIT2-mediated Cdc42 activity is associated with esophageal cancer
metastasis and poor prognosis. J Thorac Oncol. 10:189–198. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang Z, Liang G, Xiao Y, Qin T, Chen X,
Wu E, Ma Q and Wang Z: Targeting the SLIT/ROBO pathway in tumor
progression: Molecular mechanisms and therapeutic perspectives.
Ther Adv Med Oncol. 11:17588359198552382019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lucas B and Hardin J: Mind the
(sr)GAP-roles of Slit-Robo GAPs in neurons, brains and beyond. J
Cell Sci. 130:3965–3974. 2017.PubMed/NCBI
|
|
13
|
Blockus H and Chédotal A: Slit-Robo
signaling. Development. 143:3037–3044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu H, Zhu J, Yu J, Pu H and Dong R: SLIT2
is epigenetically silenced in ovarian cancers and suppresses growth
when activated. Asian Pac J Cancer Prev. 12:791–795.
2011.PubMed/NCBI
|
|
15
|
Dai CF, Jiang YZ, Li Y, Wang K, Liu PS,
Patankar MS and Zheng J: Expression and roles of Slit/Robo in human
ovarian cancer. Histochem Cell Biol. 135:475–485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong R, Yu J, Pu H, Zhang Z and Xu X:
Frequent SLIT2 promoter methylation in the serum of patients with
ovarian cancer. J Int Med Res. 40:681–686. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dickinson RE and Duncan WC: The SLIT-ROBO
pathway: A regulator of cell function with implications for the
reproductive system. Reproduction. 139:697–704. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sirohi VK, Popli P, Sankhwar P, Kaushal
JB, Gupta K, Manohar M and Dwivedi A: Curcumin exhibits anti-tumor
effect and attenuates cellular migration via Slit-2 mediated
down-regulation of SDF-1 and CXCR4 in endometrial adenocarcinoma
cells. J Nutr Biochem. 44:60–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang PH, Hwang-Verslues WW, Chang YC,
Chen CC, Hsiao M, Jeng YM, Chang KJ, Lee EY, Shew JY and Lee WH:
Activation of Robo1 signaling of breast cancer cells by Slit2 from
stromal fibroblast restrains tumorigenesis via blocking
PI3K/Akt/β-catenin pathway. J Cancer Res. 72:4652–4661. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yeh YM, Chuang CM, Chao KC and Wang LH:
MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis
by targeting SOX4 and HIF-1α. Int J Cancer. 133:867–878. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marchini S, Mariani P, Chiorino G,
Marrazzo E, Bonomi R, Fruscio R, Clivio L, Garbi A, Torri V,
Cinquini M, et al: Analysis of gene expression in early-stage
ovarian cancer. Clin Cancer Res. 14:7850–7860. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Makii C, Oda K, Sone K, Sone K, Hasegawa
K, Uehara Y, Nishijima A, Asada K, Koso T, Fukuda T, et al: MDM2 is
a potential therapeutic target and prognostic factor for ovarian
clear cell carcinomas with wild type TP53. Oncotarget.
7:75328–75338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaguchi K, Huang Z, Matsumura N, Mandai
M, Okamoto T, Baba T, Konishi I, Berchuck A and Murphy SK:
Epigenetic determinants of ovarian clear cell carcinoma biology.
Int J Cancer. 135:585–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang J, Shin SH, Yoon H, Huh J, Shin HW,
Chun YS and Park JW: FIH is an oxygen sensor in ovarian cancer for
G9a/GLP-Driven epigenetic regulation of metastasis-related genes.
Cancer Res. 78:1184–1199. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang JM, Tsai AC, Huang WR and Tseng RC:
The alteration of CTNNBIP1 in lung cancer. Int J Mol Sci.
20:56842019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Puisieux A, Brabletz T and Caramel J:
Oncogenic roles of EMT-inducing transcription factors. Nat Cell
Biol. 16:488–494. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang LH, Wu CF, Rajasekaran N and Shin YK:
Loss of tumor suppressor gene function in human cancer: An
overview. Cell Physiol Biochem. 51:2647–2693. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dickinson RE, Myers M and Duncan WC: Novel
regulated expression of the SLIT/ROBO pathway in the ovary:
Possible role during luteolysis in women. Endocrinology.
149:5024–5034. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dickinson RE, Fegan KS, Ren X, Hillier SG
and Duncan WC: Glucocorticoid regulation of SLIT/ROBO tumour
suppressor genes in the ovarian surface epithelium and ovarian
cancer cells. PLoS One. 6:e277922011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeon MJ, Lim S, You MH, Park Y, Song DE,
Sim S, Kim TY, Shong YK, Kim WB and Kim WG: The role of Slit2 as a
tumor suppressor in thyroid cancer. Mol Cell Endocrinol. 483:87–96.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Göhrig A, Detjen KM, Hilfenhaus G, Körner
JL, Welzel M, Arsenic R, Schmuck R, Bahra M, Wu JY, Wiedenmann B
and Fischer C: Axon guidance factor SLIT2 inhibits neural invasion
and metastasis in pancreatic cancer. Cancer Res. 74:1529–1540.
2014. View Article : Google Scholar
|
|
34
|
Tseng RC, Lee SH, Hsu HS, Chen BH, Tsai
WC, Tzao C and Wang YC: SLIT2 attenuation during lung cancer
progression deregulates beta-catenin and E-cadherin and associates
with poor prognosis. Cancer Res. 70:543–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vergara D, Merlot B, Lucot JP, Collinet P,
Vinatier D, Fournier I and Salzet M: Epithelial-mesenchymal
transition in ovarian cancer. Cancer Lett. 291:59–66. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stella MC, Trusolino L and Comoglio PM:
The Slit/Robo system suppresses hepatocyte growth factor-dependent
invasion and morphogenesis. Mol Biol Cell. 20:642–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|