Introduction
Gallbladder carcinoma (GBC) is the most common malignancy of the biliary tract (1). GBC is estimated to have an incidence rate of 2.5 per 10,000 (based on the Surveillance, Epidemiology and End Results database), with the seventh-highest incidence rate among all digestive tract cancers (1,2). Given that GBC cells can spread rapidly in the early stages through direct infiltration, lymph node metastasis and blood metastasis, most patients are already in the middle or late stages of disease at the time of diagnosis (3). In addition, the prognosis of patients with GBC is very poor and the 5-year survival rate is <5% (4). Currently, surgery is the only option to cure GBC; however, the local recurrence rate following radical surgery is high (3). There is no clear evidence for improving the long-term prognosis and survival rate of patients with GBC with other treatments, including radiotherapy and chemotherapy (5). Therefore, in-depth studies of the molecular mechanisms of the growth and metastasis of GBC, determining key molecules involved the development of GBC, and screening effective diagnosis and treatment targets are vital to the future development of diagnosis and treatment of GBC.
Long non-coding RNAs (lncRNAs), which are functional transcripts >200 nucleotides in length, have been highly implicated in the development of different types of cancer by functioning as oncogenes or tumor suppressor genes (6–8). In recent years, with the development of lncRNA research, key lncRNAs have been reported to be abnormally expressed in GBC and are involved in the malignancy and progression of GBC, which may be novel biomarkers for middle and late diagnosis and prognosis prediction of GBC (9–11). For example, lncRNA plasmacytoma variant translocation 1 (PVT1) is upregulated in GBC tissues and cells. Knockdown of lncRNA PVT1 significantly suppresses cell proliferation, migration and invasion in vitro, and inhibits tumor growth in vivo by regulating the microRNA (miRNA/miR)-143/hexokinase-2 axis (12). Furthermore, lncRNA DILC drives the self-renewal, tumorigenicity, proliferation and metastasis of GBC stem cells by activating the Wnt/β-catenin signaling pathway (13). lncRNA SPRY4 intronic transcript 1 (SPRY4-IT1) is upregulated in GBC tissues, and SPRY4-IT1 knockdown significantly inhibits GBC cell proliferation, and is partly associated with epithelial-to-mesenchymal transition (EMT) (14). Taken together, these findings suggest that lncRNAs may serve as potential therapeutic targets for novel therapies in human GBC.
lncRNA thymopoietin antisense transcript 1 (TMPO-AS1) is in chromosome 12q21.2 (15). Abnormal expression of TMPO-AS1 notably affects the occurrence and development of different types of cancer, such as lung adenocarcinoma, hepatocellular carcinoma and prostate cancer (15,16). For example, TMPO-AS1 is upregulated in hepatocellular carcinoma (HCC) tissues and cells (16). TMPO-AS1 knockdown downregulates HCC cell proliferation, invasion, migration and EMT processes in vitro and tumor growth in vivo by regulating the miR-329-3p/forkhead box protein K1 (FOXK1) axis mediated by the AKT/mTOR signaling pathway (16). TMPO-AS1 is highly expressed in cervical cancer cells, and overexpression of TMPO-AS1 promotes cervical cancer cell proliferation, migration and invasion by regulating the miR-143-3p/zinc finger E-box-binding homeobox 1 axis (17). TMPO-AS1 is also highly expressed in bladder cancer tissues and cell lines, and overexpression of TMPO-AS1 facilitates cell proliferation, migration and invasion by sponging the miR-98-5p/EBF transcription factor 1 axis (18). However, to the best of our knowledge, the exact functions and corresponding mechanisms of TMPO-AS1 in the occurrence and development of GBC have rarely been reported. Thus, the present study aimed to determine whether TMPO-AS1 exerts its functional role in the viability, proliferation, migration, invasion and EMT of GBC cells, and investigate the potential molecular mechanisms.
Materials and methods
Tissue samples
A total of 30 GBC tissues, including 13 stage I+II tissues and 17 stages III+IV tissues and 30 corresponding adjacent normal tissues (2 cm away from GBC tissues) were collected from patients who underwent surgical resection or biopsy between January 2015 and December 2020 at the People's Hospital of Danyang and Affiliated Danyang Hospital of Nantong University (Jiangsu, China). The patients included 16 women and 14 men. Patients <18 or >75 years old and who had received radiotherapy or chemotherapy before surgery were excluded from the study. Specimens were frozen at −80°C until subsequent experimentation. The present study was approved by the Ethics Committee of the People's Hospital of Danyang and Affiliated Danyang Hospital of Nantong University (Jiangsu, Chain; approval no. 20190621) and written informed consent was provided by all patients prior to the study start. All tissues were examined using a tissue microarray (TMA). Anatomic and histological grades were established according to the American Joint Committee on Cancer (AJCC) tumor node metastasis (TNM) grading scale (19).
Cell lines and culture
The GBC cell lines, SGC-996, GBC-SD, EH-GB1 and NOZ, and the human gallbladder epithelial cell line, H69 were purchased from the American Type Culture Collection. Cells were routinely cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS, Invitrogen; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Sigma-Aldrich; Merck KGaA) in a humidified atmosphere with 5% CO2 at 37°C.
Cell transfection
For cell transfection, short hairpin (sh)-TMPO-AS1, sh-E2F transcription factor 2 (E2F2) and sh-negative control (NC), miR-1179 mimics, miR-1179 inhibitor and mimic or inhibitor NCs were purchased from GenScript. GBC cells were seeded into 6-well plates at a density of 3×105 cells/well and cultured to 80% confluence. Plasmids (1 µg) and shRNAs (20 nm) or miRNA mimic/inhibitor (20 nm) were transfected into cells, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at 37°C. The sequences used were as follows: miR-1179 mimics, 5′-AAGCATTCTTTCATTGGTTGG-3′; miR-1179 inhibitor, 5′-TCAACCAATAAGAGGATGCCA-3′; mimic NC, 5′-GTGCGAGGCAATGTCGGTGGA-3′; and inhibitor NC, 5′-CGTCCACCGTGTAAGCCTGAA-3′. Transfection efficiency was assessed via reverse transcription-quantitative (RT-q)PCR analysis 48 h post-transfection.
Cell Counting Kit-8 (CCK-8) assay
The viability of GBC-SD and NOZ cells was indirectly assessed via the CCK-8 assay. Cells were seeded into 96-well plates at a density of 1×104 cells/well and incubated for 0, 24, 48 and 72 h. Cell viability was assessed using the CCK-8 kit (Beyotime Institute of Biotechnology), according to the manufacturer's instructions. CCK-8 reagent (10 µl) was added to each well and cells were incubated at 37°C for 2 h. Optical density was detected at a wavelength of 490 nm, using a microplate reader (BioTek Instruments Inc.) and analyzed using GraphPad Prism 5.0 (GraphPad Software, Inc.).
Colony formation assay
The colony formation assay was performed to assess the proliferative potential of transfected GBC cells. Briefly, transfected GBC-SD and NOZ cells were seeded into 6-well plates at a density of 1×103 cells/well. The media was replaced with fresh culture medium (Procell Life Science & Technology Co., Ltd.) every 2–3 days for a total of 2 weeks. Subsequently, colonies were washed twice with PBS, fixed with 70% methanol for 30 min and stained with 0.1% crystal violet for 1 h at room temperature. Colonies (>50 cells) were observed and counted under a light microscope (magnification, ×40).
Wound healing assay
The wound healing assay was performed to assess the migratory ability of transfected GBC cells. Briefly, transfected GBC-SD and NOZ cells were seeded into 6-well plates at a density of 5×105 cells/well. Once the cells reached 80% confluence, a 200 µl pipette tip was used to make a scratch, and serum-free medium was used. Following incubation for 0 and 48 h, GBC-SD and NOZ cells were observed under an inverted microscope (Olympus Corporation) and the distance between the wounds was recorded.
Transwell migration and invasion assays
Transwell migration and invasion assays (Corning, Inc.) were performed to assess the migratory and invasive abilities of transfected GBC cells. For the Transwell invasion assay, Matrigel (Becton, Dickinson and Company) was dissolved overnight at 4°C, diluted with serum-free medium (Procell Life Science & Technology Co., Ltd.) and added to the upper chambers to form a gel for 30 min at 37°C, with 5% CO2. For the Transwell migration and invasion assays, transfected GBC-SD and NOZ cells were seeded into the upper chamber at a density of 1×106 cells/well with serum-free DMEM, while DMEM supplemented with 20% FBS was added to the lower chamber. Following incubation for 48 h at 37°C, the migratory and invasive cells were treated with 70% ethanol, stained with 0.5% crystal violet for 20 min at room temperature, and counted under an IX70 inverted optical microscope (magnification, ×100; Olympus Corporation).
RT-qPCR
Total RNA was extracted from clinical GBC tissues and cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using the TaqMan Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following temperature protocol was used for RT: 42°C for 50 min, 99°C for 5 min, then 4°C until further use. qPCR was subsequently performed on the ABI 7300-Fast RT PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the SYBR Green PCR kit (Qiagen China Co., Ltd), according to the manufacturer's instructions. The thermocycling conditions were as follows: 10 min at 95°C for 1 cycle, followed by denaturation at 95°C for 30 sec, annealing at 56°C for 1 min, and final extension at 72°C for 30 sec for 40 cycles. The relative expression levels of miRNAs and mRNAs were evaluated using the 2−ΔΔCq method (20), with U6 or GAPDH as the internal references, respectively. The following primer sequences were used: TMPO-AS1 forward, 5′-AGCCCACACACTACAGGCA-3′ and reverse, 5′-GCACAAAAGCAGTACGACCT-3′; miR-1179 forward, 5′-GCGGAAGCATTCTTTCATT-3′ and reverse, 5′-CAAGGGCTCGACTCCTGTTC-3′; E2F2 forward, 5′-TCCACAAACAGAACAAGATG-3′ and reverse, 5′-TCAATTAATCAACAGGTCCCC-3′; U6 forward, 5′-GGCCTTTTGGCTAAGATCAA-3′, and reverse, 5′-TATTCCATCTCCCTGCTCCA-3′; and GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
Subcellular fractionation analysis
Cytoplasmic and nuclear fractions were extracted from transfected GBC-SD and NOZ cells using NE-PER Nuclear and Cytoplasmic Extraction reagent (Thermo Fisher Scientific, Inc.). RNA from each fraction was measured via RT-qPCR analysis. GAPDH and U6 were used as internal references for TMPO-AS1 in cytoplasmic and nuclear fractions, respectively.
Western blotting
Protein from clinical GBC tissues and cell lines was extracted using RIPA lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.) and quantified using the BCA kit (Beyotime Institute of Biotechnology). Subsequently, 25 µg protein/lane were separated via 12% SDS-PAGE, transferred onto PVDF membranes (MilliporeSigma) and blocked with 5% non-fat milk for 1 h at room temperature. The membranes were incubated with primary antibodies against E-cadherin (1:1,000; cat. no. ab1416), vimentin (1:1,000; cat. no. ab92547), α-smooth muscle actin (SMA; 1:1,000; cat. no. ab32575), E2F2 (1:1,000; cat. no. ab138515) and β-actin (1:2,000; cat. no. ab8227) overnight at 4°C (all purchased from Abcam). Following the primary incubation, membranes were washed three times with TBST (0.05% Tween-20) and probed with HRP-conjugated secondary antibody (1:2,000; cat. no. ab6728; Abcam) for 1 h at room temperature. Protein bands were visualized using enhanced chemiluminescence (MilliporeSigma) and quantified using ImageJ software (version 4.3; National Institutes of Health).
Immunofluorescence assay
Transfected GBC-SD and NOZ cells were seeded into 6-well plates at a density of 1×105 cells/well. After fixing in 4% paraformaldehyde for 24 h at room temperature, and permeabilization with 0.2% Triton X-100 in PBS, cells were incubated with primary antibody against α-SMA (1:1,000; cat. no. ab32575; Abcam) overnight at 4°C. Subsequently, cells were incubated with goat anti-rabbit IgG H&L secondary antibody (1:1,000; cat. no. ab150077; Abcam) at 37°C for 1 h. The nuclei were counterstained with DAPI. Positive staining was observed under a fluorescence microscope (Carl Zeiss AG) at ×200 magnification.
RNA pull-down analysis
RNA pull-down assays were performed using the pierce magnetic RNA protein pull-down kit (cat. No. 20164; Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. GBC-SD and NOZ cells were incubated with biotin-labeled miR-1179-wild-type (WT) or mutant (MUT) for 48 h and lysed in RNase-free cell lysis solution (1 mM HEPES, 200 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 200 U/ml RNase Inhibitor) at 4°C. Cell lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen; Thermo Fisher Scientific, Inc.) overnight at 4°C, according to the manufacturer's protocol. The beads were subsequently washed with high salt buffer (1% Triton X-100; 0.1% SDS; 20 mM Tris-HCl, pH 8.0; 2 mM EDTA; 500 mM NaCl). The bound RNA was purified using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). TMPO-AS1 enrichment was detected via RT-qPCR analysis.
Dual-luciferase reporter assay
The StarBase database (version 2.0; http://starbase.sysu.edu.cn/starbase2) was used to predict the potential miRNA and miR-1179 target genes. The sequences of WT TMPO-AS1 (TMPO-AS1-WT) and E2F2 (E2F2-WT) and MUT TMPO-AS1 (TMPO-AS1-Mut) and E2F2 (E2F2-MUT) were inserted into pmirGLO reporter vector (GenScript). Subsequently, pmirGLO-TMPO-AS1-WT/MUT and pmirGLO-E2F2-WT/MUT were transfected with miR-1179 mimics or NC into GBC-SD and NOZ cells, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation for 48 h at 37°C, Luciferase activity was measured using a dual-luciferase reporter assay system (Promega Corporation) and normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, Inc.) and data are presented as the mean ± SD. Each experiment was repeated ≥3 times. Paired and unpaired Student's t-test was used to compare differences between tumor and adjacent normal tissues and two independent groups, respectively. The differences between multiple groups for all in vitro analyses were compared using one-way ANOVA followed by Tukey's post hoc test. Pearson's correlation coefficient analysis was performed to assess the correlations between TMPO-AS1 and miR-1179, and E2F2 and miR-1179. Survival rates were assessed using the Kaplan-Meier method and log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
TMPO-AS1 expression is upregulated in GBC tissues and cell lines, and aberrant TMPO-AS1 expression is independently associated with the overall survival of patients with GBC
To investigate the potential role and underlying molecular mechanisms of TMPO-AS1 in GBC, RT-qPCR analysis was performed to detect TMPO-AS1 expression in clinical GBC tissues. As presented in Fig. 1A, TMPO-AS1 expression was significantly upregulated (P<0.01) in GBC tissues compared with adjacent normal tissues. In addition, TMPO-AS1 expression was measured in patients with GBC with different TNM stages. The results demonstrated that TMPO-AS1 expression was significantly higher (P<0.01) in patients with GBC at advanced stages III+IV compared with patients with GBC at advanced stages I+II (Fig. 1B). The clinical significance of aberrant TMPO-AS1 expression in the prognosis of patients with GBC was further investigated. As presented in Fig. 1C, patients with GBC with low TMPO-AS1 expression exhibited higher overall survival rates compared with those with high TMPO-AS1 expression. RT-qPCR was performed to detect TMPO-AS1 expression in GBC cells. As presented in Fig. 1D, TMPO-AS1 expression was significantly upregulated (P<0.01) in GBC cells compared with H69 cells, particularly in GBC-SD and NOZ cells. Taken together, these results suggest that TMPO-AS1 is upregulated in GBC tissues and cell lines, and may function as an oncogene in the occurrence and development of GBC.
 |
Figure 1.
TMPO-AS1 expression is upregulated in GBC tissues and cell lines and aberrant TMPO-AS1 expression is independently associated with the overall survival of patients with GBC. (A) RT-qPCR analysis was performed to detect TMPO-AS1 expression in GBC and adjacent normal tissues, and (B) GBC tissues at different stages. (C) Patients with low TMPO-AS1 expression had a higher survival rate than those with high TMPO-AS1 expression. (D) RT-qPCR analysis was performed to detect TMPO-AS1 expression in GBC cell lines and H69 cells. Data are presented as the mean ± SD (n=3). **P<0.01 vs. adjacent normal tissues, tumor tissues at stages I+II or H69 cells. TMPO-AS1, thymopoietin antisense transcript 1; GBC, gallbladder carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
|
TMPO-AS1 knockdown inhibits the proliferation, migration, invasion and EMT of GBC cells
To determine the potential role of TMPO-AS1 in GBC, GBC-SD and NOZ cells were transfected with sh-TMPO-AS1. As presented in Fig. 2A, TMPO-AS1 expression significantly decreased (P<0.01) in sh-TMPO-AS1-transfected cells, whereas no changes were observed in the sh-NC group. The CCK-8 assay was performed to assess the effects of sh-TMPO-AS1 on the viability of GBC-SD and NOZ cells. As presented in Fig. 2B, TMPO-AS1 knockdown significantly inhibited (P<0.05, P<0.01) the viability of GBC-SD and NOZ cells compared with the sh-NC group, in a time-dependent manner. Furthermore, the colony formation assay was performed to assess the effects of sh-TMPO-AS1 on the proliferation of GBC-SD and NOZ cells. As presented in Fig. 2C, transfection with sh-TMPO-AS1 significantly suppressed (P<0.01) the proliferation of GBC-SD and NOZ cells. The wound healing assay was performed to determine the role of sh-TMPO-AS1 on the migratory ability of GBC-SD and NOZ cells. As presented in Fig. 2D, transfection with sh-TMPO-AS1 significantly inhibited the migratory ability of GBC-SD (P<0.05) and NOZ (P<0.01) cells compared with the sh-NC group. Similarly, the results of the Transwell migration and invasion assays demonstrated that transfection with sh-TMPO-AS1 significantly decreased (P<0.01) the migratory and invasive abilities of GBC-SD and NOZ cells compared with the sh-NC group (Fig. 2E). In addition, the immunofluorescence assay was performed to evaluate the expression of α-SMA associated with EMT. The results demonstrated that transfection with sh-TMPO-AS1 significantly inhibited (P<0.01) α-SMA expression in the sh-NC group (Fig. 2F). Western blot analysis was performed to assess the effects of sh-TMPO-AS1 on the expression levels of EMT-related proteins, including E-cadherin, vimentin and α-SMA. As presented in Fig. 2G, transfection with sh-TMPO-AS1 significantly decreased the protein expression levels of vimentin and α-SMA, and increased E-cadherin protein expression in GBC-SD and NOZ cells compared with the sh-NC group. Collectively, these results suggest that TMPO-AS1 knockdown inhibits the viability, proliferation, migration, invasion and EMT progression of GBC cells.
 |
Figure 2.
TMPO-AS1 knockdown inhibits the proliferation, migration, invasion and EMT of GBC cells. (A) Reverse transcription-quantitative PCR analysis was performed to detect TMPO-AS1 expression in GBC cells transfected with sh-TMPO-AS1. (B) The Cell Counting Kit-8 assay was performed to assess the viability of GBC cells transfected with sh-TMPO-AS1 at the indicated times. (C) The colony formation assay was performed to assess the proliferation of GBC cells transfected with sh-TMPO-AS1. (D) The wound healing assay was performed to assess the migratory ability of GBC cells transfected with sh-TMPO-AS1. (E) Transwell migration and invasion assays were performed to assess the migratory and invasive abilities of GBC cells transfected with sh-TMPO-AS1. (F) The immunofluorescence assay was performed to detect α-SMA protein expression in GBC cells transfected with sh-TMPO-AS1. (G) Western blot analysis was performed to detect the protein expression levels of EMT markers, including E-cadherin, vimentin and α-SMA, in GBC cells transfected with sh-TMPO-AS1. Data are presented as the mean ± SD (n=3). *P<0.05, **P<0.01 vs. sh-NC. TMPO-AS1, thymopoietin antisense transcript 1; EMT, epithelial-to-mesenchymal transition; GBC, gallbladder carcinoma; sh, short hairpin; SMA, smooth muscle actin; NC, negative control; OD, optical density.
|
TMPO-AS1 acts as a competing endogenous RNA (ceRNA) by sponging miR-1179
To determine the potential molecular mechanisms of TMPO-AS1 in GBC, the distribution of TMPO-AS1 was initially investigated via subcellular fraction analysis. As presented in Fig. 3A, TMPO-AS1 was primarily found in the cytoplasm, suggesting that TMPO-AS1 may function at the post-transcriptional level. In addition, to verify whether TMPO-AS1 acta as a ceRNA, bioinformatics analysis was performed to identify miRNAs that potentially bind to TMPO-AS1. As presented in Fig. 3B, miR-1179 was predicted to possess the target domain for TMPO-AS1. Subsequently, miR-1179 mimics and mimic NC were transfected into GBC-SD and NOZ cells, and RT-qPCR analysis was performed. As presented in Fig. 3C, miR-1179 expression was significantly upregulated (P<0.01) in GBC-SD and NOZ cells transfected with miR-1179 mimics. The dual-luciferase reporter assay was performed to validate the interaction between TMPO-AS1 and miR-1179. As expected, exogenous miR-1179 expression significantly decreased (P<0.01) the luciferase intensity of GBC-SD and NOZ cells transfected with TMPO-AS1-WT (Fig. 3D). Similarly, the RNA pull-down assay determined that TMPO-AS1 was highly enriched in GBC-SD and NOZ cells treated with WT miR-1179 (Fig. 3E), which confirmed the association between TMPO-AS1 and miR-1179. Furthermore, miR-1179 expression was downregulated (P<0.01) in clinical GBC tissues and cell lines (Fig. 3F and G), and was significantly upregulated (P<0.01) in GBC-SD and NOZ cells transfected with sh-TMPO-AS-1 (Fig. 3H). Pearson's correlation coefficient analysis demonstrated a negative correlation between TMPO-AS1 and miR-1179 expression levels in clinical GBC tissues (Fig. 3I). Taken together, these results suggest that TMPO-AS1 acts as a ceRNA for miR-1179 in GBC.
 |
Figure 3.
TMPO-AS1 acts as a competing endogenous RNA by sponging miR-1179. (A) Subcellular fractionation analysis was performed to determine the cellular location of TMPO-AS1. (B) Binding sites between TMPO-AS1 and miR-1179. (C) RT-qPCR analysis was performed to detect miR-1179 expression in GBC cells transfected with miR-1179 mimics. (D) The dual-luciferase reporter assay was performed to verify the interaction between TMPO-AS1 and miR-1179. (E) The RNA pull-down assay was performed to measure relative enrichment of TMPO-AS1 in GBC cells. (F) RT-qPCR analysis was performed to detect miR-1179 expression in GBC tissues. (G) RT-qPCR analysis was performed to detect miR-1179 expression in GBC cell lines. (H) RT-qPCR analysis was performed to detect miR-1179 expression in GBC cell lines transfected with sh-TMPO-AS1. (I) Pearson's correlation coefficient analysis was performed to assess the correlation between TMPO-AS1 and miR-1179 expression in GBC tissues. Data are presented as the mean ± SD (n=3). **P<0.01 vs. cytoplasmic fraction, NC mimic, adjacent normal tissues, H69 cells or sh-NC; ***P<0.001 vs. IgG. TMPO-AS1, thymopoietin antisense transcript 1; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; GBC, gallbladder carcinoma; sh, short hairpin; NC, negative control; WT, wild-type; MUT, mutant
|
Overexpression of miR-1179 inhibits the proliferation, migration, invasion and EMT of GBC cells
To determine the potential role of miR-1179 in GBC, miR-1179 mimics was transfected into GBC-SD and NOZ cells (Fig. 4A). The CCK-8 assay was performed to assess the effects of miR-1179 mimics on the viability of GBC-SD and NOZ cells. As presented in Fig. 4B, overexpression of miR-1179 significantly inhibited (P<0.05) the viability of GBC-SD and NOZ cells compared with the NC mimic group, in a time-dependent manner. The colony formation assay was performed to assess the effects of miR-1179 mimics on the proliferation of GBC-SD and NOZ cells. As presented in Fig. 4C, overexpression of miR-1179 significantly suppressed (P<0.01) the proliferation of GBC-SD and NOZ cells. The wound healing assay was performed to assess the effects of miR-1179 mimics on the migratory ability of GBC-SD and NOZ cells. As presented in Fig. 4D, transfection with miR-1179 mimics significantly inhibited (P<0.05) the migratory ability of GBC-SD and NOZ cells compared with the NC mimic group. The immunofluorescence assay was performed to detect α-SMA expression associated with EMT. The results demonstrated that transfection with miR-1179 mimics significantly inhibited α-SMA expression compared with the NC mimic group (Fig. 4E). The results of the Transwell migration and invasion assays demonstrated that the migratory and invasive abilities of GBC-SD and NOZ cells significantly decreased (P<0.01) following transfection with miR-1179 mimics compared with the NC mimic group (Fig. 4F). Western blot analysis was performed to assess the effects of miR-1179 mimics on the expression levels of EMT-related proteins, including E-cadherin, vimentin and α-SMA. As presented in Fig. 4G, transfection with miR-1179 mimics significantly decreased the protein expression levels of vimentin and α-SMA, and increased E-cadherin protein expression in GBC-SD and NOZ cells compared with the NC mimic group. Taken together, these results suggest that overexpression of miR-1179 inhibits the viability, proliferation, migration, invasion and EMT progression of GBC cells.
 |
Figure 4.
Overexpression of miR-1179 inhibits the proliferation, migration, invasion and EMT of GBC cells. (A) Reverse transcription-quantitative PCR analysis was performed to detect miR-1179 expression in GBC cells transfected with miR-1179 mimics. (B) The Cell Counting Kit-8 assay was performed to assess the viability of GBC cells transfected with miR-1179 mimics at the indicated times. (C) The colony formation assay was performed to assess the proliferation of GBC cells transfected with miR-1179 mimics. (D) The wound healing assay was performed to assess the migratory ability of GBC cells transfected with miR-1179 mimics. (E) The immunofluorescence assay was performed to detect α-SMA protein expression in GBC cells transfected with miR-1179 mimics. (F) The Transwell migration and invasion assays were performed to assess the migratory and invasive abilities of GBC cells transfected with miR-1179 mimics. (G) Western blot analysis was performed to detect the protein expression levels of the EMT markers, including E-cadherin, vimentin and α-SMA. Data are presented as the mean ± SD (n=3). *P<0.05, **P<0.01 vs. NC mimic. miR, microRNA; EMT, epithelial-to-mesenchymal transition; GBC, gallbladder carcinoma; TMPO-AS1, thymopoietin antisense transcript 1; SMA, smooth muscle actin; NC, negative control; OD, optical density.
|
E2F2 is a direct target gene of miR-1179 and is negatively correlated with miR-1179
miRNAs function as oncogenes or tumor suppressors by regulating downstream target genes (20). A complementary sequence of miR-1179 was identified in the 3′-untranslated region (UTR) of E2F2 (Fig. 5A). The results of the dual-luciferase reporter assay confirmed that E2F2 is a direct target gene of miR-1179 (Fig. 5B). In addition, RT-qPCR analysis was performed to detect E2F2 expression in clinical GBC tissues and cell lines. As presented in Fig. 5C and D, E2F2 was highly expressed (P<0.01) in clinical GBC tissues and cell lines. Furthermore, RT-qPCR and western blot analyses were performed to detect the mRNA and protein expression levels of E2F2 in GBC-SD and NOZ cells transfected with miR-1179 mimics or NC mimic, respectively. As presented in Fig. 5E and F, the mRNA and protein expression levels of E2F2 significantly decreased (P<0.01) in GBC-SD and NOZ cells transfected with miR-1179 mimics. Notably, a negative correlation was observed between miR-1179 and E2F2 expression in clinical GBC tissues (Fig. 5G). Collectively, these results suggest that E2F2 is direct target gene of miR-1179, and is negatively correlated with miR-1179 expression in GBC.
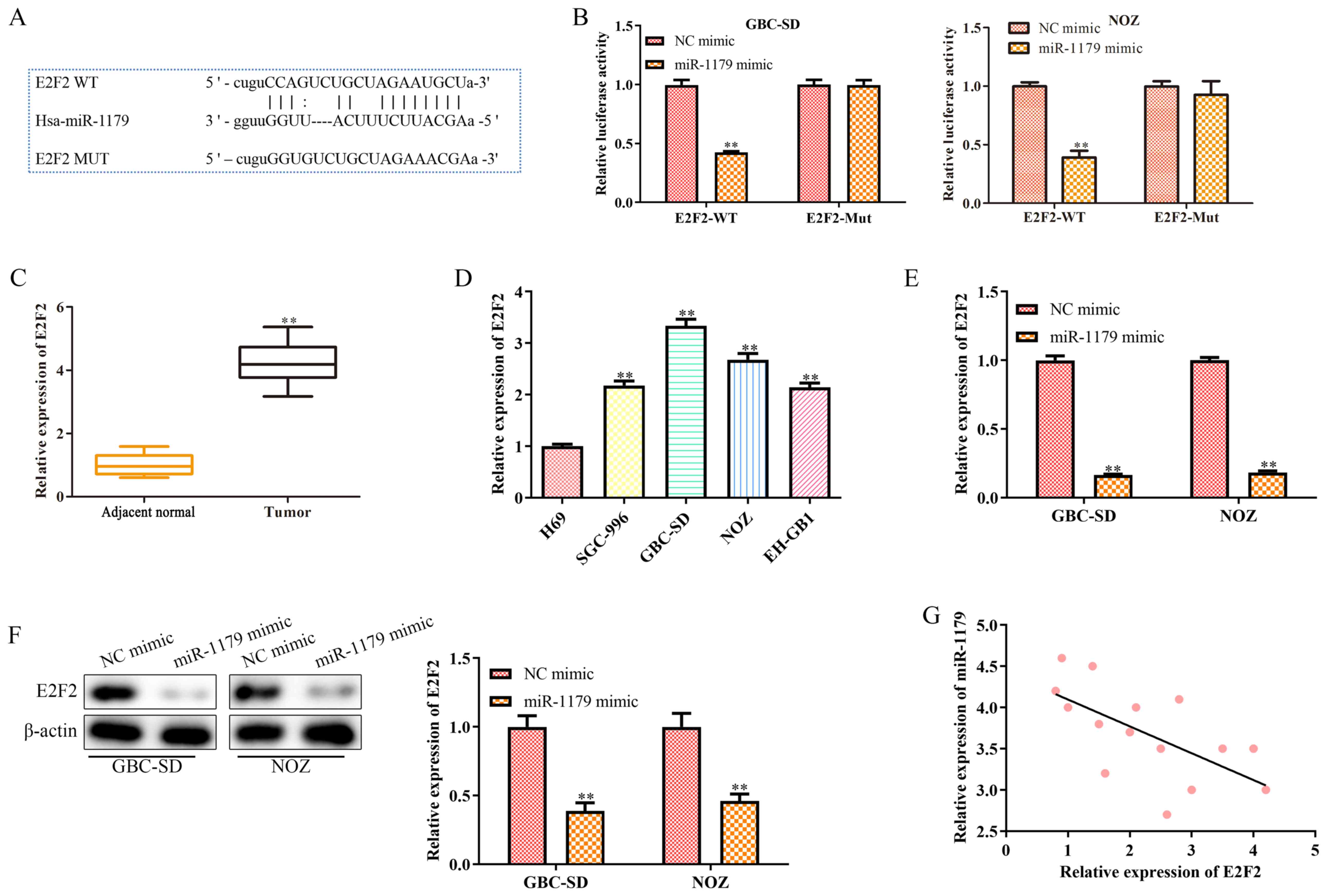 |
Figure 5.
E2F2 is a direct target gene of miR-1179 and is negatively correlated with miR-1179 expression. (A) Binding sites between miR-1179 and E2F2. (B) The dual-luciferase reporter assay was performed to verify the interaction between miR-1179 and E2F2. (C) RT-qPCR analysis was performed to detect E2F2 expression in GBC tissues. (D) RT-qPCR analysis was performed to detect E2F2 expression in GBC cell lines. (E) RT-qPCR and (F) western blot analyses were performed to detect E2F2 expression in GBC cells transfected with miR-1179 mimics. (G) Pearson's correlation coefficient analysis was performed to assess the correlation between miR-1179 and E2F2 expression in GBC tissues. Data are presented as the mean ± SD (n=3). **P<0.01 vs. NC mimic, adjacent normal tissues, H69 cells or NC mimic. E2F2, E2F transcription factor 2; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; GBC, gallbladder carcinoma; NC, negative control; WT, wild-type; MT, mutant.
|
TMPO-AS1 regulates the proliferation, migration, invasion and EMT of GBC cells by modulating the miR-1179/E2F2 pathway
To further determine whether TMPO-AS1 exhibits its role by regulating the miR-1179/E2F2 pathway, the present study performed rescue assays via transfection with miR-1179 inhibitor or sh-E2F2 (Fig. 6A). CCK-8 and colony formation assays were performed. As presented in Fig. 6B and C, TMPO-AS1 knockdown inhibited the viability and proliferation of GBC-SD and NOZ cells, respectively. However, transfection with miR-1179 inhibitor promoted GBC cell viability and proliferation, while E2F2 knockdown exhibited inhibitory effects on the viability and proliferation of GBC cells. The wound healing and Transwell migration and invasion assays were performed, and the results demonstrated that TMPO-AS1 knockdown inhibited the migration and invasion of GBC-SD and NOZ cells. However, transfection with miR-1179 inhibitor promoted GBC cell migration and invasion, while E2F2 knockdown suppressed the migratory and invasive abilities of GBC cells (Fig. 6D and E). Immunofluorescence and western blot analysis were performed. As presented in Fig. 6F and G, TMPO-AS1 knockdown inhibited the EMT progression of GBC-SD and NOZ cells, the effects of which were reversed following transfection with miR-1179 inhibitor. Notably, E2F2 knockdown suppressed the effects of EMT progression on GBC cells. Taken together, these results suggest that TMPO-AS1 and E2F2 act as oncogenes, while miR-1179 exerts tumor suppressive roles in GBC and TMPO-AS1 exerts its roles by the modulating the miR-1179/E2F2 signaling pathway.
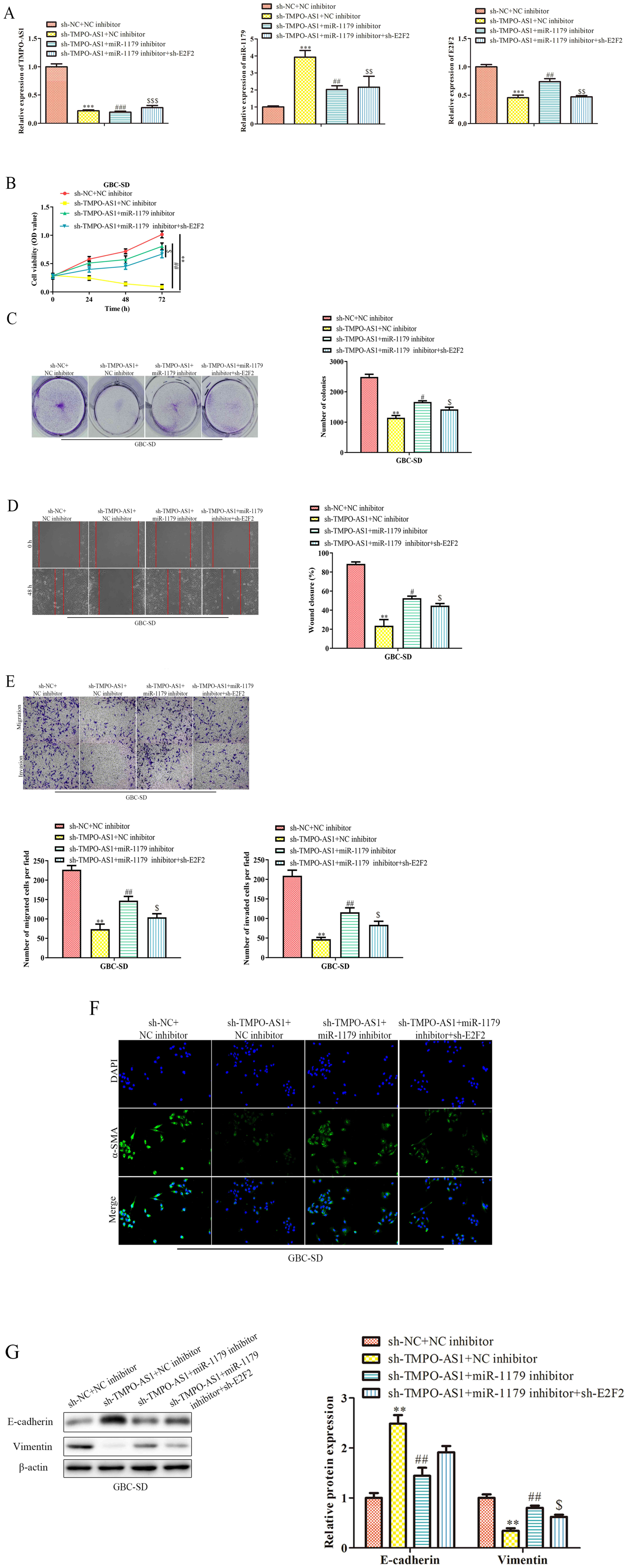 |
Figure 6.
TMPO-AS1 regulates the proliferation, migration, invasion and EMT of GBC cells by modulating the miR-1179/E2F2 pathway. (A) Reverse transcription-quantitative PCR analysis was performed to detect the expression levels of TMPO-AS1, miR-1179 and E2F2 in GBC cells following transfection. (B) The Cell Counting Kit-8 assay was performed to assess the viability of GBC cells following transfection at the indicated times. (C) The colony formation assay was performed to assess the proliferation of GBC cells following transfection. (D) The wound healing assay was performed to assess the migration of GBC cells following transfection. (E) The Transwell migration and invasion assays were performed to assess the migratory and invasive abilities of GBC cells following transfection. (F) The immunofluorescence assay was performed to detect α-SMA protein expression in GBC cells following transfection. (G) Western blot analysis was performed to detect the protein expression levels of EMT markers, including E-cadherin and vimentin. Data are presented as the mean ± SD (n=3). **P<0.01 and ***P<0.001 vs. sh-NC + NC inhibitor; #P<0.05, ##P<0.01 and ###P<0.001 vs. sh-TMPO-AS1 + NC inhibitor; $P<0.05, $$P<0.01 and $$$P<0.001 vs. sh-TMPO-AS1 + miR-1179 inhibitor. TMPO-AS1, thymopoietin antisense transcript 1; EMT, epithelial-to-mesenchymal transition; miR, microRNA; E2F2, E2F transcription factor 2; GBC, gallbladder carcinoma; sh, short hairpin; NC,negative control; OD, optical density.
|
Discussion
GBC is the most common type of biliary tract cancer (BTC) and is more prevalent in women (21). Due to GBC atypical early symptoms, the poor results of late surgical treatment and early metastasis and diffusion, the 5-year survival rate across all tumor stages range from 5–20% (21,22). Thus, targeted treatment of GBC has become a focus of research investigations (23). lncRNAs, as a group of non-coding RNAs, are abnormally expressed in GBC tissues (24). lncRNAs can further promote or inhibit the proliferation, invasion and metastasis of GBC by regulating upstream genes or directly binding with miRNAs to regulate downstream proteins (24). Therefore, an in-depth study of lncRNAs may provide a new research direction for the early diagnosis and targeted treatment of GBC in the future.
Increasing evidence has found that aberrant expression of lncRNAs function as oncogenes or tumor suppressive genes (25). Previous studies have reported that TMPO-AS1 was highly expressed in different types of cancer, including breast, cervical and non-small cell lung cancers (26–28). TMPO-AS1 could acts as an oncogene in the occurrence and development of different types of cancer. For example, Yang et al (27) found that TMPO-AS1 was highly expressed in cervical cancer cell lines, while Mu et al (29) reported that TMPO-AS1 was highly expressed in lung adenocarcinoma tissues and cell lines. The results of the present study demonstrated that TMPO-AS1 was significantly upregulated in clinical GBC tissues and cell lines. Furthermore, patients with GBC with low TMPO-AS1 expression exhibited higher overall survival rates than those with high TMPO-AS1 expression. Notably, Mitobe et al (26) found that low TMPO-AS1 expression predicts a better prognosis in patients with cancer. This is consistent with the results of the present study, suggesting that TMPO-AS1 may act as an oncogene in the occurrence and progression of GBC.
In addition, Mu et al (29) reported that TMPO-AS1 knockdown significantly inhibits lung adenocarcinoma cell proliferation and invasion and induces apoptosis and G1/S arrest by negatively regulating miR-383-5p expression. Guo et al (16) reported that TMPO-AS1 knockdown significantly inhibits HCC cell proliferation, invasion, migration and EMT processes, as well as tumor growth in vivo by modulating miR-329-3p/FOXK1. Cui et al (30) found that TMPO-AS1 is highly expressed in osteosarcoma tissues and cells, and TMPO-AS1 knockdown suppresses cell proliferation and induces apoptosis of osteosarcoma cells by regulating the miR-199a-5p/WNT7B axis. These findings provide a potential therapeutic target for patients with cancer. As expected, in the present study, TMPO-AS1 knockdown inhibited the viability, proliferation, migration, invasion and EMT progression of GBC cells. These results are consistent with those observed in previous studies, suggesting that abnormal TMPO-AS1 expression in GBC may play an important role in GBC progression.
lncRNAs serve as ceRNAs to induce the expression of target mRNAs by sequestering miRNAs in several cellular process, including cancer progression (24). TMPO-AS1, as an oncogene, has been reported to exert its role by regulating miRNAs. For example, TMPO-AS1 is abnormally upregulated in retinoblastoma tissues and regulates the proliferation and migration of retinoblastoma cells by sponging miR-199a-5p expression (31). Furthermore, TMPO-AS1 knockdown attenuates ovarian cancer progression by inhibiting invasion and metastasis by regulating miR-200c expression (32). In the present study, bioinformatics analysis was performed to retrieve miRNAs that interact with TMPO-AS1. Among the candidates, miR-1179 was selected due to its inhibitory role in different types of tumors, including gastric, breast and pancreatic cancer (33–36). Li and Qin (33), Li et al (34), Lin et al (35) and Zhang et al (36) found that miR-1179 was expressed at low levels in gastric, breast, pancreatic and ovarian cancers, respectively. The present study demonstrated that miR-1179 expression was decreased in GBC tissues and cells, and could be negatively post transcriptionally regulated by TMPO-AS1. In addition, mechanistic analysis revealed that overexpression of miR-1179 inhibited the viability, proliferation, migration, invasion and EMT progression of GBC cells. Notably, miR-1179 knockdown reversed the inhibitory effects of TMPO-AS1 knockdown on the proliferation and metastasis of GBC cells, suggesting that TMPO-AS1 knockdown exerts an inhibitory role in the occurrence and progression of GBC, potentially by regulating miR-1179.
The present study predicted that the 3′-UTR of E2F2 contained complementary sites to miR-1179, suggesting that E2F2 may be a target gene of miR-1179. E2F2 is a member of the E2F transcription factor family (37). In mammals, E2F2 mainly regulates cell proliferation, DNA replication, DNA repair, differentiation, apoptosis in a cell cycle-dependent manner (38). Previous studies have reported that E2F2 has strong carcinogenic abilities, maintains cell homeostasis together with p53 protein and plays a precise regulatory role in cell transformation and cell cycle progression (38,39). Currently, high E2F2 expression is associated with low overall survival rates in patients with cancer, including ovarian cancer, colon cancer and glioblastoma multiforme and acts as a tumor suppressor (40–43). Furthermore, E2F2 is involved in the occurrence and progression of cancers meditated by a lncRNA/miRNA axis. For example, differentiation antagonizing non-protein coding RNA (DANCR) is significantly upregulated in clinical pancreatic cancer tissues and cell lines, and DANCR knockdown inhibits the proliferation and metastasis of pancreatic cancer cells by positively modulating E2F2 expression via miR-214-5p (44). NNT-antisense RNA 1 (NNT-AS1) is upregulated in lung cancer tissues and cells, and NNT-AS1 knockdown attenuates proliferation and invasion but enhances the apoptosis of lung cancer cells by regulating the miR-3666/E2F2 axis (45). In the present study, E2F2 expression was upregulated in GBC tissues and cells, and it was revealed to be a target of miR-1179, as confirmed via the dual-luciferase reporter assay, which was consistent with bioinformatics prediction results. In addition, the mechanisms of the TMPO-AS1/miR-1179/E2F2 axis were further assessed via rescue experiments. As expected, E2F2 knockdown partly restored the effects of miR-1179 inhibitor on the viability, proliferation, migration, invasion and EMT progression of GBC cells transfected with sh-TMPO-AS1.
However, the results of the present study are limited to in vitro studies. At present, there is no evidence to show the effect of TMPO-AS1 on tumor growth or metastasis in vivo and its clinical significance in GBC. Therefore, more in-depth research is required.
In conclusion, the results of the present study demonstrated that TMPO-AS1 was significantly upregulated in GBC tissues and cell lines, and high TMPO-AS1 expression predicted the poor prognosis of patients with GBC. In addition, TMPO-AS1 knockdown inhibited the viability, proliferation, migration, invasion and EMT progression of GBC cells by regulating the miR-1179/E2F2 axis, suggesting that TMPO-AS1 may be used as an effective target for treating patients with GBC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZS and XS conceived and designed the present study. Both authors performed the literature review, data extraction, the experiments and analyzed the data, drafted the initial manuscript and confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The People's Hospital of Danyang and Affiliated Danyang Hospital of Nantong University (Jiangsu, Chain; approval no. 20190621) and written informed consent was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Sharma A, Sharma KL, Gupta A, Yadav A and Kumar A: Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol. 23:3978–3998. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baiu I and Visser B: Gallbladder Cancer. JAMA. 320:12942018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hickman L and Contreras C: Gallbladder Cancer: Diagnosis, Surgical Management, and Adjuvant Therapies. Surg Clin North Am. 99:337–355. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yifan T, Zheyong L, Miaoqin C, Liang S and Xiujun C: A predictive model for survival of gallbladder adenocarcinoma. Surg Oncol. 27:365–372. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vega EA, Sanhueza M and Viñuela E: Minimally Invasive Surgery for Gallbladder Cancer. Surg Oncol Clin N Am. 28:243–253. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhan A, Soleimani M and Mandal SS: Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 77:3965–3981. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y and Tang L: The Application of lncRNAs in Cancer Treatment and Diagnosis. Recent Patents Anticancer Drug Discov. 13:292–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Castro-Oropeza R, Melendez-Zajgla J, Maldonado V and Vazquez-Santillan K: The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol (Dordr). 41:585–603. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Liu H, Shen X, Wang Y, Zhang D, Shen S, Suo T, Pan H, Ming Y, Ding K, et al: Long non-coding RNA expression profiles in gallbladder carcinoma identified using microarray analysis. Oncol Lett. 13:3508–3516. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu YP, Jin YP, Wu XS, Yang Y, Li YS, Li HF, Xiang SS, Song XL, Jiang L, Zhang YJ, et al: LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol Cancer. 18:1672019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji D, Zhong X, Jiang X, Leng K, Xu Y, Li Z, Huang L, Li J and Cui Y: The role of long non-coding RNA AFAP1-AS1 in human malignant tumors. Pathol Res Pract. 214:1524–1531. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y, Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 18:332019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang C, Yang P, Han T, Wang RY, Xing XL, Si AF, Ma QY, Chen Z, Li HY and Zhang B: Long non-coding RNA DILC promotes the progression of gallbladder carcinoma. Gene. 694:102–110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Cheng X, Ge N, Guo W, Feng F and Wan F: Long non-coding RNA SPRY4-IT1 promotes gallbladder carcinoma progression. Oncotarget. 8:3104–3110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang W, Su X, Yan W, Kong Z, Wang D, Huang Y, Zhai Q, Zhang X, Wu H, Li Y, et al: Overexpression of AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and poor prognosis in prostate cancer. Prostate. 78:1248–1261. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo X and Wang Y: LncRNA TMPO-AS1 promotes hepatocellular carcinoma cell proliferation, migration and invasion through sponging miR-329-3p to stimulate FOXK1-mediated AKT/mTOR signaling pathway. Cancer Med. 9:5235–5246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gang X, Yuan M and Zhang J: Long Non-Coding RNA TMPO-AS1 Promotes Cervical Cancer Cell Proliferation, Migration, and Invasion by Regulating miR-143-3p/ZEB1 Axis. Cancer Manag Res. 12:1587–1599. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo H, Yang L, Liu C, Wang X, Dong Q, Liu L and Wei Q: TMPO-AS1/miR-98-5p/EBF1 feedback loop contributes to the progression of bladder cancer. Int J Biochem Cell Biol. 122:1057022020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edge SB and Compton CC: The American Joint Committee on Cancer: the 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann Surg Oncol. 17:1471–1474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hua CB, Song SB, Ma HL and Li XZ: MiR-1-5p is down-regulated in gallbladder carcinoma and suppresses cell proliferation, migration and invasion by targeting Notch2. Pathol Res Pract. 215:200–208. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nepal C, Zhu B, O'Rourke CJ, Bhatt DK, Lee D, Song L, Wang D, Van Dyke AL, Choo-Wosoba H, Liu Z, et al: Integrative molecular characterisation of gallbladder cancer reveals micro-environment-associated subtypes. J Hepatol. 74:1132–1144. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khan MR and Begum S: Extended resection for xanthogranulomatous cholecystitis mimicking gallbladder carcinoma: Cases and review of diagnostic approach. J Pak Med Assoc. 69:256–260. 2019.PubMed/NCBI
|
|
23
|
Yadav S, DE Sarkar N, Kumari N, Krishnani N, Kumar A and Mittal B: Targeted Gene Sequencing of Gallbladder Carcinoma Identifies High-impact Somatic and Rare Germline Mutations. Cancer Genomics Proteomics. 14:495–506. 2017.PubMed/NCBI
|
|
24
|
Wang SH, Zhang WJ, Wu XC, Weng MZ, Zhang MD, Cai Q, Zhou D, Wang JD and Quan ZW: The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J Cell Mol Med. 20:2299–2308. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng WX, Koirala P and Mo YY: LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mitobe Y, Ikeda K, Suzuki T, Takagi K, Kawabata H, Horie-Inoue K and Inoue S: ESR1-Stabilizing Long Noncoding RNA TMPO-AS1 Promotes Hormone-Refractory Breast Cancer Progression. Mol Cell Biol. 39:e00261–e00269. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang J, Liang B and Hou S: TMPO-AS1 promotes cervical cancer progression by upregulating RAB14 via sponging miR-577. J Gene Med. 21:e31252019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qin Z, Zheng X and Fang Y: Long noncoding RNA TMPO-AS1 promotes progression of non-small cell lung cancer through regulating its natural antisense transcript TMPO. Biochem Biophys Res Commun. 516:486–493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mu X, Wu H, Liu J, Hu X, Wu H, Chen L, Liu W, Luo S and Zhao Y: Long noncoding RNA TMPO-AS1 promotes lung adenocarcinoma progression and is negatively regulated by miR-383-5p. Biomed Pharmacother. 125:1099892020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cui H and Zhao J: LncRNA TMPO-AS1 serves as a ceRNA to promote osteosarcoma tumorigenesis by regulating miR-199a-5p/WNT7B axis. J Cell Biochem. 121:2284–2293. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng X, Yan J and Cheng F: LncRNA TMPO-AS1 up-regulates the expression of HIF-1α and promotes the malignant phenotypes of retinoblastoma cells via sponging miR-199a-5p. Pathol Res Pract. 216:1528532020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mitobe Y, Ikeda K, Sato W, Kodama Y, Naito M, Gotoh N, Miyata K, Kataoka K, Sasaki H, Horie-Inoue K, et al: Proliferation-associated long noncoding RNA, TMPO-AS1, is a potential therapeutic target for triple-negative breast cancer. Cancer Sci. 111:2440–2450. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Y and Qin C: MiR-1179 inhibits the proliferation of gastric cancer cells by targeting HMGB1. Hum Cell. 32:352–359. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li WJ, Xie XX, Bai J, Wang C, Zhao L and Jiang DQ: Increased expression of miR-1179 inhibits breast cancer cell metastasis by modulating Notch signaling pathway and correlates with favorable prognosis. Eur Rev Med Pharmacol Sci. 22:8374–8382. 2018.PubMed/NCBI
|
|
35
|
Lin C, Hu Z, Yuan G, Su H, Zeng Y, Guo Z, Zhong F, Jiang K and He S: MicroRNA-1179 inhibits the proliferation, migration and invasion of human pancreatic cancer cells by targeting E2F5. Chem Biol Interact. 291:65–71. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang ZH, Chen RB, Lu LP, Mao AP, Guo H, Wu YT and Shan ZX: MicroRNA-1179 regulates proliferation and chemosensitivity of human ovarian cancer cells by targeting the PTEN-mediated PI3K/AKT signaling pathway. Arch Med Sci. 16:907–914. 2019. View Article : Google Scholar
|
|
37
|
Wang S, Wang L, Wu C, Sun S and Pan JH: E2F2 directly regulates the STAT1 and PI3K/AKT/NF-κB pathways to exacerbate the inflammatory phenotype in rheumatoid arthritis synovial fibroblasts and mouse embryonic fibroblasts. Arthritis Res Ther. 20:2252018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Clijsters L, Hoencamp C, Calis JJA, Marzio A, Handgraaf SM, Cuitino MC, Rosenberg BR, Leone G and Pagano M: Cyclin F Controls Cell-Cycle Transcriptional Outputs by Directing the Degradation of the Three Activator E2Fs. Mol Cell. 74:1264–1277.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Iglesias-Ara A, Zenarruzabeitia O, Buelta L, Merino J and Zubiaga AM: E2F1 and E2F2 prevent replicative stress and subsequent p53-dependent organ involution. Cell Death Differ. 22:1577–1589. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou Q, Zhang F, He Z and Zuo MZ: E2F2/5/8 Serve as Potential Prognostic Biomarkers and Targets for Human Ovarian Cancer. Front Oncol. 9:1612019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yao H, Lu F and Shao Y: The E2F family as potential biomarkers and therapeutic targets in colon cancer. Peer J. 8:e85622020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Y, Han D, Wei W, Cao W, Zhang R, Dong Q, Zhang J, Wang Y and Liu N: MiR-218 Inhibited Growth and Metabolism of Human Glioblastoma Cells by Directly Targeting E2F2. Cell Mol Neurobiol. 35:1165–1173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li X, Zhang Z, Jiang H, Li Q, Wang R, Pan H, Niu Y, Liu F, Gu H, Fan X, et al: Circular RNA circPVT1 Promotes Proliferation and Invasion Through Sponging miR-125b and Activating E2F2 Signaling in Non-Small Cell Lung Cancer. Cell Physiol Biochem. 51:2324–2340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yao Z, Chen Q, Ni Z, Zhou L, Wang Y, Yang Y and Huang H: Long Non-Coding RNA Differentiation Antagonizing Nonprotein Coding RNA (DANCR) Promotes Proliferation and Invasion of Pancreatic Cancer by Sponging miR-214-5p to Regulate E2F2 Expression. Med Sci Monit. 25:4544–4552. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang JW, Luo XY, Li ZH and Lang BP: LncRNA NNT-AS1 regulates the progression of lung cancer through the NNT-AS1/miR-3666/E2F2 axis. Eur Rev Med Pharmacol Sci. 24:238–248. 2020.PubMed/NCBI
|




















