Introduction
Glioblastoma (GBM) is a common malignant primary
brain tumor that accounts for ~57% of all gliomas and 48% of all
malignant primary tumors of the central nervous system in the
United States (1). The prevalence of
GBM is highest in North America and Australia, as well as Northern
and Western Europe (2). Caucasian
patients have the poorest survival rates and highest incidence of
GBM (3), and treatment strategies
such as surgery, radiotherapy and chemotherapy appear to be
ineffective. Moreover, poor prognosis and low long-term survival
remain challenging issues (4),
though precision oncology and targeted gene therapy herald much
promise in developing more efficacious treatments for patients with
GBM.
MicroRNAs (miRNAs/miRs) are abundant small
non-coding RNAs, ~20-24 nucleotides in length, that regulate gene
expression by inhibiting translation or degrading messenger RNA
(5,6). A cluster of miRNAs has been shown to
affect tumor cellular biological processes and to serve as
potential diagnostic targets in various human cancers (7,8). For
example, miR-744-5p has been associated with numerous cancers and
plays an important role in tumor progression characteristics,
including cellular proliferation, invasiveness and apoptosis
(9–11). Accumulating evidence suggests that
miR-744-5p may also be a promising therapeutic target for cancer
treatment (12,13), and has also been reported to suppress
cellular proliferation and invasiveness in ovarian (14) and lung cancer (15). Further studies have shown that
miR-744-5p is abnormally expressed in GBM, regulating its
occurrence and development (16,17).
Nevertheless, the underlying mechanisms by which miR-744-5p acts on
GBM require further investigation.
The replication factor C subunit 2 (RFC2) gene is
located on chromosome 7q11.23 and consists of 11 exons, encoding a
member of the activator 1 small subunit family. RFC2 plays an
important role in the elongation of primed DNA templates by DNA
polymerase δ and ε (18).
Upregulated RFC2 expression has been associated with a number of
cancers, such as nasopharyngeal (19) and esophageal squamous cell carcinoma
(20). However, to the best of our
knowledge, only a single study has reported the involvement of RFC2
in GBM, in which RFC2 was indicated to enhance temozolomide (TMZ)
cytotoxicity (21). Notably, RFC2
was found to be a direct target of miR-744-5p in the progression of
colorectal cancer cell tumorigenicity (22). However, little is known about the
expression pattern and biological functions of RFC2 in GBM,
including its interaction with miR-744-5p. Therefore, it may be
beneficial to elucidate the biological role of RFC2 and its
potential interaction with miR-744-5p therein.
Thus, the aim of the present study was to explore a
novel interactome, miR-744-5p-RFC2, and to determine the effects of
miR-744-5p and RFC2 in GBM. We hypothesized that miR-744-5p
suppressed GBM by inhibiting RFC2, which may provide novel targets
for the clinical treatment of GBM, and improve future patient
prognosis.
Materials and methods
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia2.cancer-pku.cn/#index), including gene
expression profiling data from The Cancer Genome Atlas project, was
performed to screen differentially expressed genes (DEGs, criteria:
|log2FC| ≥2 and P<0.01) and survival-related genes
(criteria: P<0.01) associated with GBM. Then, the DEGs and
survival-associated genes were overlapped using Venny 2.1.0
(https://bioinfogp.cnb.csic.es/tools/venny/), and
STRING (https://string-db.org/) was used to
predict the protein-protein interactions between the overlapping
genes. Finally, the ENCORI starBase (http://starbase.sysu.edu.cn/index.php), TargetScan
Human 7.2 (http://www.targetscan.org/vert_71/) and miRDB.org
algorithms (http://mirdb.org/) were used to predict
the upstream miRNAs of key genes.
Tissue samples
GBM tissues (n=39) and normal adjacent tissues
(n=39) were collected from Union Hospital, Tongji Medical College,
Huazhong University of Science and Technology (Wuhan, China)
between April 2015 and October 2019. Inclusion criteria included:
i) Postoperative pathology diagnosed as GBM; and ii) Complete
patient information obtained. Exclusion criteria included: i)
Patients who underwent radiotherapy and chemotherapy before
surgery; ii) Patients with a history of another kind of malignant
cancer; and iii) Patients with serious heart, liver, spleen, lung
or kidney diseases. The study protocol was approved by the Ethics
Committee of Union Hospital on September 12, 2018 [approval. no.
(2018) (216)], and all patients provided written informed consent.
The clinical characteristics of the GBM subjects are presented in
Table I. The patient aged 45–70
years old, with a mean age of 52.6±9.3 years.
 | Table I.Association of RFC2 or miR-744-5p
expression with the epidemiologic features of 39 patients with
glioblastoma. |
Table I.
Association of RFC2 or miR-744-5p
expression with the epidemiologic features of 39 patients with
glioblastoma.
|
| Expression of
RFC2 | Expression of
miR-744-5p |
|---|
|
|
|
|
|---|
| Clinical features
(n) | Low | High | P-Value | Low | High | P-value |
|---|
| Age |
|
|
|
|
|
|
| <60
(23) | 10 | 13 | 0.4325 | 11 | 12 | 0.7491 |
| ≥60
(16) | 9 | 7 |
| 7 | 9 |
|
| Sex |
|
|
|
|
|
|
| Male
(20) | 9 | 11 | 0.6337 | 10 | 10 | 0.6211 |
| Female
(19) | 10 | 9 |
| 8 | 11 |
|
| Grade |
|
|
|
|
|
|
| +II
(15) | 12 | 3 | 0.0020 | 3 | 12 | 0.0096 |
| III+IV
(24) | 7 | 17 |
| 15 | 9 |
|
| Tumor size, cm |
|
|
|
|
|
|
| <5
(18) | 13 | 6 | 0.0164 | 7 | 12 | 0.0154 |
| ≥5
(21) | 6 | 14 |
| 14 | 6 |
|
| IDH status |
|
|
|
|
|
|
|
Wild-type (20) | 12 | 8 |
| 7 | 13 | 0.3373 |
| Mutated
(19) | 7 | 12 |
| 11 | 8 |
|
| Distant
metastasis |
|
|
|
|
|
|
|
Metastasis (21) | 5 | 16 | 0.0008 | 15 | 6 | 0.0006 |
| No
metastasis (18) | 14 | 4 |
| 3 | 15 |
|
|
|
| RFC2
expression | miR-744-5p
expression |
|
|
|
|
| Clinical feature
(n) | Low | High | P-Value | Low | High | P-value |
|
| Age, years |
|
|
|
|
|
|
| <60
(23) | 10 | 13 | 0.4325 | 11 | 12 | 0.7491 |
| ≥60
(16) | 9 | 7 |
| 7 | 9 |
|
| Sex |
|
|
|
|
|
|
| Male
(20) | 9 | 11 | 0.6337 | 10 | 10 | 0.6211 |
| Female
(19) | 10 | 9 |
| 8 | 11 |
|
| Grade |
|
|
|
|
|
|
| +II
(15) | 12 | 3 | 0.0020 | 3 | 12 | 0.0096 |
| III+IV
(24) | 7 | 17 |
| 15 | 9 |
|
| Tumor size, cm |
|
|
|
|
|
|
| <5
(18) | 13 | 6 | 0.0164 | 7 | 12 | 0.0154 |
| ≥5
(21) | 6 | 14 |
| 14 | 6 |
|
| IDH status |
|
|
|
|
|
|
|
Wild-type (20) | 12 | 8 | 0.1481 | 7 | 13 | 0.3373 |
| Mutated
(19) | 7 | 12 |
| 11 | 8 |
|
| Distant
metastasis |
|
|
|
|
|
|
|
Metastasis (21) | 5 | 16 | 0.0008 | 15 | 6 | 0.0006 |
| No
metastasis (18) | 14 | 4 |
| 3 | 15 |
|
Cell culture and transfection
The human GBM cell lines, U87 (cat. no. HTB-14; GBM
of unknown origin) and A172 (cat. no. CRL-1620) were purchased from
the American Type Culture Collection. U251 (cat. no. TCHu 58) and
SHG44 (cat. no. TCHu 48) cells were purchased from the cell bank of
the Chinese Academy of Sciences (Shanghai, China). Normal human
astrocyte (NHA) cells (cat. no. 1800) were purchased from ScienCell
Research Laboratories, Inc. U251, SHG44, A172 and NHA cells were
maintained in DMEM and U87 cells were maintained in MEM, both with
10% fetal bovine serum (FBS) (Invitrogen; Thermo Fisher Scientific,
Inc.), at 37°C (5% CO2 and 90% humidity). All cell lines
were verified by short tandem repeat authentication.
For transfection, the RFC2-overexpression (RFC2-OE)
vector, pcDNA 3.1 empty vector [used as the RFC2-OE negative
control (NC)], si-NC, si-RFC2, miR-744-5p inhibitor, miR-744-5p
mimics, mimic-NC and inhibitor-NC were all purchased from Shanghai
Tuoran Co., Ltd. A total of 3×105/well U251 and U87
cells were cultured in 24-well plates and transfected with either
si-NC, si-RFC2, RFC2-OE, miR-744-5p inhibitor, miR-744-5p mimic or
pcDNA 3.1, mimic-NC or inhibitor-NC, at a concentration of 50 nM
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and maintained at 37°C for 48 h before
subsequent experiments were performed. The sequences of RFC2-OE,
si-RFC2, miR-744-5p inhibitor, the miR-744-5p mimics and their
corresponding NC are presented in Table
SI.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA from the tissue specimens and cell lines
was isolated using TRIzol® reagent following the
manufacturer's instructions (Thermo Fisher Scientific, Inc.).
Subsequently, total RNA was reverse transcribed into cDNA using the
PrimeScript First Strand cDNA Synthesis kit (Takara Biotechnology
Co., Ltd.). The targeted reverse transcription of miRNA was
performed using the Hairpin-it miRNA qPCR Quantitation Kit
(Shanghai GenePharma Co., Ltd.). Then, SYBR Premix Ex Taq (Takara
Biotechnology Co., Ltd.) was used for qPCR, performed on an ABI
Prism 7900 Detector System (Thermo Fisher Scientific, Inc.). The
qPCR conditions were 1 cycle at 95°C for 20 sec, followed by 40
cycles of 1 min each at 95°C, and 20 sec at 60°C. Relative
expression levels were normalized to that of U6 and β-actin,
corresponding to miRNA and mRNA, respectively. The expression
levels were calculated using the 2−ΔΔCq method (23). The primer sequences are listed in
Table II.
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Primer | Sequence |
|---|
| RFC2 |
|
Forward |
5′-CCTGAGGTCCTTCTGGTGGT-3′ |
|
Reverse |
5′-CAACGTCAATGCCCCTGTCA-3′ |
| miR-744-5p |
|
Forward |
5′-TGCGGGGCTAGGGCTA-3′ |
|
Reverse |
5′-CGGCCCAGTGTTCAGACTAC-3′ |
| miR-2355-5p |
|
Forward |
5′-ATTGTCCTTGCTGTTTGGAGAT-3′ |
|
Reverse |
5′-GCGAGCACAGAATTAATACGAC-3′ |
| miR-122-5p |
|
Forward |
5′-TATTCGCACTGGATACGACACAAC-3′ |
|
Reverse |
5′-GCCCGTGGAGTGTGACAATGGT-3′ |
| GAPDH |
|
Forward |
5′-CCAGGTGGTCTCCTCTGA-3′ |
|
Reverse |
5′-GCTGTAGCCAAATCGTTGT-3′ |
| U6 |
|
Forward |
5′-CTCGCTTCGGCAGCACA-3′ |
|
Reverse |
5′-AACGCTTCACGAATTTGCGT-3′ |
Western blotting
Total proteins from the U251 and U87 cells were
extracted using radioimmunoprecipitation assay lysis buffer
(MilliporeSigma), and the total protein content was quantified
using a BCA detection kit (Thermo Fisher Scientific, Inc.). Then,
the proteins (30 µg per well) were separated by 10% SDS-PAGE and
subsequently transferred onto PVDF membranes (EMD Millipore). The
membranes were blocked with 5% skim milk for 1 h at 25°C, and then
incubated with the following primary antibodies at 4°C for 12 h:
Anti-RFC2 (cat. no. ab174271; 1:10,000; Abcam), goat anti-rabbit
Bax (cat. no. ab32503; 1:2,000; Abcam), goat anti-rabbit Bcl-2
(cat. no. ab182858; 1:2,000; Abcam) and anti-β-actin (cat. no.
ab8226; 1:10,000; Abcam). After washing three times with TBS
containing 0.05% Tween 20 (TBST), the membranes were incubated then
with the following secondary antibodies: Goat anti-rabbit for RFC2
(cat. no. ab205718; 1:2,000; Abcam) and goat anti-mouse for β-actin
(cat. no. ab175783; 1:2,000; Abcam) for 1 h at 25°C. Finally,
positive bands were visualized with the Immobilon™ Western
Chemiluminescent horseradish peroxidase (HRP) substrate (EMD
MilliporeSigma). The densities of the bands was determined using
ImageJ software version 1.53 (National Institutes of Health).
Cell Counting Kit 8 (CCK-8) assay
The CCK-8 (Sangon Biotech Co., Ltd.) was used to
determine the viability of U251 and U87 cells. Transfected cells
(2×103/well) in 96-well plates were incubated for 0, 24,
48 and 72 h. Then, 10 µl CCK-8 reagent was added to each well and
the cells were incubated for a further 2 h. The absorbance was
measured at an optical density of 450 nm using a multimode
microplate reader (Tecan Group, Ltd.).
5-bromo-2-deoxyuridine (BrdU)
assay
Cellular proliferation was evaluated using the BrdU
Cell Proliferation Assay Kit (Cell Signaling Technology, Inc.) per
the manufacturer's instructions. U251 and U87 cells were seeded
into 96-well plates (2×104 cells per well) and cultured
in FBS-free medium for 24 h to synchronize the cell cycle. Then,
the medium was removed and the cells were labeled with 1X BrdU
solution (prepared in cell culture medium) for 6 h at 37°C to
induce proliferation. The labeling medium was removed, and the
cells were fixed and denatured using the supplied
fixation/denaturation solution. BrdU mouse mAb was then added and
the cells were incubated for 2 h at 37°C. Anti-mouse immunoglobulin
G and HRP-linked antibody were used to detect the binding antibody,
and HRP substrate (3,3,5,5-tetramethylbenzidine) was added for
color development. Finally, the absorbance at 450 nm was determined
using a multimode microplate reader (Tecan Group, Ltd.).
Wound-healing assay
The migratory ability of U251 and U87 cells was
evaluated by wound-healing assay. Cells were seeded into 6-well
plates (2×105/well), cultured to 80% density and
subsequently, 200-µl pipette tips were used to create wounds in the
center of each well. After incubation with FBS-free medium for 24 h
at 37°C, the wound was photographed using an optical microscope
(Olympus Corporation) at ×100 magnification. The wound closure was
measured using the following formula: (W0 h-W24 h)/W0 h ×100%,
where W is the width.
Cellular adhesion Assay
A 96-well plate was coated with collagen I solution
(40 µg/ml; Sigma-Aldrich; Merck KGaA) and stored at 4°C for 12 h.
U251 and U87 cells (2×105 cells/ml) were cultured in
FBS-free DMEM for 8 h before harvesting by trypsinization. Then,
the cells were collected and suspended in 100 µl DMEM with 0.1% BSA
in the coated 96-well plate, and incubated at 37°C for 20 min.
Subsequently, the cells were incubated with medium containing 10%
FBS for 4 h at 37°C. Then, 10 µl MTT substrate (MilliporeSigma) was
added to each well and the plate was incubated for a further 2 h at
30°C. The cells were then washed twice with PBS and 100 µl DMSO was
added to each well. Absorbance was measured at a wavelength of 570
nm using a multimode microplate reader (Tecan Group, Ltd.).
Caspase activity assay
U251 and U87 cells were seeded into 96-well plates
(3×105 cells per well). Caspase-3 assay loading solution
(Cell Signaling Technology, Inc.) was prepared by adding the
caspase-3 substrate to DL-dithiothreitol reagent according to the
manufacturer's instructions. Cells were collected at 80% density
and lysed with ice-cold cell lysis buffer for 10 min. Subsequently,
loading solution (100 µl/well) was added to each sample and
incubated at 37°C for 2 h. The absorbance was then measured at a
wavelength of 405 nm using a multimode microplate reader (Tecan
Group, Ltd.).
Dual-luciferase reporter assay
The psiCHECK2-RFC2-3′ untranslated region (3′-UTR)
wild-type vectors and psiCHECK2 RFC2-3′-UTR mutated vectors were
purchased from Weizhen Bio (Shanghai, China). U251 and U87 cells
were co-transfected with the vectors and negative control (NC) or
miR-744-5p mimics using Lipofectamine® 3000, and
cultured in a 24-well plate. After 48 h, the dual-luciferase
reporter assay System (Promega Corporation) was used to detect the
firefly and Renilla luciferase activities. Relative
luciferase activity was normalized to the activity of
Renilla luciferase (internal control).
RNA pull-down analysis
The biotin-labeled miR-2355-5p (Bio-miR-2355-5p,
5′-AUCCCCAGAUACAAUGGACAA-biotin-3′), miR-122-5p (Bio-miR-122-5p,
5′-AACGCCAUUAUCACACUAAAUA-biotin-3′) and miR-744-5p
(Bio-miR-744-5p, 5′-UGCGGGGCUAGGGCUAACAGCA-biotin-3′), as well as
negative controls (Bio-NC, 5′-GUGCACGAAGGCUCAUCAUU-biotin-3′) were
purchased from Guangzhou RiboBio Co., Ltd, and were used to
transfect both U251 and U87 cells for 48 h at 37°C using
Lipofectamine® 3000. Next, the cells were collected and
lysed using 0.5 ml lysis buffer, containing 25 mM Tris-HCl (pH
7.5), 70 mM KCl, 2.5 mM EDTA, 0.05% NP-40, protease inhibitor
cocktail (Sigma-Aldrich; Merck KGaA) and RNase inhibitors (Thermo
Fisher Scientific, Inc.), for 20 min on ice. The mixture was
centrifuged at 1,400 × g at 4°C for 20 min, and 0.5 ml supernatant
lysate was collected. The streptavidin beads (50 µl; cat. no.
88816; Thermo Fisher Scientific, Inc.) were washed and added to the
supernatant lysates, and then incubated at 4°C on a rotating
platform for 12 h. The following day, the beads were washed twice
with cold lysis buffer, three times with low salt buffer solution,
and once with high salt buffer solution; the specifically-bound
RNAs were purified using the RNeasy Mini Kit (QIAGEN). Finally, the
enrichment of RFC2 was detected by performing RT-qPCR.
Statistical analysis
All analyses were performed using GraphPad Prism 8.0
software (GraphPad Software, Inc.). Paired Student's t-test
(two-tailed) was used to compare the differences between two
groups, while the differences between three or more groups were
determined by one-way ANOVA with Dunnett's (comparisons with one
group) or Tukey's (comparisons with more than one group) post hoc
test. The correlation between miR-744-5p and RFC2 expression levels
was determined by Pearson's correlation analysis. Data are
presented as the mean ± standard deviation, and P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of study objects of
interest
DEGs and survival-related genes for GBM were
acquired from the GEPIA database, which is a gene expression
profiling interactome analysis tool that includes gene expression
profiling data from The Cancer Genome Atlas project. GEPIA yielded
1,384 DEGs with the criteria of |log2FC| ≥2 and
P<0.01, and 547 GBM survival-related genes with the criterial of
P<0.01. Among these genes, 47 were both DEGs and GBM
survival-related (Fig. 1A). By
performing STRING analysis of the 47 genes, 22 were found to be
closely related (Fig. 1B). The
STRING network indicated that RFC2 had been studied in glioma once,
and it was reported to be related to drug cytotoxicity;
furthermore, the knockdown of RFC2 led to reduced cell viability
(21). Nonetheless, how RFC2 affects
other GBM cell phenotypes has not been extensively studied.
Therefore, RFC2 was chosen as a gene of interest. The ENCORI
starBase, TargetScan Human 7.2 and miRDB.org algorithms were used
to predict the upstream miRNAs of RFC2, and three overlapping
miRNAs (miR-2355-5p, miR-122-5p and miR-744-5p) were identified
(Fig. 1C). After constructing
bio-miR-2355-5p, bio-miR-122-5p and bio-744-5p, these plasmids
showed high transfection efficiency in U251 and U87 (Fig. S1). After RNA pull-down analysis, it
was observed that the association between RFC2 and miR-744-5p was
stronger than that with miR-2355-5p or miR-122-5p (Fig. 1D). Therefore, the involvement of RFC2
mRNA and miR-744-5p miRNA in GBM was investigated further.
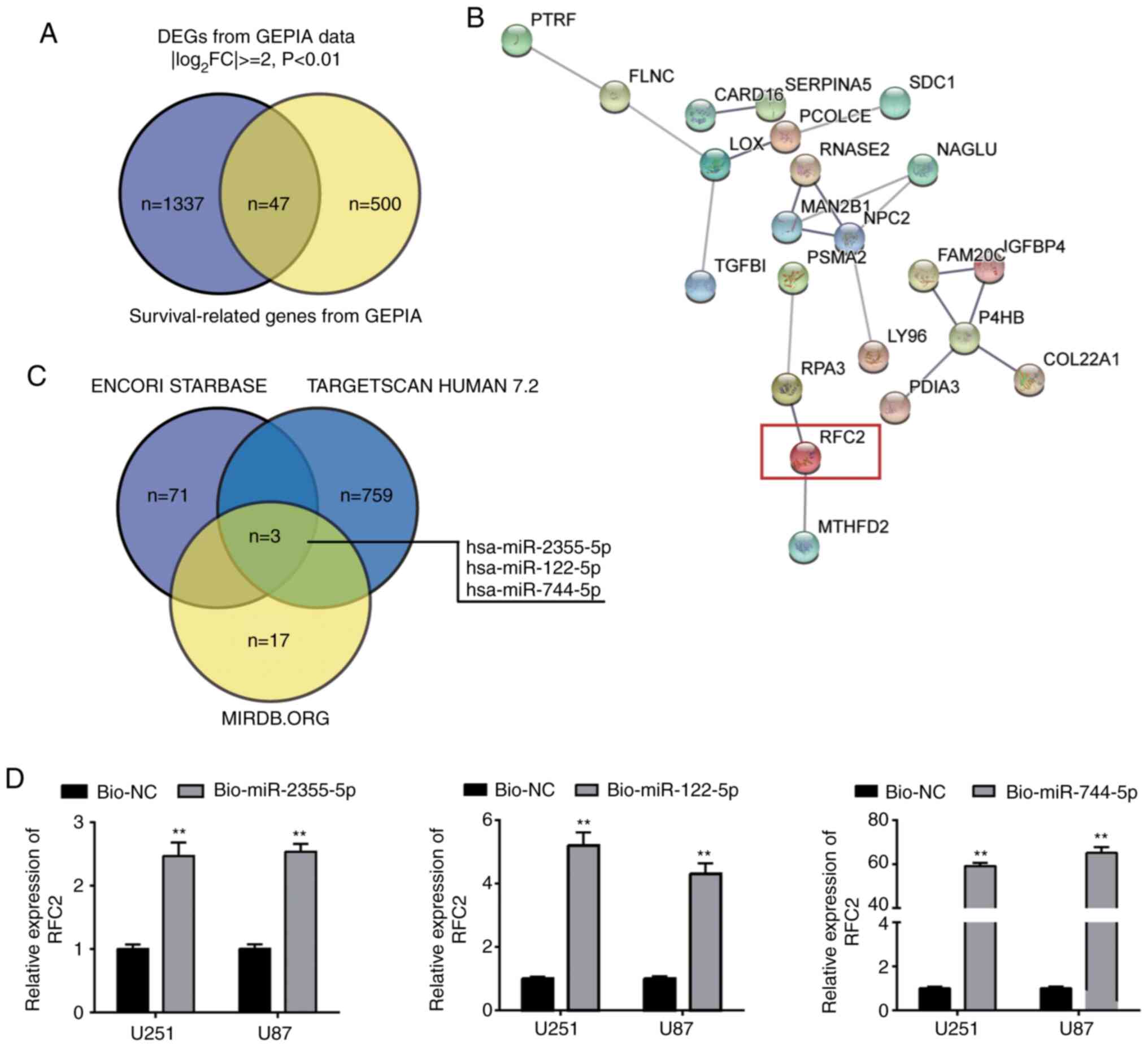 | Figure 1.Identification of study objects of
interest. (A) Venn diagram demonstrating the intersection of DEGs
and survival-related genes in GBM from GEPIA database. (B) STRING
results demonstrating the interaction network of the 47 intersected
genes from (A). (C) Intersection of upstream miRNAs of RFC2 from
the ENCORI starBase, TargetScan Human 7.2 and miRDB.org algorithms;
log2FC: log2fold change. (D) RFC2 was mainly
pulled down in the GBM cells transfected with bio-miR-744-5p.
Bio-NC, Biotin-labelled NC; Bio-miR-2355-5p, Biotin-labeled
miR-2355-5p; Bio-miR-122-5p, Biotin-labeled miR-122-5p;
Bio-miR-744-5p, Biotin-labeled miR-744-5p. **P<0.001, compared
with Bio-NC using one-way ANOVA with Tukey's test. DEG,
differentially expressed gene; GBM, glioblastoma; GEPIA, Gene
Expression Profiling Interactive Analysis; RFC2, replication factor
C subunit 2; miR, microRNA; NC, negative control. |
RFC2 expression is upregulated in GBM
tissues and cell lines
To elucidate whether RFC2 is involved in GBM, RFC2
expression was first detected in tumor tissues and corresponding
normal tissues from patients with GBM. The results showed that RFC2
expression increased 1.5-fold compared to normal control tissues
(Fig. 2A). RFC2 gene expression was
also detected in NHAs and GBM cells (U251, U87, SHG44 and A172),
and was significantly increased in the GBM cell lines compared to
NHA cells. In particular, U251 and U87 cells showed more than a
3-fold increase in RFC2 expression compared with NHA cells.
Therefore, U251 and U87 cells were chosen for subsequent
experiments due to having the highest expression of RFC2 (Fig. 2B). To further explore the effects of
RFC2 in GBM, U251 and U87 cells were transfected with si-NC, OE-NC
(pcDNA 3.1 empty vector), Co-NC (si-NC+OE-NC), RFC2-overexpression
(OE),or si-RFC2 constructs. Cells transfected with RFC2-OE
displayed ~4-fold upregulated RFC2 levels compared with the control
cells, while cells transfected with si-RFC2 exhibited an ~70%
decrease in RFC2 levels compared with the control cells (Fig. 2C). Furthermore, the protein
expression levels of RFC2 increased more than 1.4-fold in cells
transfected with RFC2-OE, with an ~60% decrease in protein levels
observed in si-RFC2 cells compared with the control cells (Fig. 2D). As the Co-NC exerted no obvious
effect on RFC2 expression, Co-NC was used as the NC group in
subsequent experiments.
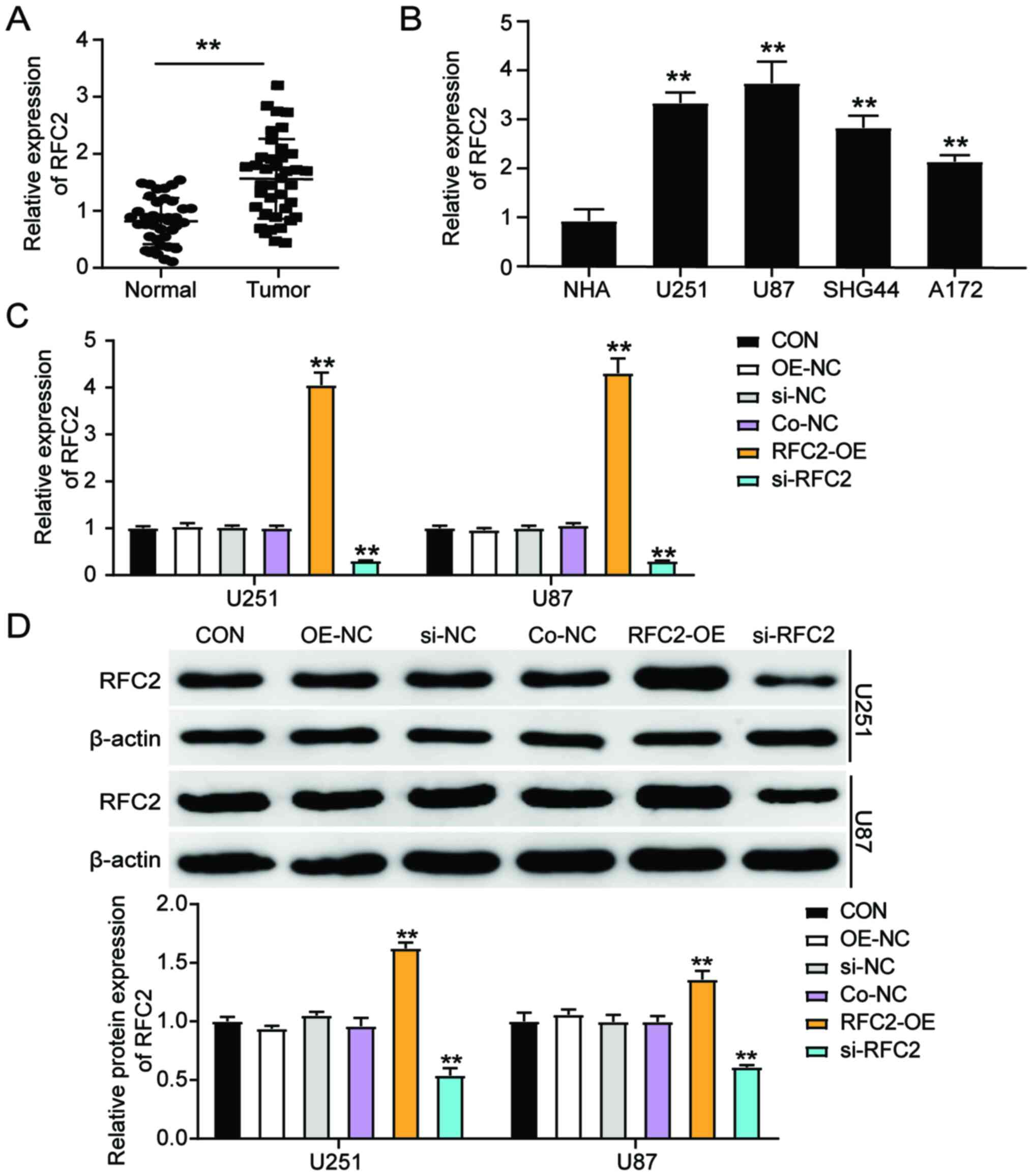 | Figure 2.RFC2 expression is upregulated in GBM
tissues and cells. (A) RT-qPCR analysis of gene expression of RFC2
in GBM tumor tissues (n=39) and adjacent controls (n=39) from
patients with GBM. **P<0.001, compared with Normal group using
paired Student's t-test. (B) RT-qPCR analysis of the RFC2
expression in GBM cell lines (U251, U87, SHG44 and A172) and the
normal astrocyte NHA cell line. **P<0.001, compared with NHA
using one-way ANOVA with Dunnett's test. (C) RT-qPCR analysis of
gene expression of RFC2 in U251 and U87 cells transfected with NC,
RFC2-OE and si-RFC2. (D) Western blot analysis of RFC2 protein
expression in U251 and U87 cells transfected with NC, RFC2-OE and
si-RFC2. (C and D) **P<0.001, compared with CON using one-way
ANOVA with Dunnett's test. CON, blank control; si-NC, si-RFC2
negative control; OE-NC, pcDNA 3.1 empty vector; Co-NC,
si-NC+OE-NC; si-RFC2, siRNA-RFC2; RFC2-OE, RFC2-overexpression;
RFC2, replication factor C subunit 2; GBM, glioblastoma; RT-q,
reverse transcription-quantitative; si(RNA), small interfering; OE,
overexpression; NC, negative control. |
RFC2 promotes cellular proliferation,
migration and adhesion, and suppresses apoptosis in GBM
To determine whether RFC2 promotes GBM
tumorigenesis, a CCK-8 assay was performed using transfected U251
and U87 cells. Cells transfected with RFC2-OE exhibited higher
proliferative capacity, while cells transfected with si-RFC2
proliferated to a lower degree than the control cells (Fig. 3A). Moreover, according to the results
of the BrdU assay, cells transfected with RFC2-OE exhibited an ~30%
enhancement in proliferation, while cells transfected with si-RFC2
indicated reduced proliferation (~50%) compared with the control
cells (Fig. 3B). A wound-healing
assay was performed to assess the migratory capacity of the
transfected cells. The results showed a 30% increase in the
migratory ability of cells transfected with RFC2-OE, and a 50%
decrease in the migratory ability of cells transfected with si-RFC2
compared with the control cells (Fig.
3C). Furthermore, adhesion ability was elevated by ~30% in
cells transfected with RFC2-OE, while cells transfected with
si-RFC2 showed ~30% reduced cell adhesion ability compared with the
control cells (Fig. 3D).
Additionally, the levels of caspase-3 activity were elevated 6-fold
in cells transfected with si-RFC2, while cells transfected with
RFC2-OE indicated reduced levels of caspase-3 activity by 70%
compared to control cells (Fig. 3E).
Furthermore, western blot analysis showed that compared with the
control group, Bax protein expression was decreased and Bcl-2
expression was increased in U251 and U87 cells overexpressing RFC2,
while the trend was opposite in cells with low expression of RFC2
(Fig. 3F). Therefore, these results
demonstrated that RFC2 promoted cell proliferation, migration, and
adhesion, and suppressed cell apoptosis in GBM.
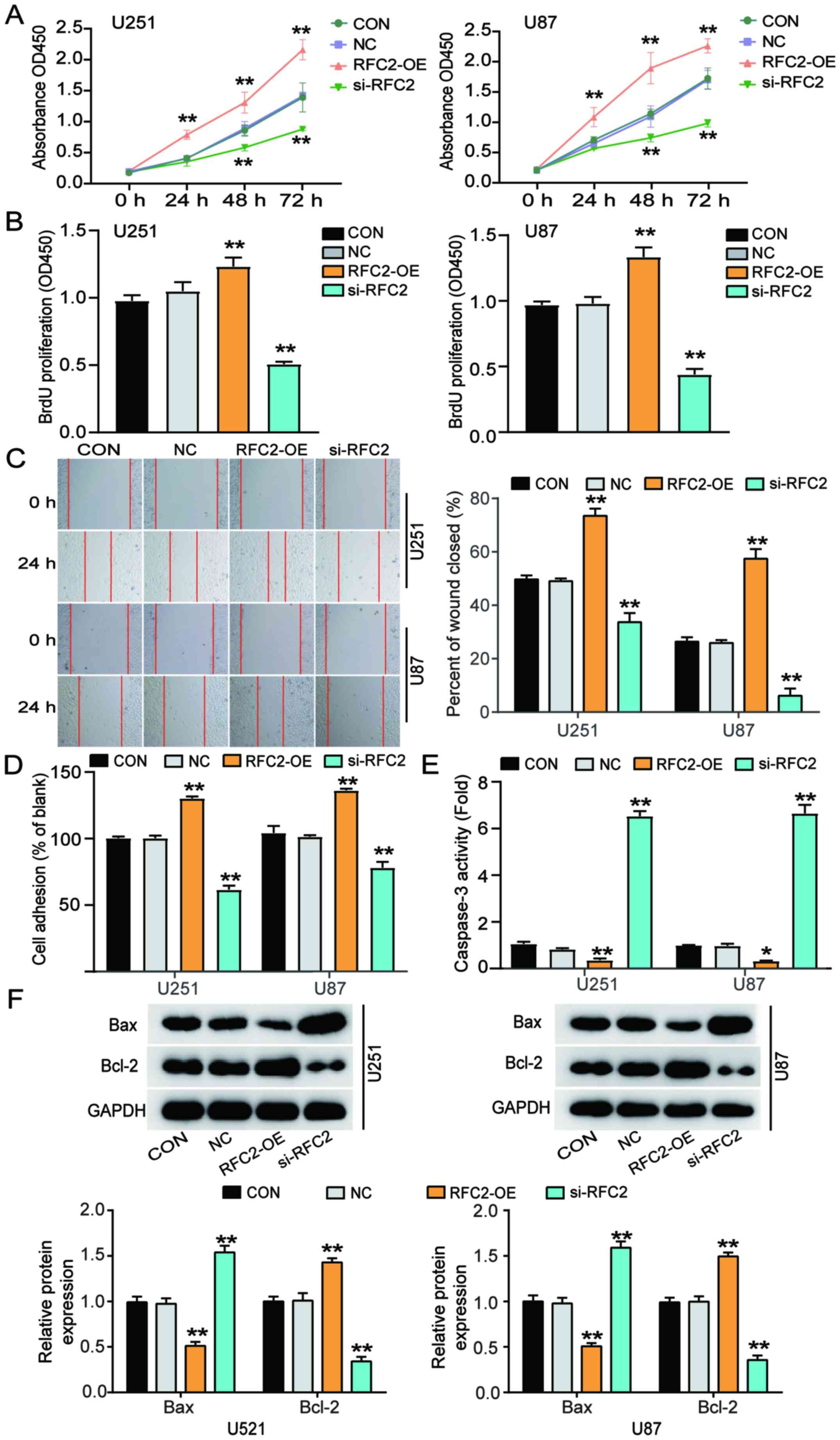 | Figure 3.RFC2 promotes cellular proliferation,
migration and adhesion, and suppresses cell apoptosis in
glioblastoma. (A) Viability of U251 and U87 cells transfected with
NC, RFC2-OE and Si-RFC2 was determined by Cell Counting Kit 8
assay. (B) Cellular proliferation was detected in U251 and U87
cells transfected with NC, RFC2-OE and si-RFC2 by BrdU assay. (C)
Wound-healing assay was performed in U251 and U87 cells transfected
with NC, RFC2-OE and si-RFC2. (D) Adhesion ability was detected in
U251 and U87 cells transfected with NC, RFC2-OE and si-RFC2. (E)
Caspase3 activity was determined in U251 and U87 cells transfected
with NC, RFC2-OE and si-RFC2 by caspase3 activity assay kit. (F)
Protein expression levels of Bax and Bcl-2 were determined in U251
and U87 cells transfected with NC, RFC2-OE and si-RFC2 by western
blotting. *P<0.05 and **P<0.001, compared with CON using
one-way ANOVA with Dunnett's test. CON, blank control; NC, negative
control; si-RFC2, siRNA-RFC2; RFC2-OE, RFC2-overexpression; RFC2,
replication factor C subunit 2. |
RFC2 is a target of miR-744-5p in
GBM
The TargetScan Human 7.2 database was used to
identify potential miRNAs targeting RFC2. The analysis identified
miR-744-5p as a potential miRNA that could interact with the 3′UTR
of RFC2 mRNA; the binding sequences are displayed in Fig. 4A. Subsequently, U251 and U87 cells
were successfully transfected with miR-744-5p mimics and inhibitor
(Fig. S2). The results showed that
U251 and U87 cells co-transfected with wild-type RFC2 3′-UTR
plasmid and miR-744-5p mimic showed ~50% reduced luciferase
activity compared with cells co-transfected with wild-type RFC2
3′-UTR plasmid and miR-NC. However, no change was observed in cells
co-transfected with the mutant RFC2 3′-UTR plasmid (Fig. 4B). This suggested that miR-744-5p
directly bound to the 3′-UTR of RFC2. RT-qPCR analysis demonstrated
that miR-744-5p expression in GBM tissues was ~50% lower than that
in normal-adjacent tissues (Fig.
4C). A significant negative correlation was found between
miR-744-5p and RFC2 expression in GBM tissues (Fig. 4D). Additionally, the results revealed
that miR-744-5p levels were downregulated by ~50% in U87 cells and
25% in U251 cells compared to NHAs (Fig.
4E). Overall, RFC2 acts as a negative downstream target of
miR-744-5p in GBM. U251 and U87 cells were transfected with RFC2-OE
and miR-744-5p inhibitor, or si-RFC2 together with miR-744-5p
inhibitor. RT-qPCR analysis confirmed that cells transfected with
RFC2-OE and miR-744-5p inhibitor showed more than 2.5-fold higher
expression of RFC2, and 50% lower expression of RFC2 in cells
transfected with si-RFC2 than in the control cells. Cells
transfected with si-RFC2 and miR-744-5p inhibitor showed the same
level as control cells (Fig. 4F).
RFC2 protein expression was markedly increased in cells transfected
with miR-744-5p inhibitor, and significantly reduced by in si-RFC2
cells, compared with the controls. Cells transfected with si-RFC2
and miR-744-5p inhibitor showed the same levels of RFC2 expression
as the control cells (Fig. 4G).
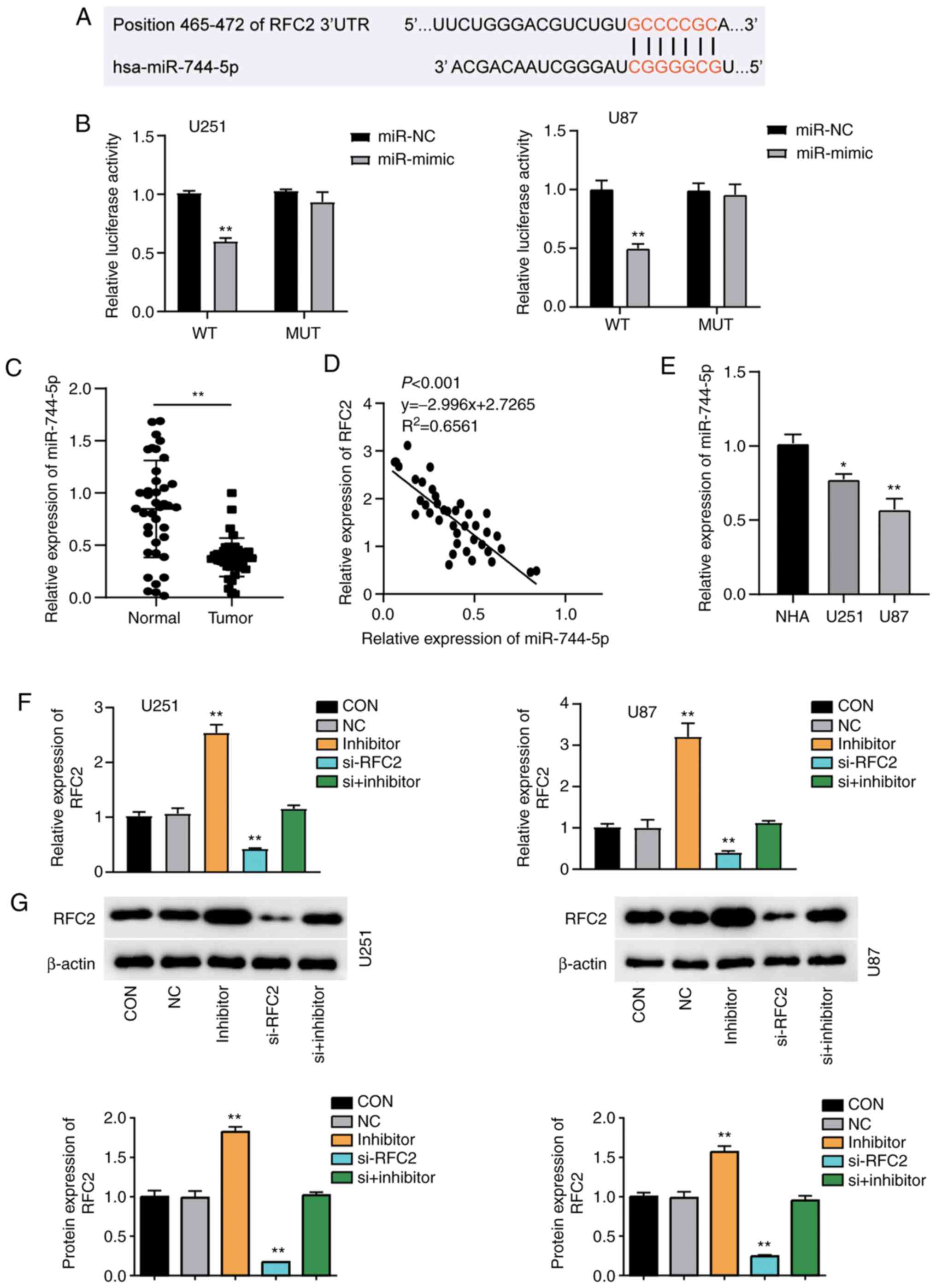 | Figure 4.RFC2 is a direct target of miR-744-5p
in GBM. (A) Bioinformatics analysis showed the predicted binding
sequence of RFC2 3′-UTR. (B) Dual luciferase assay was performed in
cells co-transfected with WT RFC2 plasmid or MUT RFC2 plasmid and
miR-NC or miR-744-5p mimic in U251 and U87 cells. **P<0.001,
one-way ANOVA with Tukey's test. (C) Expression of miR-744-5p in
GBM tumor tissues (n=39) and adjacent controls (n=39) from patients
with GBM was analyzed by RT-qPCR. **P<0.001, compared with
Normal group using paired Student's t-test. (D) Correlation between
RFC2 and miR-744-5p expression in GBM tissues. (E) RT-qPCR
detection of RFC2 expression in NHA, U251 and U87 cells. *P<0.05
and **P<0.001 compared with NHA using one-way ANOVA with
Dunnett's test. (F) RT-qPCR analysis of the mRNA expression of RFC2
in U251 and U87 cells transfected with NC, miR-744-5p inhibitor,
si-RFC2, and Si-RFC2+ miR-744-5p inhibitor. (G) Western blot
analysis of RFC2 protein expression in U251 and U87 cells
transfected with NC, miR-744-5p inhibitor, si-RFC2, and si-RFC2+
miR-744-5p inhibitor. (F-G) CON, blank control; NC, negative
control. **P<0.001 compared with CON using one-way ANOVA with
Dunnett's test. RFC2, replication factor C subunit 2; GBM,
glioblastoma; RT-q, reverse transcription-quantitative; si(RNA),
small interfering; OE, overexpression; NC, negative control; WT,
wild-type; MUT, mutant; miR, microRNA. |
miR-744-5p targets RFC2 to suppress
GBM progression
To assess the important role of miR-744-5p and RFC2
in GBM, NC and si-RFC2, miR-744-5p inhibitor, and Si-RFC2 together
with miR-744-5p inhibitor were transfected into U251 and U87 cells.
CCK-8 assay analysis showed that after 48 h, cells transfected with
miR-744-5p inhibitor exhibited significantly enhanced proliferation
capacity, whereas cells transfected with si-RFC2 showed
significantly reduced capacity, compared with the control cells.
Cells transfected with si-RFC2 and miR-744-5p inhibitor showed the
same level as control cells (Fig.
5A). Furthermore, proliferation was enhanced by ~50% in cells
transfected with miR-744-5p inhibitor but decreased by ~30% in
si-RFC2 cells compared to control cells. Cells transfected with
si-RFC2 and miR-744-5p inhibitor showed the same level as control
cells (Fig. 5B). In addition, cells
transfected with miR-744-5p inhibitor showed ~30% enhanced
migratory ability, while cells transfected with si-RFC2 showed ~50%
decrease in migratory ability compared to control cells as observed
by wound healing assay. Cells transfected with si-RFC2 and
miR-744-5p inhibitor showed the same level as control cells
(Fig. 5C). Moreover, cells
transfected with si-RFC2 exhibited a 40% decrease in cell adhesion,
while cells transfected with miR-744-5p inhibitor showed a 40%
increase in cell adhesion levels compared to control cells. Cells
transfected with si-RFC2 and miR-744-5p inhibitor showed the same
level as control cells (Fig. 5D).
Additionally, cells transfected with si-RFC2 showed >4-fold
caspase-3 activity, while cells transfected with miR-744-5p
inhibitor showed ~50% reduced caspase-3 activity compared to
control cells. Cells transfected with si-RFC2 and miR-744-5p
inhibitor showed the same level as control cells (Fig. 5E). Furthermore, western blotting
showed that in U251 and U87 cells, with low expression of
miR-744-5p, the expression levels of Bax protein, were lower than
those in the control group, while the expression levels of Bcl-2
protein was higher than that in the control group (Fig. 5F). Moreover, the protein expression
levels of Bax and Bcl-2 in RFC2 and miR-744-5p knockdown cells were
the same as that in the control group (Fig. 5F). Collectively, these results
revealed that miR-744-5p suppressed cellular proliferation,
migration and adhesion, and promoted apoptosis in GBM by inhibiting
RFC2.
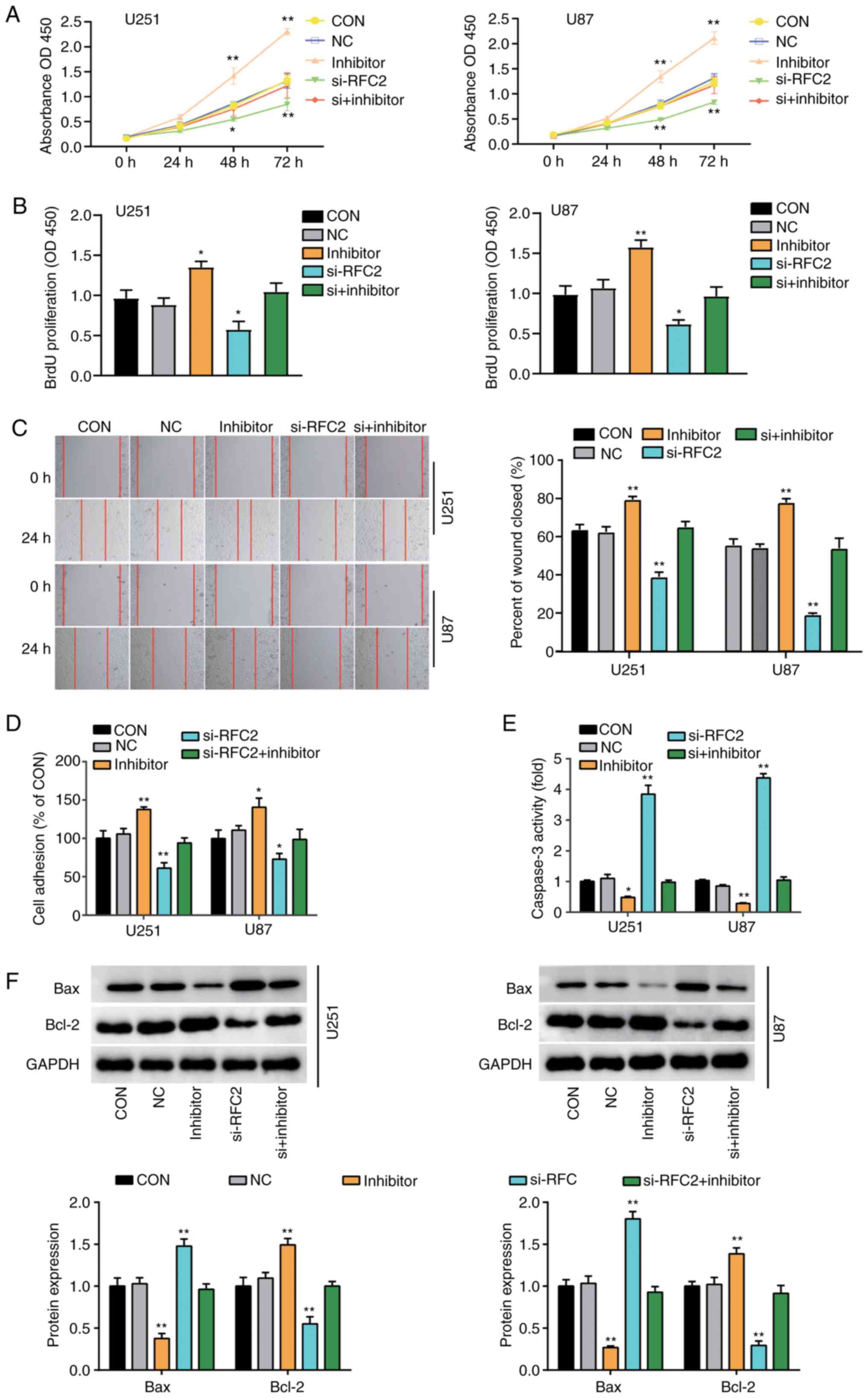 | Figure 5.miR-744-5p targeting to RFC2
suppresses glioblastoma progression. (A) Viability of U251 and U87
cells transfected with NC, miR-744-5p inhibitor, Si-RFC2, and
Si-RFC2+ miR-744-5p inhibitor was determined by Cell Counting Kit 8
analysis. (B) Proliferation was detected in U251 and U87 cells
transfected with NC, miR-744-5p inhibitor, Si-RFC2, and Si-RFC2+
miR-744-5p inhibitor by BrdU assay. (C) Wound-healing assay was
performed in U251 and U87 cells transfected with NC, miR-744-5p
inhibitor, Si-RFC2, and Si-RFC2+ miR-744-5p inhibitor. (D) Adhesion
ability was detected in U251 and U87 cells transfected with NC,
miR-744-5p inhibitor, Si-RFC2, and Si-RFC2+ miR-744-5p inhibitor.
(E) Caspase-3 activity was determined in U251 and U87 cells
transfected with NC, miR-744-5p inhibitor, Si-RFC2, and Si-RFC2+
miR-744-5p inhibitor by caspase-3 activity assay kit. (F) Protein
expression levels of Bax and Bcl-2 were determined in U251 and U87
cells transfected with NC, miR-744-5p inhibitor, Si-RFC2, and
Si-RFC2+ miR-744-5p inhibitor by western blot assay. *P<0.05 and
**P<0.001, compared with CON using one-way ANOVA with Dunnett's
test. CON, blank control; NC, negative control; Si-RFC2,
SiRNA-RFC2; Si-RFC2+ miR-744-5p inhibitor, SiRNA-RFC2+ miR-744-5p
inhibitor; RFC2, replication factor C subunit 2. |
Discussion
GBM is the most lethal form of primary glioma
(24). The standard therapy for GBM
consists of surgery followed by radiation therapy with adjuvant
chemotherapy (25,26). The highly infiltrative, heterogeneous
and mutable nature of GBM frequently contributes to tumor
recurrence and treatment failure (27). Therefore, despite advances in
multimodal therapies, the 5-year survival rate of patients with GBM
is only 5% after diagnosis (28).
Hence, it is necessary to investigate the therapeutic methods for
GBM at the molecular level. In the present study, the expression of
RFC2 was found to be significantly upregulated in GBM tumor tissues
and cell lines, while the expression of miR-744-5p was
downregulated. In addition, miR-744-5p targeted RFC2 and
functionally repressed its expression, thereby suppressing the
progression of GBM cells.
Recently, an increasing number of reports has
emphasized the molecular mechanism and significance of miR-744-5p
expression in different human tumors. Studies have shown that
abnormal expression of miRNAs may be one of the primary reasons for
the occurrence of GBM, and therefore, miRNAs can be used as
predictive biomarkers for the diagnosis of GBM (29,30). The
expression of miRNA-744-5p in GBM has also been investigated in
recent years. For instance, miRNA-744-5p inhibited tumorigenesis by
inhibiting NIN1/RPN12 binding protein 1 homolog (NOB1) in GBM
(17). The study demonstrated that
miRNA-744-5p expression was significantly reduced in GBM tissues
and cell lines, and that it inhibited the proliferation, colony
formation, migration and invasiveness, and promoted apoptosis in
GBM cells by targeting NOB1. Another study revealed that the
expression of miR-744-5p was markedly decreased in GBM specimens
and primary GBM cell lines. miR-744-5p inhibited GBM migration by
suppressing transforming growth factor β1 and Dishevelled 2, which
further functionally downregulated MAP2K4 signaling (17). Consistent with their results, the
present study indicated that miR-744-5p was significantly decreased
in GBM specimens and cell lines. Downregulation of miR-744-5p
enhanced proliferation and repressed apoptosis in GBM cells, as
observed by CCK-8, wound-healing, and caspase-3 activity
assays.
RFC2, a member of the RFC family, acts as a primer
recognition factor for DNA polymerase, and regulates cellular
proliferation, migration, invasiveness and metastasis in various
cancers (18,22). Previous studies have reported the
dysregulation of RFC2 in various human cancer types (19,31). For
instance, high expression of RFC2 has been considered as a
molecular marker of nasopharyngeal carcinoma (19). Ho et al (21) observed that miR-4749-5p inhibited
RFC2 expression, which further enhanced TMZ cytotoxicity in GBM.
They further clarified that a higher level of RFC2 was observed in
GBM patients with poor survival. Upregulated miR-4749-5p targeting
RFC2 decreased cell viability, increased apoptosis and enhanced TMZ
cytotoxicity in GBM cells. In the present study, abnormally
increased expression of RFC2 was observed in both GBM tissues and
cells. Upregulation of RFC2 enhanced cellular proliferation,
migration and adhesion, and suppressed apoptosis in GBM.
Furthermore, it was also identified that the impact of RFC2 on GBM
cell proliferation, migration, adhesion and apoptosis was regulated
by miR-744-5p, which directly targets RFC2.
During the advanced stages of cancer, synaptosomal,
associated protein of 25 kDa (SNAP-25), a membrane-binding protein
in neurons, is thought to promote tumor development through
autophagy, and to play a role in the process of tumor pain
(32,33). Furthermore, studies have shown that
the expression of SNAP-25 alters the characteristics of synaptic
transmission, leading to pathological changes in neuronal circuits
in psychiatric diseases (34).
Therefore, the effect of miR-744-5p/RFC2 on SNAP-25 is a focus of
future research. In addition, the detection of apoptosis-related
factors in cancer cells is important when exploring the genesis and
development of GBM. For example, E3 ubiquitin-protein ligase XIAP,
belonging to the inhibitor of apoptosis protein (IAP) family, is
highly expressed in various tumor types, such as malignant glioma
(35). p53 mutation may not only
lead to the development of GBM tumors (as an early event), but also
lead to its malignant progression (as a late event) (36). It has been speculated that XIAP or
p53 may be highly expressed in GBM and regulated by the
miR-744-5p/RFC2 axis, and this hypothesis will be verified in
future studies. Various types of cells exist in the
microenvironment of primary, invasive and metastatic tumors
(37). The expression levels of
miR-744-5p and RFC2 in GBM tissues do not fully reflect the true
status of cancer cells. Therefore, the effect of the
miR-744-5p/RFC2 axis in GBM requires clinical investigation in
greater detail. TMZ resistance is a common cause of treatment
failure in GBM (38). A previous
study revealed that miR-4749-5p inhibited RFC2 expression, which
could further enhance TMZ cytotoxicity in GBM (21). Therefore, we hypothesize that
miR-744-5p enhances the chemosensitivity of TMZ in GBM by targeting
RFC2, which is also a focus of our future research. Besides,
Weighted Gene Co-expression Network Analysis (WGCNA) is a tool
often used to identify the key biological processes and hub genes
in GBM by constructing gene co-expression networks (39–41),
which is different from GEPIA used in the present study. Due to the
lack of WGCNA technology, we will learn WGCNA to help us identify
the biological processes and genomic networks to develop effective
treatment strategies for GBM by targeting the miR-744-5p/RFC2 axis.
Moreover, this study only observed these tumor characteristics at
the cellular level, detected cell apoptosis by western blot assay
and detected cell adhesion by spectrophotometry; however, these
findings need to be further determined by establishing animal
models, performing flow cytometry and using atomic force
microscope.
In conclusion, the results of the present study
demonstrated that miR-744-5p targeted RFC2 and suppressed the
progression of GBM through repressing cell proliferation and
migration, and effectively promoting cell apoptosis. Consequently,
our study demonstrated that the miR-744-5p/RFC2 interaction could
be a potential candidate for the treatment of GBM.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fundamental
Research Funds for the Central Universities (grant. no.
2019kfyXKJC071).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FF and DY performed the experiments and data
analysis. PY conceived and designed the study. XJ and JH acquired
the data, and are responsible for the authenticity of the raw data.
FF and DY conducted the analysis and interpretation of data. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Union Hospital, Tongji Medical College, Huazhong
University of Science and Technology. The processing of clinical
tissue samples is in strict compliance with the ethical standards
of the Declaration of Helsinki. All patients signed written
informed consent.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. New Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Gittleman H, Truitt G, Boscia
A, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical report:
Primary brain and other central nervous system tumors diagnosed in
the United States in 2011–2015. Neuro Oncol. 20 (Suppl 4):iv1–iv86.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patel NP, Lyon KA and Huang JH: The effect
of race on the prognosis of the glioblastoma patient: A brief
review. Neurol Res. 41:967–971. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Taphoorn MJ, Sizoo EM and Bottomley A:
Review on quality of life issues in patients with primary brain
tumors. Oncologist. 15:618–626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li XT, Wang HZ, Wu ZW, Yang TQ, Zhao ZH,
Chen GL, Xie XS, Li B, Wei YX, Huang YL, et al: miR-494-3p
regulates cellular proliferation, invasion, migration, and
apoptosis by PTEN/AKT signaling in human glioblastoma cells. Cell
Mol Neurobiol. 35:679–687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chae DK, Park J, Cho M, Ban E, Jang M, Yoo
YS, Kim EE, Baik JH and Song EJ: MiR-195 and miR-497 suppress
tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-β
receptor I ubiquitination. Mol Oncol. 13:2663–2678. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen J and Li M: MicroRNA-744 inhibits
cellular proliferation and invasion of colorectal cancer by
directly targeting oncogene notch1. Oncol Res. 26:1401–1409. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen XF and Liu Y: MicroRNA-744 inhibited
cervical cancer growth and progression through apoptosis induction
by regulating Bcl-2. Biomed Pharmacother. 81:379–387. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Cai H, Dai Z and Wang G:
Down-regulation of lncRNA XIST inhibits cell proliferation via
regulating miR-744/RING1 axis in non-small cell lung cancer. Clin
Sci (Lond). 133:1567–1579. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo Q, Zhang Q, Lu L and Xu Y: Long
noncoding RNA RUSC1-AS1 promotes tumorigenesis in cervical cancer
by acting as a competing endogenous RNA of microRNA-744 and
consequently increasing Bcl-2 expression. Cell Cycle. 19:1222–1235.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miyamae M, Komatsu S, Ichikawa D,
Kawaguchi T, Hirajima S, Okajima W, Ohashi T, Imamura T, Konishi H
and Shiozaki A: Plasma microRNA profiles: Identification of miR-744
as a novel diagnostic and prognostic biomarker in pancreatic
cancer. Br J Cancer. 113:1467–1476. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kleemann M, Schneider H, Unger K, Sander
P, Schneider EM, Fischer-Posovszky P, Handrick R and Otte K:
MiR-744-5p inducing cell death by directly targeting HNRNPC and
NFIX in ovarian cancer cells. Sci Rep. 8:90202018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen S, Shi F, Zhang W, Zhou Y and Huang
J: miR-744-5p inhibits non-small cell lung cancer proliferation and
invasion by directly targeting PAX2. Technol Cancer Res Treat.
18:15330338198769132019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hübner M, Hinske CL, Effinger D, Wu T,
Thon N, Kreth FW and Kreth S: Intronic miR-744 inhibits
glioblastoma migration by functionally antagonizing its host gene
MAP2K4. Cancers (Basel). 25:4002018. View Article : Google Scholar
|
|
17
|
Deng Y, Li Y, Fang Q, Luo H and Zhu G:
microRNA-744 is downregulated in glioblastoma and inhibits the
aggressive behaviors by directly targeting NOB1. Am J Cancer Res.
8:2238–2253. 2018.PubMed/NCBI
|
|
18
|
Li Y, Gan S, Ren L, Yuan L, Liu J, Wang W,
Wang X, Zhang Y, Jiang J, Zhang F and Qi X: Multifaceted regulation
and functions of replication factor C family in human cancers. Am J
Cancer Res. 8:1343–1355. 2018.PubMed/NCBI
|
|
19
|
Xiong S, Wang Q, Zheng L, Gao F and Li J:
Identification of candidate molecular markers of nasopharyngeal
carcinoma by tissue microarray and in situ hybridization. Med
Oncol. 28 (Suppl 1):S341–S348. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Wang G, Gong W, Guo S, Li D and Zhan
Q: The noncoding function of NELFA mRNA promotes the development of
oesophageal squamous cell carcinoma by regulating the Rad17-RFC2-5
complex. Mol Oncol. 14:611–624. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ho KH, Kuo TC, Lee YT, Chen PH, Shih CM,
Cheng CH, Liu AJ, Lee CC and Chen KC: Xanthohumol regulates
miR-4749-5p-inhibited RFC2 signaling in enhancing temozolomide
cytotoxicity to glioblastoma. Life Sci. 254:1178072020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu T, Shen H, Li J, Yang P, Gu Q and Fu Z:
RFC2, a direct target of miR-744, modulates the cell cycle and
promotes the proliferation of CRC cells. J Cell Physiol.
235:8319–8333. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kato T, Natsume A, Toda H, Iwamizu H,
Sugita T, Hachisu R, Watanabe R, Yuki K, Motomura K, Bankiewicz K
and Wakabayashi T: Efficient delivery of liposome-mediated
MGMT-siRNA reinforces the cytotoxity of temozolomide in
GBM-initiating cells. Gene Ther. 17:1363–1371. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. Jama. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gilbert MR, Wang M, Aldape KD, Stupp R,
Hegi ME, Jaeckle KA, Armstrong TS, Wefel JS, Won M, Blumenthal DT,
et al: Dose-dense temozolomide for newly diagnosed glioblastoma: A
randomized phase III clinical trial. J Clin Oncol. 31:4085–4091.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cloughesy TF, Cavenee WK and Mischel PS:
Glioblastoma: From molecular pathology to targeted treatment. Ann
Rev Pathol. 9:1–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Johnson DR and O'Neill BP: Glioblastoma
survival in the United States before and during the temozolomide
era. J Neurooncol. 107:359–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo JW, Wang X, Yang Y and Mao Q: Role of
micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur Rev Med
Pharmacol Sci. 19:1630–1639. 2015.PubMed/NCBI
|
|
30
|
Saadatpour L, Fadaee E, Fadaei S, Mansour
RN, Mohammadi M, Mousavi SM, Goodarzi M, Verdi J and Mirzaei H:
Glioblastoma: Exosome and microRNA as novel diagnosis biomarkers.
Cancer Gene Ther. 23:415–418. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui JQ, Shi YF and Zhou HJ: Expression of
RFC2 and PCNA in different gestational trophoblastic diseases. Ai
Zheng. 23:196–200. 2004.(In Chinese). PubMed/NCBI
|
|
32
|
Olbrich K, Costard L, Möser CV, Syhr KMJ,
King-Himmelreich TS, Wolters MC, Schmidtko A, Geisslinger G and
Niederberger E: Cleavage of SNAP-25 ameliorates cancer pain in a
mouse model of melanoma. Eur J Pain. 21:101–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mu Y, Yan X, Li D, Zhao D, Wang L, Wang X,
Gao D, Yang J, Zhang H, Li Y, et al: NUPR1 maintains autolysosomal
efflux by activating SNAP25 transcription in cancer cells.
Autophagy. 14:654–670. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Antonucci F, Corradini I, Morini R,
Fossati G, Menna E, Pozzi D, Pacioni S, Verderio C, Bacci A and
Matteoli M: Reduced SNAP-25 alters short-term plasticity at
developing glutamatergic synapses. EMBO Rep. 14:645–651. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fulda S, Wick W, Weller M and Debatin KM:
Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced
apoptosis and induce regression of malignant glioma in vivo. Nat
Med. 8:808–815. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shiraishi S, Tada K, Nakamura H, Makino K,
Kochi M, Saya H, Kuratsu Ji and Ushio Y: Influence of p53 mutations
on prognosis of patients with glioblastoma. Cancer. 95:249–257.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu T and Dai Y: Tumor microenvironment and
therapeutic response. Cancer Lett. 387:61–68. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G,
Xiang J, Wu M and Li G: miR-128 and miR-149 enhance the
chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal
remodeling in glioblastoma. Oncol Rep. 32:957–964. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu P, Yang J, Liu J, Yang X, Liao J, Yuan
F, Xu Y, Liu B and Chen Q: Identification of glioblastoma gene
prognosis modules based on weighted gene co-expression network
analysis. BMC Med Genomics. 11:962018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiang Y, Zhang CQ and Huang K: Predicting
glioblastoma prognosis networks using weighted gene co-expression
network analysis on TCGA data. BMC Bioinformatics. 13 (Suppl
2):S122012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kong Y, Feng ZC, Zhang YL, Liu XF, Ma Y,
Zhao ZM, Huang B, Chen AJ, Zhang D, Thorsen F, et al:
Identification of immune-related genes contributing to the
development of glioblastoma using weighted gene co-expression
network analysis. Front Immunol. 11:12812020. View Article : Google Scholar : PubMed/NCBI
|



















