Introduction
Cervical cancer is one of the most common malignant
tumors to occur in women (1). An
estimated 527,600 new cases of cervical cancer were diagnosed
worldwide and 265,700 women succumbed to this disease in 2012
(2). The majority of these cases
occured in developing countries (3).
Currently, recurrence, metastasis and drug resistance are the major
obstacles encountered in the treatment of cervical cancer (4). Therefore, the pathogenesis of cervical
cancer requires further investigations to improve current treatment
options.
MicroRNAs (miRNAs/miRs) have been demonstrated to
serve an important role in tumorigenesis (5,6). miRNAs
are a group of small, non-coding RNAs ~22 nucleotides in length
(7). miRNAs function as guide
molecules in gene silencing and translational repression by binding
to the 3′-untranslated region (3′-UTR) of their target mRNAs
(8). Abnormal expression of miRNAs
is closely associated with tumor initiation, progression and
prognosis (9). miR-148a is a novel
tumor suppressor gene, which is involved in various biological
functions, including cell apoptosis, cell cycle arrest and cell
senescence (10,11). The expression levels of miR-148a have
been reported to be dysregulated in various cancer types, such as
prostate, pancreatic (12) and
colorectal cancer (13). However, to
the best of our knowledge, the effects and underlying molecular
mechanism of miR-148a-3p in cervical cancer remain unclear.
Therefore, the present study aimed to investigate the effects and
the mechanism of miR-148a-3p in cervical cancer. The findings
provide potential therapeutic targets for cervical cancer.
Materials and methods
Patient tissue samples
A total of 20 cervical cancer (mean ± SD age,
56±10.05 years; age range, 39–68 years old, all female) and 8
normal cervical samples (mean ± SD age, 53±9.13 years; age range,
40–65 years old, all female) were collected from patients that
underwent surgical resection at The Second Affiliated Hospital of
Xi'an Jiaotong University (Xi'an, China) between February 2017 and
February 2018. In cases with histologically confirmed cervical
cancer, only patients who underwent diagnostic procedures, such as
biopsy were included. Any patients with non-epithelial cervical
cancer, recurrent disease and other malignancies were excluded from
the present study. The normal cervical tissues were obtained from
patients with uterine leiomyoma. None of the patients had received
chemotherapy, immunotherapy or radiotherapy prior to specimen
collection. All tissue samples were frozen in liquid nitrogen at
−80°C until required for further experiments. The present study was
approved by the Ethics Committee of The Second Affiliated Hospital
of Xi'an Jiaotong University (Xi'an, China) and the patients
provided written informed consent prior to sample collection.
Cell lines and culture
The HeLa and SiHa human cervical cancer cell lines
and human embryonic kidney cell line 293T were purchased from
American Type Culture Collection and cultured in DMEM
(Sigma-Aldrich; Merck KGaA) supplemented with 10% heat-inactivated
FBS (Invitrogen; Thermo Fisher Scientific, Inc.), 80 U/ml
penicillin and 80 ug/ml streptomycin. The cells were maintained at
37°C with 5% CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from frozen samples and cell
lines using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RT reactions were performed using the
PrimeScript RT reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocol. Subsequently, qPCR was performed using the
SYBR-Green Master mix (Takara Bio, Inc.) according to the
manufacturer's protocol. GAPDH and U6 spliceosomal RNA were used as
an internal control for the quantification of mRNAs and miRNAs,
respectively. The primer sequences are shown in Table I. The thermocycling conditions were
as follows: Pre-denaturation at 50°C for 2 min, denaturation at
95°C for 10 min, annealing at 95°C for 30 sec and extension at 60°C
for 30 sec (40 cycles). The relative gene expression was quantified
using the 2−ΔΔCq method (14).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Name | Primer | Sequence (5′-3′) |
|---|
| GAPDH | Forward |
TCAAGAAGGTGGTGAAGCAGG |
|
| Reverse |
TCAAAGGTGGAGGAGTGGGT |
| DNMT1 | Forward |
TACCACGCAGACATCAACCT |
|
| Reverse |
GCCCTTCCCTTTGTTTCCAG |
| UTF1 | Forward |
ATGGGGCTGCTGGGCGACAACG |
|
| Reverse |
GGGGAGGCGTCCGCAGACTTCG |
| miR-148a-3p | Forward |
TGCGCTCAGTGCACTACAGAAC |
|
| Reverse |
CCAGTGCAGGGTCCGAGGTATT |
| U6 | Forward |
CGCTTCGGCAGCACATATAC |
|
| Reverse |
AAATATGGAACGCTTCACGA |
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was measured using a CCK-8 assay
(Beyotime Institute of Biotechnology). Briefly, 1×103
cells/well were cultured in 96-well plates and assessed the
following day. The assessment was carried out for 6 days in total.
At the same time point on 2, 4, 6 days, 10 µl CCK-8 solution was
added to each well and the samples were incubated for 4 h at 37°C.
The absorbance was measured at a wavelength of 450 nm using a plate
reader. Each experiment was performed in triplicate.
Western blot analysis
Total protein was extracted from frozen samples and
cell lines using RIPA lysis buffer (Beyotime Institute of
Biotechnology). The protein concentration was estimated using a BCA
assay and 20 µg protein/lane was separated via 10% SDS-PAGE, and
then transferred onto PVDF membranes (MilliporeSigma). The
membranes were blocked with 5% skimmed milk suspended in TBST at
room temperature for 2 h. The membranes were incubated with primary
antibodies against UTF1 (1:100; cat. no. ab65453; Abcam); DNMT1
(1:200; cat. no. SC-20701; Santa Cruz Biotechnology, Inc.) or GAPDH
(1:1,000; cat. no. AB-P-R 001; Hangzhou Xianzhi Biological Co.,
Ltd.) at 4°C overnight. Following the primary antibody incubation,
the membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies (1:10,000; cat. no. BA1054; Wuhan Boster
Biological Technology Co. Ltd.) at 37°C for 2 h. The membranes were
briefly incubated with an enhanced chemiluminescence reagent
(MilliporeSigma) at room temperature for 2 min and visualized using
X-ray films. GAPDH was used to normalize the expression of the
genes. Protein level was quantified using Quantity One software
v.4.6, (Bio-Rad Laboratories, Inc.).
Cell transfection
miR-148a-3p mimic (5′-UCAGUGCACUACAGAACUUUGU-3′),
miRNA mimic control (5′-TTCTCCGAACGTGTCACGT-3′), DNMT1-short
hairpin (sh) RNA plasmid expression vector (pGPU6/GFP/Neo,
5′-GGAUGAGUCCAUCAAGGAATT-3′) and control-shRNA
(5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Shanghai
GenePharma Co., Ltd. Cells were incubated (1×105) in a
6-well plate for at 37°C for 24 h before transfection and
transfected with either miR-148a-3p mimic, control mimic,
DNMT1-shRNA and control-shRNA at a final concentration of 50 nM
using Lipofectamine® 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 5 h
according to the manufacturer's protocol. After 48 h, the cells
were used for subsequent experiments.
Luciferase reporter assay
293T cells were seeded into a 24 well plate at a
density of 5×104 cells/well. Following incubation at
37°C for 24 h, a wild type or mutated DNMT1 3′-UTR luciferase
reporter vector (Promega Corporation), combined with miR-148a-3p
mimics (5′-UCAGUGCACUACAGAACUUUGU-3′; Shanghai GenePharma Co.,
Ltd.) or miRNA mimic control (5′-TTCTCCGAACGTGTCACGT-3′, Shanghai
GenePharma Co., Ltd.), were transfected into the cells at a final
concentration of 20 nM using a Vigofect transfection reagent
[Weiglas Biotechnology (Beijing) Co., Ltd.] according to the
manufacturer's protocol. At 48 h post-transfection, the firefly and
Renilla luciferase activities were detected using the
Dual-Luciferase Reporter assay system (Promega Corporation).
Renilla luciferase activity was used as the internal
control.
Bisulfite sequencing
Bisulfite sequencing was carried out as previously
described (15). Genomic DNA was
extracted from SiHa and HeLa cells using the Universal Genomic DNA
Extraction kit (cat. no. DV811A; Takara Bio, Inc.) according to the
manufacturer's protocol. Genomic DNA (250 ng) of each sample was
bisulfite converted using EpiTect Bisulfite kit (cat. no. 59104;
Qiagen, Inc.) according to the manufacturer's protocol. A 360 bp
segment (nucleotides −977 to −617, transcriptional start site, +1)
from bisulfate-modified DNA was amplified using MSP DNA polymerase
(TIANGEN Biotech Co., Ltd.) with the following primer sequences:
Forward, 5′-TGATTAGAGTAGGGATGGAAAG-3′ and reverse,
5′-TACAACCAACATCCCTAAAAA−3′. The thermocycling conditions were as
follows: 1 cycle at 95°C for 10 min, followed by amplification for
40 cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec
and final extension at 72°C for 10 min. The PCR products were
recovered and purified by 1.0% agarose gel electrophoresis and
quantified using the ImageJ software v.1.53a (National Institutes
of Health), then subcloned by TA cloning using the pEASY-T1 Cloning
kit (cat. no. CT101-01; TransGen Biotech Co., Ltd.) and then
transformed into Escherichia coli strain DH5α (Invitrogen;
Thermo Fisher Scientific Inc.) using standard procedures.
Recombinant plasmids positive for inserts of correct size (559 bp)
were identified by colony PCR with Taq DNA polymerase (Takara Bio,
Inc.). At least 10 positive inserted clones were selected and
sequenced by Wuhan Biofavor Biotech Service Co., Ltd. using Sanger
sequencing method (POP-7™ Polymer for 3730/3730×l DNA Analyzers,
cat. no. 4332241; Thermo Fisher Scientific Inc.) with M13 primers
(M13 forward, 5′-GTAAAACGACGGCCAGT-3′ and reverse,
5′-CAGGAAACAGCTATGAC-3′). The quality of processed samples was
estimated according to the optical density (OD) 260/280 ratio; the
ratio between 1.8–2.0 meet the experimental requirements. The OD
260/280 ratio of DNA was estimated using a microspectrophotometer
and the concentration of DNA was calculated according to the
formula: Total DNA concentration
(µg/µl)=OD260×50×200×10−3. The concentration
requirement: >50 ng/µl. The methylation density was quantified
using BiQ Analyzer software v.2.0 (16) (Max Planck Institute Informatik).
Statistical analysis
Statistical analysis was performed using SPSS
version 16.0 software (SPSS, Inc.). Data are presented as the mean
± SD. An independent sample unpaired t-test and one-way ANOVA
followed by Tukey's post hoc test were used for group comparisons.
Correlation analysis was performed using Pearson's correlation
analysis. The experiments were performed in triplicate and repeated
three times independently. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression levels of miR-148a-3p in
cervical cancer
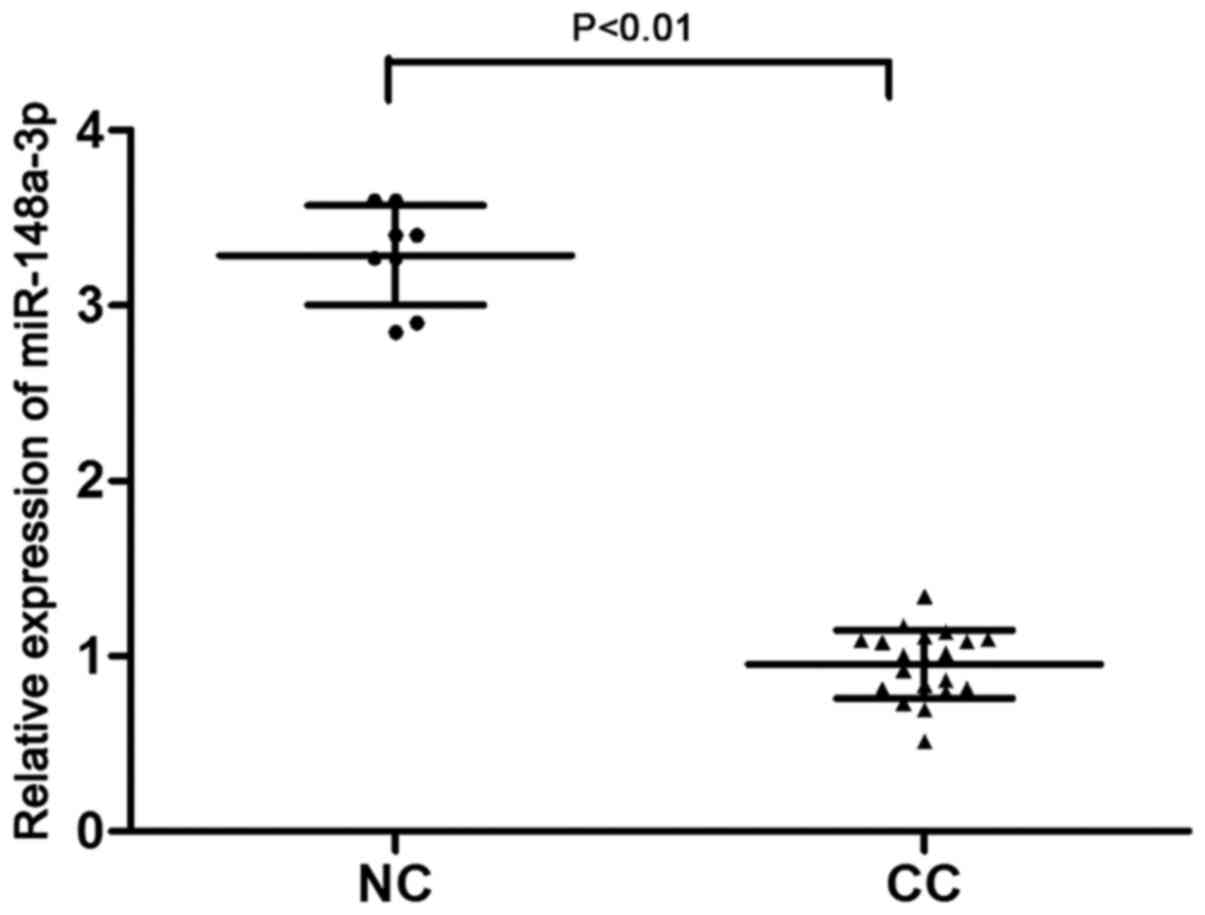
To explore the role of miR-148a-3p in cervical
cancer, its expression levels were assessed in cervical cancer and
normal cervical tissues by RT-qPCR analysis. miR-148a-3p expression
was significantly decreased in cervical cancer tissues compared
with in normal cervical tissues (P<0.01; Fig. 1). These data suggested that
miR-148a-3p may be associated with the progression of cervical
cancer.
miR-148a-3p inhibits the proliferation
of cervical cancer cells
To assess the effects of miR-148a-3p on the
proliferation of cervical cancer cells, miR-148a-3p mimics were
successfully transfected into HeLa and SiHa cells (Fig. 2A and B) and cell proliferation was
evaluated using a CCK-8 assay. The cell proliferation curve
revealed that the viability of miR-148a-3p overexpressing HeLa and
SiHa cells were significantly decreased compared with the control
mimics (P<0.01; Fig. 2C and D).
These results demonstrated that miR-148a-3p inhibited the
proliferation of cervical cancer cells.
miR-148a-3p inhibits DNMT1 expression
by targeting the 3′-UTR
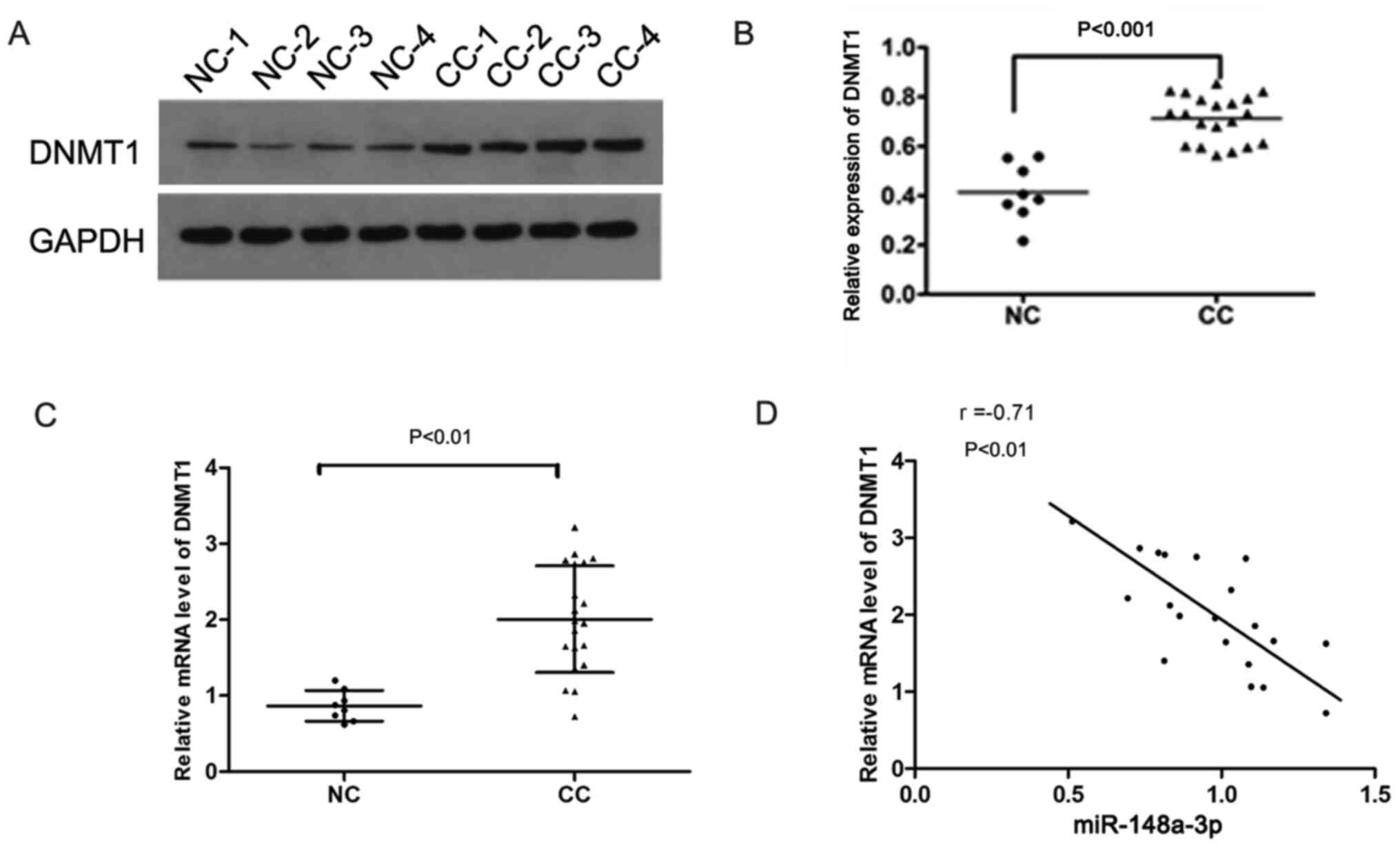
The protein expression levels of DNMT1 were
significantly increased in cervical cancer tissues compared with in
normal cervical tissues (P<0.001; Fig. 3A and B). In addition, the mRNA
expression levels of DNMT1 in cervical cancer were significantly
higher than those in normal cervical tissues (P<0.01; Fig. 3C). Correlation analysis indicated
that the expression levels of DNMT1 were negatively correlated with
miR-148a-3p expression (P<0.01; Fig.
3D), suggesting that DNMT1 expression may be regulated by
miR-148a-3p. Using bioinformatics analysis, it was identified that
DNMT1 was a potential target gene of miR-148a-3p (Fig. 4A). To verify this hypothesis,
luciferase reporter vectors containing the potential binding
sequences of the 3′-UTR of DNMT1 (wt and mut) were constructed and
co-transfected with miR-148a-3p or mimic control into 293T cells
(Fig. 4A). The luciferase activity
levels in the wt DNMT1 + miR-148a-3p group were significantly
decreased compared with those of the wt DNMT1 + control mimic
group, whereas the luciferase activity in the mut DNMT1 +
miR-148a-3p group was not significantly altered compared with that
of the mut DNMT1 + control mimic group (Fig. 4B). These findings demonstrated that
miR-148a-3p targeted the 3′-UTR of DNMT1.
In addition, to further verify the regulatory effect
of miR-148a-3p on DNMT1 expression, the expression of DNMT1 in
miR-148a-3p overexpressing HeLa and SiHa cells were measured. DNMT1
mRNA (Fig. 5A and B) and protein
expression levels (Fig. 5C-F) were
significantly decreased in miR-148a-3p-overexpressing HeLa and SiHa
cells compared with those of the control mimics (P<0.01). These
data further demonstrated that miR-148a-3p regulated DNMT1
expression by targeting its 3′-UTR in cervical cancer cells.
DNMT1 regulates the expression levels
of UTF1 via methylation in cervical cancer
In our previous study, UTF1 was demonstrated to
serve an important tumor suppressive role in cervical
carcinogenesis (15). However, UTF1
is highly methylated in cervical cancer (15). Based on the important role of DNMT1
in DNA methylation (17), it was
hypothesized that UTF1 expression may be regulated by DNMT1 in
cervical cancer. Correlation analysis indicated that the protein
(r=−0.55; P<0.05, Fig. 6A) and
mRNA expression levels (r=−0.53; P<0.05, Fig. 6B) of UTF1 were negatively correlated
with the protein expression levels of the DNMT1 in cervical cancer.
Additional experiments indicated that DNMT1 knockdown led to a
significant increase in the mRNA and protein expression levels of
UTF1 in HeLa and SiHa cells (P<0.01 or P<0.05; Fig. 7). Furthermore, compared with the
control knockdown group, the methylation levels of the UTF1
promoter were significantly attenuated following DNMT1 knockdown
(P<0.05; Fig. 8). These findings
demonstrated the important role of DNMT1 in the regulation of UTF1
expression.
Discussion
Numerous miRNAs have been reported to serve
important roles in the pathogenesis of tumors by regulating cell
proliferation, apoptosis and invasion (18,19). It
has been reported that miR-148a exhibits antitumor effects in
various cancer types including gastric, colorectal, pancreatic,
liver and breast cancers (13,20). In
the present study, the data indicated that miR-148a-3p expression
was reduced in cervical cancer tissues compared with in normal
cervical tissues. Furthermore, miR-148a-3p overexpression
significantly inhibited the proliferation of HeLa and SiHa cells.
These findings suggested that miR-148a-3p exerted inhibitory
effects in cervical cancer, which was consistent with a previous
study that demonstrated that miR-148a acts as a tumor suppressor
gene in colorectal cancer (21).
The antiproliferative mechanism of action of
miR-148a-3p was investigated by identifying its target gene, DNMT1,
using bioinformatic analysis. Furthermore, a previous study has
reported that miR-148a-3p directly represses the expression levels
of DNMT1 in human colon cancer cells (22). Therefore, DNMT1 expression was
analyzed in cervical cancer tissues and a negative correlation was
observed between the expression levels of DNMT1 and miR-148a-3p,
implying that miR-148a-3p may regulate DNMT1 expression in cervical
cancer. In addition, the present study demonstrated that
miR-148a-3p targeted the 3′-UTR of DNMT1. Additional experiments
demonstrated that overexpression of miR-148a-3p inhibited the
protein and mRNA expression of DNMT1 in HeLa and SiHa cells.
Collectively, these data demonstrated that miR-148a-3p regulated
DNMT1 expression by targeting its 3′-UTR sequence in cervical
cancer.
UTF1 is a stem cell-associated transcription factor,
which serves a critical role in cell differentiation and
development (23). In our previous
study, UTF1 functioned as a tumor suppressor gene and its
expression was downregulated in cervical cancer (15). In addition, the promoter region of
UTF1 was hypermethylated in cervical cancer (15,24).
DNMT enzymes typically mediate global hypermethylation of the
genome (25). DNMT1 is an important
member of the DNMT superfamily (26). Notably, In the present study, DNMT1
was highly expressed in cervical cancer and the association between
the expression levels of UTF1 and DNMT1 was analyzed in cervical
cancer tissues. UTF1 expression was negatively correlated with
DNMT1 expression, implying that the latter may regulate UTF1
expression in cervical cancer. DNMT1 knockdown significantly
increased the expression levels of UTF1 in HeLa and SiHa cells,
which demonstrated that DNMT1 regulated UTF1 expression in cervical
cancer. The modification of DNA methylation is considered the main
pattern of epigenetic regulation (27). DNMT1 serves a key role in DNA
methylation (28). It has been
demonstrated that the overexpression of DNMT1 increases DNA
methylation (29). In the present
study, methylation analysis indicated that DNMT1 knockdown
significantly reduced the methylation levels of the UTF1 promoter
in cervical cancer cells. These findings are consistent with a
previous report that demonstrated that promoter hypermethylation
contributes to the decreased expression of tumor suppressor genes
(30).
In summary, the results of the present study
demonstrated that miR-148a-3p inhibited the proliferation of
cervical cancer cells. The mechanism of action was associated with
the regulation of the expression of DNMT1 and UTF1, which may
provide potential therapeutic targets for cervical cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81702578).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QC and YW performed the experiments and data
analysis. XW conceived and designed the study. QC and HD confirmed
the authenticity of all the raw data. XW and HD reviewed and
revised the manuscript for important intellectual content. HD
participated in data analysis and draft writing. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Second Affiliated Hospital of Xi'an Jiaotong
University (approval no. 2017-113; Xi'an, China). All the patients
signed written informed consent prior to participation in the
present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Lv H, Xue Z, Wang L and Bai Z:
Temporal trends of common female malignances on breast, cervical,
and ovarian cancer mortality in japan, republic of Korea, and
Singapore: Application of the age-period-cohort model. Biomed Res
Int. 2018:53074592018.PubMed/NCBI
|
|
4
|
Gupta S, Kumar P and Das BC: HPV:
Molecular pathways and targets. Curr Probl Cancer. 42:161–174.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Drusco A and Croce CM: MicroRNAs and
cancer: A long story for short RNAs. Adv Cancer Res. 135:1–24.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fanini F and Fabbri M: Cancer-derived
exosomic microRNAs shape the immune system within the tumor
microenvironment: State of the art. Semin Cell Dev Biol. 67:23–28.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Costello A, Lao N, Clynes M and Barron N:
Conditional knockdown of endogenous MicroRNAs in CHO cells using
TET-ON-SanDI sponge vectors. Methods Mol Biol. 1603:87–100. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Höck J and Meister G: The Argonaute
protein family. Genome Biol. 9:2102008. View Article : Google Scholar
|
|
9
|
Lin X, Xiaoqin H, Jiayu C, Li F, Yue L and
Ximing X: Long non-coding RNA miR143HG predicts good prognosis and
inhibits tumor multiplication and metastasis by suppressing
mitogen-activated protein kinase and Wnt signaling pathways in
hepatocellular carcinoma. Hepatol Res. 49:902–918. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang M, Wang C and Mu L: Mir-148a induces
apoptosis by upregulating bim expression in gastric cancer cells.
Int J Clin Exp Med. 10:2791–2799. 2017.PubMed/NCBI
|
|
11
|
Liu J, Si L and Tian H: MicroRNA-148a
inhibits cell proliferation and cell cycle progression in lung
adenocarcinoma via directly targeting transcription factor E2F3.
Exp Ther Med. 16:5400–5409. 2018.PubMed/NCBI
|
|
12
|
Xiao WD, Ao J, Huang Z, Lin SR, Peng L and
Li Y: The effects of up-regulation of miR-148a on the expression of
anti-oncogene ppENK, p16 and RASSF1A in pancreatic carcinoma AsPC-1
cells. Pancreatology. 16 (Suppl):S152016. View Article : Google Scholar
|
|
13
|
Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng
H and Lai M: MiR-148a promotes apoptosis by targeting Bcl-2 in
colorectal cancer. Cell Death Differ. 18:1702–1710. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu XL and Zheng PS: Undifferentiated
embryonic cell transcription factor-1 (UTF1) inhibits the growth of
cervical cancer cells by transactivating p27 Kip1. Carcinogenesis.
34:1660–1668. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bock C, Reither S, Mikeska T, Paulsen M,
Walter J and Lengauer T: BiQ Analyzer: Visualization and quality
control for DNA methylation data from bisulfite sequencing.
Bioinformatics. 21:4067–4068. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han M, Li J, Cao Y, Huang Y, Li W, Zhu H,
Zhao Q, Han JJ, Wu Q, Li J, et al: A role for LSH in facilitating
DNA methylation by DNMT1 through enhancing UHRF1 chromatin
association. Nucleic Acids Res. 48:12116–12134. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei Y, Peng S, Wu M, Sachidanandam R, Tu
Z, Zhang S, Falce C, Sobie EA, Lebeche D and Zhao Y: Multifaceted
roles of miR-1s in repressing the fetal gene program in the heart.
Cell Res. 24:278–292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu X and Li Z: The role of microRNAs
expression in laryngeal cancer. Oncotarget. 6:23297–23305. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Deng X, Zeng X and Peng X: The role
of Mir-148a in cancer. J Cancer. 7:1233–1241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao W, Zheng J, Wei G, Yang K, Wang G and
Sun X: miR-148a inhibits cell proliferation and migration through
targeting ErbB3 in colorectal cancer. Oncol Lett. 18:2530–2536.
2019.PubMed/NCBI
|
|
22
|
Zuo J, Xia J, Ju F, Yan J, Zhu A, Jin S,
Shan T and Zhou H: MicroRNA-148a can regulate runt-related
transcription factor 3 gene expression via modulation of DNA
methyltransferase 1 in gastric cancer. Mol Cells. 35:313–319. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pantazis G, Harter PN, Capper D, Kohlhof
P, Mittelbronn M and Schittenhelm J: The embryonic stem cell factor
UTF1 serves as a reliable diagnostic marker for germinomas.
Pathology. 46:225–229. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Okuda A, Fukushima A, Nishimoto M, Orimo
A, Yamagishi T, Nabeshima Y, Kuro-o M, Nabeshima Yi, Boon K,
Keaveney M, et al: UTF1, a novel transcriptional coactivator
expressed in pluripotent embryonic stem cells and extra-embryonic
cells. EMBO J. 17:2019–2032. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jin B and Robertson KD: DNA
Methyltransferases, DNA damage repair, and cancer. Adv Exp Med
Biol. 754:3–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Foltz G, Yoon JG, Lee H, Ryken TC,
Sibenaller Z, Ehrich M, Hood L and Madan A: DNA
methyltransferase-mediated transcriptional silencing in malignant
glioma: A combined whole-genome microarray and promoter array
analysis. Oncogene. 28:2667–2677. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scopa CD, Zolota V and Kourea HP: Targeted
pathways in breast cancer: Molecular and protein markers guiding
therapeutic decisions. Curr Mol Pharmacol. 7:4–21. 2014.PubMed/NCBI
|
|
28
|
Kumar R and Rao DN: Role of DNA
Methyltransferases in epigenetic regulation in bacteria. Subcell
Biochem. 61:81–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Subramaniam D, Thombre R, Dhar A and Anant
S: DNA Methyltransferases: A novel target for prevention and
therapy. Front Oncol. 4:80. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
You Y, Yang W, Wang Z, Zhu H, Li H, Lin C
and Ran Y: Promoter hypermethylation contributes to the frequent
suppression of the CDK10 gene in human nasopharyngeal carcinomas.
Cell Oncol (Dordr). 36:323–331. 2013. View Article : Google Scholar : PubMed/NCBI
|