Introduction
Acute myeloid leukemia (AML) is a common
hematological malignancy, mainly caused by the clonal proliferation
of progenitor cells or primitive hematopoietic stem cells (1). AML primarily occurs in individuals
>60 years of age, but it can also affect those in other age
groups (1). With the development
of novel cancer therapies, >35% of patients with AML, aged
<35 years, can be effectively cured. However, the survival rate
decreases to ~15% in the elderly population (>60 years)
(2), mainly due to the development
of intolerable chemoresistance and side effects during chemotherapy
(3,4). Homoharringtonine (HHT) has been
widely used as a chemotherapeutic agent against numerous types of
cancer, including AML (5,6). However, the effects of HHT in AML and
the mechanisms underlying the development of HHT resistance remain
largely unknown.
A previous study identified numerous genetic
alterations involved in the development of HHT resistance (7), such as the inactivation of PTEN
(8). It has been reported that
PTEN induced cancer cell apoptosis by inhibiting the PI3K/AKT
cancer cell survival pathway (9).
In addition, another study demonstrated that microRNA
(miRNA/miR)-21, a well-characterized tumor-suppressive miRNA
(10,11), could target PTEN to induce HHT
resistance (12). Results of a
previous study demonstrated that miR-21 regulates cancer cell
proliferation and apoptosis in cervical cancer, and its expression
is negatively regulated by a long non-coding (lnc)RNA MEG3
(13). The lncRNA MIR17HG has been
recently identified as a cancer-related oncogene (14), whose roles in AML remain unclear.
Therefore, the present study aimed to determine the mRNA expression
levels of MIR17HG in AML and to evaluate the potential interaction
between MIR17HG and miR-21 in AML.
Materials and methods
Patients
A total of 40 patients with AML, including 23 men
and 17 women (age range, 52–78 years; mean age, 64.2±4.6 years) and
40 sex- and age-matched healthy individuals (age range, 52–78
years; mean age, 64.1±4.4 years) were enrolled in the present
study. Inclusion criteria: i) Newly diagnosed AML cases at the
Jiangxi Key Laboratory of Cancer Metastasis and Precision
Treatment, The Third Affiliated Hospital of Nanchang University
(Nanchang, China) between May 2015 and May 2018; and ii) patients
willing to participate. Exclusion criteria: i) Other clinical
disorders; and ii) patients with initiated therapy. Clinical
diagnosis was confirmed by histological examination, while no other
clinical disorders were recorded. Signed informed consent was
provided from all participants prior to enrollment in the present
study. The study was approved by the Ethics Committee of the Third
Affiliated Hospital of Nanchang University (Nanchang, China).
Treatment and biopsy
All the patients with AML were treated with
HHT-based chemotherapy. Bone marrow (BM) was collected from both
the patients and healthy participants prior to therapy. BM was also
collected from patients at 3 months following the initiation of
treatment (2–3 mg HHT/m2 per day for 5 days). Healthy
participants underwent BM aspiration biopsy due to suspected blood
and BM diseases, which were ruled out by BM analysis. BM
mononuclear cells (BMMNCs) were isolated using lymphocyte
separation medium (TBD Sciences) and were then immediately frozen
in liquid nitrogen and stored at −80°C.
Cells and transfection
The human AML, Kasumi-6 (American Type Culture
Collection) and the human BM stromal, HS-5, cell lines were used.
The cells were cultured at 37°C in a humidified incubator with 5%
CO2 and relative humidity of 95%. The composition of the
cell culture medium was 20% FBS and 80% RPMI-1640 medium (both
Sigma-Aldrich; Merck KGaA). After reaching 70–80% confluency, the
cells were transfected with either plasmids or miRNA mimics. The
pcDNA3.1 vector (Sigma-Aldrich; Merck KGaA) was used as the
backbone to construct the MIR17HG and PTEN overexpression plasmids.
Negative control (NC) miRNA (5′-UUGUGUCAGUGCUGUAGCCGAA-3′) and
miR-21 mimics (5′-UAGCUUAUCAGACUGAUGUUGA-3′) were synthesized by
Sigma-Aldrich (Merck KGaA). Kasumi-6 and HS-5 cells at a density of
1×106 cells/group were transfected with 50 nM miR-21/NC
mimics (NC group) or 10 nM pcDNA3.1 vector (NC
group)/pcDNA3.1-MIR17HG/PTEN plasmids using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc).
Co-transfections with MIR17HG vector and miR-21 mimic or PTEN
vector and miR-21 mimic were also performed. Untransfected cells
served as the control (C) group. Incubation with transfection
mixture was performed for 6 h at 37°C. Then, the cells were washed
with fresh medium, followed by cell culture for further 48 h prior
to the subsequent assays. To analyze the effects of HTT on MIR17HG
expression, Kasumi-6 cells were cultured in medium supplemented
with 50 nM HHT for 24 and 48 h, followed by the detection of
MIR17HG expression using qPCR.
RNA-RNA interaction prediction
The interaction between miR-21 and MIR17HG was
predicted using the IntaRNA database (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp).
The short sequence was miR-21 and the long sequence was
MIR17HG.
TCGA dataset analysis
Analysis of TCGA dataset was performed using the
GEPIA online tool (http://gepia.cancer-pku.cn/) to determine the mRNA
expression levels of MIR17HG. MIR17HG was inputted as the query and
the AML dataset was selected.
RNA and reverse
transcription-quantitative PCR (RT-qPCR) assays
For HHT treatment, the Kasumi-6 cell line was
cultured in medium supplemented with 50 nM HHT for 24 and 48 h.
Total RNA was extracted from the cells using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). To retain miRNAs in
the RNA samples, the RNA precipitation and washing steps were
completed using 85% ethanol. RNA was reverse transcribed into cDNA
using the PrimeScript RT reagent kit (Takara Biotechnology Co.,
Ltd.) with random primers. For qPCR assays, the SYBR Premix Ex Taq
kit (Takara Biotechnology Co., Ltd.) was used. The mRNA expression
levels of MIR17HG and PTEN were determined using GAPDH as the
endogenous reference gene. The expression levels of mature miR-21
were measured with the All-in-OneTM miRNA RT-qPCR kit
(GeneCopoeia, Inc.). All steps, including polyadenylation, RT and
qPCR assays were performed according to the manufacturer's
instructions. U6 served as the endogenous control for miRNA. Fold
changes of gene expression across the samples were calculated using
the 2−ΔΔCq method (15). Each PCR reaction was repeated three
times. The PCR primer sequences were: MIR17HG forward,
5′-TCAGGAGTTCGAGACCAACC-3′ and reverse, 5′-TGCCTCAGCCTCCAGAGTAG-3′;
GAPDH forward, 5′-ACAGTCAGCCGCATCTTCT-3′ and reverse,
5′-CCCAATACGACCAAATCC-3′; PTEN forward, 5′-GAGTTCCCTCAGCCGT-3′ and
reverse, 5′-GAGGTTTCCTCTGGTCC-3′; miR-21 forward,
5′-TAGCTTATCAGACTGATGT-3′ and reverse, 5′-CAGTGCAGGGTCCGAGGT-3′,
taken from the study by Xu et al (16). Universal reverse primer and U6
forward primer were included in the kit. The PCR thermocycling
conditions were: 95°C for 1 min, followed by 40 cycles of 95°C for
10 sec and 58°C for 55 sec.
Luciferase reporter assay
The MIR17HG cDNA was sub-cloned into the psiCHECK-2
vector (Promega Corporation) and the Kasumi-6 and HS-5 cells were
then co-transfected with MIR17HG plasmid and miR-21 or NC mimics
using the same method as described in the Cells and
transfection section. Following cell culture, the
Dual-Luciferase reporter assay system (Promega Corporation) was
used to assess the luciferase activity at 48 h post-transfection.
The ratios of Renilla and firefly luciferase were
calculated.
Western blot analysis
Following transfection for 48 h, Kasumi-6 and HS-5
cells were harvested and washed twice with PBS. Subsequently, total
protein was extracted using a RIPA solution (Sigma-Aldrich; Merck
KGaA) and protein concentration was measured utilizing a BCA kit.
Following denaturation in boiling water for 15 min, the proteins
were separated with 10% SDS-PAGE, then transferred onto PVDF
membranes and blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) in
TBS-Tween-20 (0.1%). Subsequently, the membranes were incubated
with the primary antibodies against rabbit polyclonal GAPDH (cat.
no. ab9845; 1:1,000) and PTEN (cat. no. ab31392; 1:1,500) (both
from Abcam) at 4°C for 12 h, followed by incubation with the
corresponding HRP goat anti-rabbit (IgG) secondary antibody (cat.
no. ab6721; Abcam; 1:1,000) at 25°C for 2 h. The ECL Western
Blotting Substrate kit (cat. no. ab65623; Abcam) with X-film was
used to visualize the immunoreactive bands and data were normalized
with ImageJ v1.48 software (National Institutes of Health).
Apoptosis detection by flow
cytometry
At 48 h post-transfection, Kasumi-6 and HS-5 cells
were harvested and cultured at 37°C in the presence of 50 µM HHT
for an additional 24 h. Following trypsinization, early apoptotic
cells were analyzed by Annexin V-FITC/PI double staining using the
FACScan® flow cytometer (Becton-Dickinson and Company).
Subsequently, the flow cytometry data were analyzed using the
CellQuest software V.2 (Becton-Dickinson and Company). Each assay
was performed in triplicate.
Statistical analysis
All the data are presented as the mean ± SEM from
three independent replicates. The comparisons between two time
points of the same group were performed using paired Student's
t-test. All data were analyzed using GraphPad Prism v6 (GraphPad
Software, Inc.). The differences between two groups were compared
with an unpaired Student's t-test, while those among multiple
groups were analyzed using one-way ANOVA followed by Tukey's post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
HHT-based chemotherapy further
decreases the reduced MIR17HG mRNA expression levels in AML
TCGA dataset was analyzed using the online GEPIA
database and the analysis revealed that MIR17HG was downregulated
in AML compared with that in non-AML tissues (6.91 vs. 32.83,
respectively). To further verify the mRNA expression profile of
MIR17HG in AML, its differential expression was evaluated in BMMNCs
derived from both patients with AML (n=40) and healthy subjects
(n=40) using qPCR. Compared with that in the control group, the
mRNA expression levels of MIR17HG were significantly reduced in the
AML group (Fig. 1A; P<0.01). In
addition, the mRNA expression level of MIR17HG was also determined
using qPCR in BMMNCs from patients with AML (n=40) at 3 months
following chemotherapy. The results showed that MIR17HG was also
significantly decreased at 3 months post-treatment compared with
that in the pre-treatment group (Fig.
1B; P<0.01). The mRNA expression levels of MIR17HG in the
Kasumi-6 AML cell line and the HS-5 cell line were also detected
using RT-qPCR and the results showed that the mRNA expression level
of MIR17HG was decreased in the Kasumi-6 cell line compared with
that in the HS-5 cell line (Fig.
1C; P<0.01).
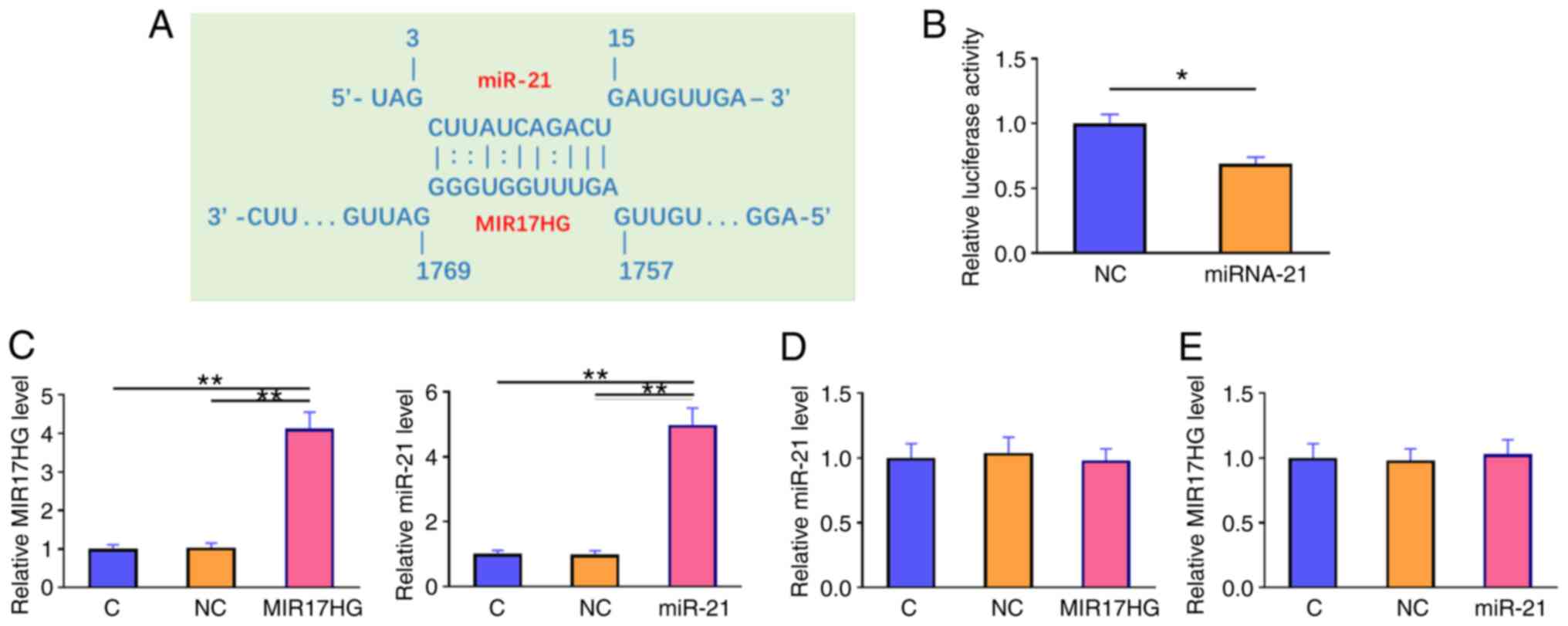
miR-21 binds to MIR17HG without
affecting its expression
The interaction between miR-21 and MIR17HG was
predicted using the IntaRNA database (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp).
The analysis revealed that miR-21 could form strong base pairing
with MIR17HG (Fig. 2A). A
dual-luciferase reporter assay was performed to further verify the
interaction between these molecules. The results showed that,
compared with that in cells transfected with NC mimics and MIR17HG
overexpression plasmid (NC group), cells transfected with miR-21
mimics and MIR17HG (miR-21 group) exhibited significantly reduced
luciferase activity (Fig. 2B;
P<0.05). Furthermore, cells were transfected with MIR17HG
overexpression plasmid or miR-21 mimics. RT-qPCR analysis confirmed
the overexpression of MIR17HG and miR-21 at 48 h post-transfection
(Fig. 2C; P<0.01). However, the
results showed that MIR17HG overexpression had no effect on the
expression level of miR-21 (Fig.
2D). Accordingly, the expression level of MIR17HG was not
altered in cells overexpressing miR-21 (Fig. 2E).
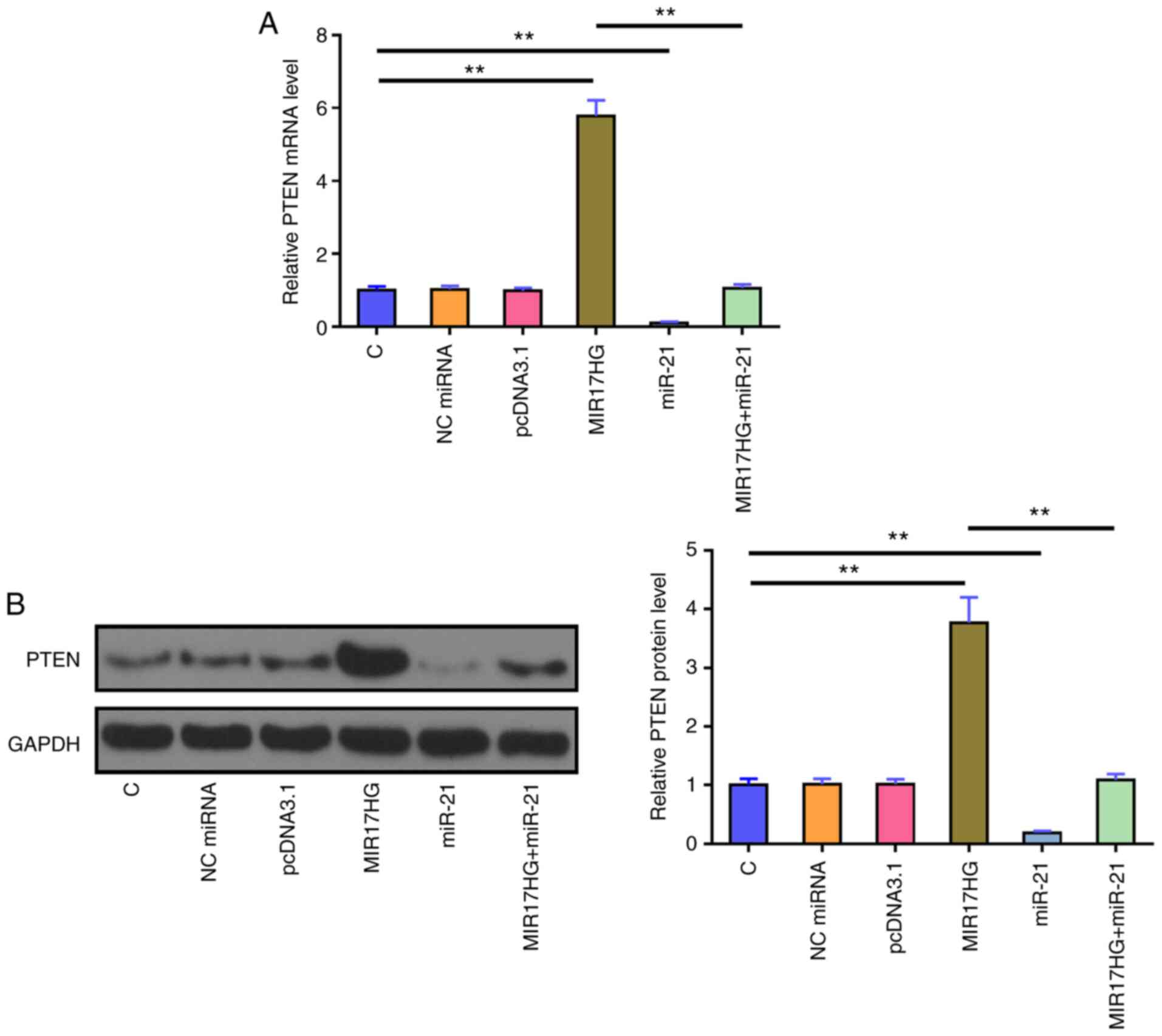
MIR17HG overexpression upregulates
PTEN
The effects of MIR17HG and miR-21 overexpression on
the mRNA (Fig. 3A) and protein
(Fig. 3B) expression levels of
PTEN were assessed using RT-qPCR and western blot analysis,
respectively. The results showed that, compared with that in the
untransfected cells (C group) and cells transfected with NC mimics
(NC group), miR-21 overexpression significantly reduced the mRNA
and protein expression level of PTEN. By contrast, MIR17HG
overexpression notably upregulated PTEN mRNA and protein expression
levels compared with that in the control groups. Furthermore,
co-transfection of miR-21 mimic and MIR17HG expression vector
showed that miR-21 overexpression restored the effect of MIR17HG
overexpression on the expression of PTEN (P<0.01).
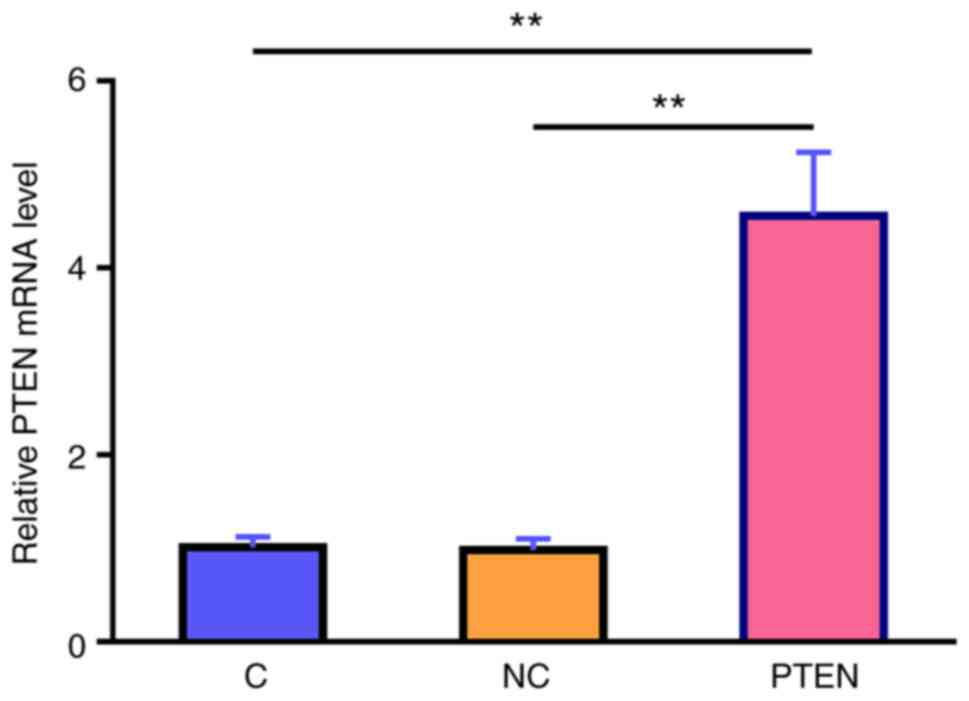
MIR17HG overexpression regulates cell
apoptosis via the miR-21/PTEN axis
Subsequently, the mRNA expression levels of MIR17HG
in the Kasumi-6 cell line, cultured in medium supplemented with 50
nM HHT for 24 and 48 h, were detected using qPCR. The results
demonstrated that, compared with that in the control group
(non-cisplatin), cell treatment with HHT markedly decreased the
mRNA expression levels of MIR17HG in a time-dependent manner
(Fig. 4A; P<0.01). PTEN
overexpression in the Kasumi-6 cell line was verified using RT-qPCR
(Fig. 5) and the effects of
MIR17HG, miR-21 and PTEN overexpression on Kasumi-6 cell apoptosis
were assessed using a cell apoptosis assay (Fig. 4B). Compared with that in the
untransfected cells (C group) and those transfected with empty
vector (NC group), HHT-induced apoptosis was further enhanced in
the Kasumi-6 cell line transfected with MIR17HG or PTEN
overexpression plasmids. However, co-transfection (MIR17HG+miR-21
or MIR17HG+miR-21) showed that miR-21 overexpression exerted the
opposite effect and restored the effect of MIR17HG and PTEN
overexpression on cell apoptosis (P<0.01 or P<0.05).
Discussion
The current study aimed to investigate the
interactions between MIR17HG and miR21, and with PTEN. The results
showed that MIR17HG mRNA expression level was decreased in AML and
could regulate the miR21/PTEN axis to promote HHT-induced apoptosis
of AML cells. MIR17HG is the host gene of senescence-associated
miRNAs (14). It has been reported
that MIR17HG-derived senescence-associated miRNAs may have
oncogenic roles (14). However,
the effect of lncRNA MIR17HG itself on tumorigenesis has not been
fully investigated. Bioinformatics analysis using TCGA datasets
revealed that MIR17HG expression was decreased in AML. Notably, the
mRNA expression levels of MIR17HG were decreased in AML compared
with that in non-AML tissues, supporting its potential
tumor-suppressive role. Consistently, the results of the present
study showed that MIR17HG mRNA expression level was decreased in
AML (Fig. 1A). In addition,
HHT-induced AML cell apoptosis was enhanced following MIR17HG
overexpression (Fig. 4B). The
aforementioned findings indicated that MIR17HG could exert a
tumor-suppressive role in HTT-treated AML by enhancing the
chemosensitivity of AML cells. However, the role of MIR17HG-derived
miRNAs was not evaluated in the present study, thus providing
future research directions.
It has been well established that miR-21 targets
PTEN to mediate the development of chemoresistance in different
types of cancer cells, such as those of ovarian and lung cancer
(10,17). Therefore, regulating the
miR-21/PTEN axis could increase the chemoresistance of cancer cells
to HHT. To the best of our knowledge, the present study was the
first to report that miR-21 could also target PTEN in AML cells to
increase their sensitivity to HHT. It is worth noting that patients
were usually treated with 2–3 mg/m2 HHT per day. An
adult usually has ~5,000 ml blood in the body. Considering drug
metabolism, it was estimated that the HHT concentration in patient
blood should be ~50 nM. The results showed that MIR17HG could
directly interact with miR-21 (Fig.
2), while miR-21 and MIR17HG overexpression had no effect on
the expression of each molecule (Fig.
2). Therefore, it was hypothesized that MIR17HG could not be a
downstream target of miR-21. By contrast, MIR17HG overexpression
attenuated the effects of miR-21 overexpression on the HHT-induced
cancer cell apoptosis and expression of PTEN (18). It has been reported that lncRNAs
may sponge miRNAs and reduce their effects on downstream targets
(19). The role of a miRNA sponge
is to suppress the function of the miRNA, but may not affect the
expression of the miRNA. Therefore, MIR17HG could sponge miR-21 to
upregulate PTEN, thereby promoting cancer cell apoptosis. In
addition to acting as a host gene for multifunctional miRNAs,
MIR17HG could also exert cancer-related functions (20,21).
It is worth noting that the cell apoptotic rate in all the groups
was low (below 15%); therefore the differences were not
notable.
Recently, it has been reported that MIR17HG was
associated with several types of cancer, such as non-small cell
lung cancer and cervical squamous cell carcinoma (22,23).
Notably, genetic variants in MIR17HG were found to be associated
with the susceptibility and prognosis of glioma in a Chinese Han
population (24). These findings
support the role of MIR17HG as a key regulator in cancer biology
and as a potential target for cancer treatment.
The present study did not further confirm the role
of role of miR-21/PTEN in chemotherapy. The mechanism that mediated
HHT-induced downregulation of MIR17HG is not clear. Future studies
are required to elucidate this mechanism. The targeting of PTEN by
miR-21 was not confirmed in the present study; however, the role of
miR-21 in targeting PTEN has been well established (12).
In conclusion, the current study demonstrated that
MIR17HG was downregulated in AML and its overexpression could
further promote the HHT-induced AML cell apoptosis by sponging
miR-21, which in turn could upregulate PTEN.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Key Project of Nanchang
Science and Technology Program, Hong Ke Zi [grant no. (2020) 133]
and The Science and Technology Program of Jiangxi Province Health
Commission (grant no. 202140001).
Availability of data and materials
The data used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JY and XL the designed the study experiments. JY and
LY performed the experiments. PL and GW analyzed the data. XL
prepared the manuscript. JY and XL confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Third Affiliated Hospital of Nanchang University.
The experiments were performed according to the World Medical
Association Declaration of Helsinki. All patients and healthy
volunteers provided written informed consent prior to their
inclusion in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Khwaja A, Bjorkholm M, Gale RE, Levine RL,
Jordan CT, Ehninger G, Bloomfield CD, Estey E, Burnett A,
Cornelissen JJ, et al: Acute myeloid leukaemia. Nat Rev Dis
Primers. 2:160102016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Estey E and Döhner H: Acute myeloid
leukaemia. Lancet. 368:1894–1907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao H, Shi P, Deng M, Jiang Z, Li Y,
Kannappan V, Wang W, Li P and Xu B: Low dose triptolide reverses
chemoresistance in adult acute lymphoblastic leukemia cells via
reactive oxygen species generation and DNA damage response
disruption. Oncotarget. 7:85515–85528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng HC: The molecular mechanisms of
chemoresistance in cancers. Oncotarget. 8:59950–59964. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan M, Zhang Q, Yuan X, Chen Y and Wu Y:
Synergistic killing effects of homoharringtonine and arsenic
trioxide on acute myeloid leukemia stem cells and the underlying
mechanisms. J Exp Clin Cancer Res. 38:3082019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lam SS, Ho ES, He BL, Wong WW, Cher CY, Ng
NK, Man CH, Gill H, Cheung AM, Ip HW, et al: Homoharringtonine
(omacetaxine mepesuccinate) as an adjunct for FLT3-ITD acute
myeloid leukemia. Sci Transl Med. 8:359ra1292016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuroda J, Kamitsuji Y, Kimura S, Ashihara
E, Kawata E, Nakagawa Y, Takeuichi M, Murotani Y, Yokota A, Tanaka
R, et al: Anti-myeloma effect of homoharringtonine with concomitant
targeting of the myeloma-promoting molecules, Mcl-1, XIAP, and
β-catenin. Int J Hematol. 87:507–515. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi X, Zhu M, Gong Z, Yang T, Yu R, Wang J
and Zhang Y: Homoharringtonine suppresses LoVo cell growth by
inhibiting EphB4 and the PI3K/AKT and MAPK/EKR1/2 signaling
pathways. Food Chem Toxicol. 136:1109602019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JF: The PTEN/PI3K/AKT signalling pathway in cancer,
therapeutic implications. Curr Cancer Drug Targets. 8:187–198.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pink RC, Samuel P, Massa D, Caley DP,
Brooks SA and Carter DR: The passenger strand, miR-21-3p, plays a
role in mediating cisplatin resistance in ovarian cancer cells.
Gynecol Oncol. 137:143–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang SM, Huang C, Li XF, Yu MZ, He Y and
Li J: miR-21 confers cisplatin resistance in gastric cancer cells
by regulating PTEN. Toxicology. 306:162–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Yao T, Wang Y, Yu J, Liu Y and
Lin Z: Long noncoding RNA MEG3 is downregulated in cervical cancer
and affects cell proliferation and apoptosis by regulating miR-21.
Cancer Biol Ther. 17:104–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lopez MF, Niu P, Wang L, Vogelsang M, Gaur
M, Krastins B, Zhao Y, Smagul A, Nussupbekova A, Akanov AA, et al:
Opposing activities of oncogenic MIR17HG and tumor suppressive
MIR100HG clusters and their gene targets regulate replicative
senescence in human adult stem cells. NPJ Aging Mech Dis. 3:72017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu XM, Qian JC, Deng ZL, Cai Z, Tang T,
Wang P, Zhang KH and Cai JP: Expression of miR-21, miR-31, miR-96
and miR-135b is correlated with the clinical parameters of
colorectal cancer. Oncol Lett. 4:339–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu ZL, Wang H, Liu J and Wang ZX:
MicroRNA-21 (miR-21) expression promotes growth, metastasis, and
chemo- or radioresistance in non-small cell lung cancer cells by
targeting PTEN. Mol Cell Biochem. 372:35–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng Y, Zou W, Hu C, Li G, Zhou S, He Y,
Ma F, Deng C and Sun L: Modulation of CASC2/miR-21/PTEN pathway
sensitizes cervical cancer to cisplatin. Arch Biochem Biophys
623-624. 20–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng B, Ye H, Chen J, Cheng D, Cai C, Chen
G, Chen X, Xin H, Tang C and Zeng J: LncRNA TUG1 sponges miR-145 to
promote cancer progression and regulate glutamine metabolism via
Sirt3/GDH axis. Oncotarget. 8:113650–113661. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Militello G, Weirick T, John D, Döring C,
Dimmeler S and Uchida S: Screening and validation of lncRNAs and
circRNAs as miRNA sponges. Brief Bioinform. 18:780–788.
2017.PubMed/NCBI
|
|
21
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing miRNA-lncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei S, Liu J, Li X and Liu X: LncRNA
MIR17HG inhibits non-small cell lung cancer by upregulating
miR-142-3p to downregulate Bach-1. BMC Pulm Med. 20:782020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Zhu C, Xu Z, Wang J, Qian L, Zhou
Q, Shen Z, Zhao W, Xiao W, Chen L and Zhou Y: lncRNA PART1 and
MIR17HG as ΔNp63α direct targets regulate tumor progression of
cervical squamous cell carcinoma. Cancer Sci. 111:4129–4141. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng J, Ouyang Y, Xu D, He Q, Liu D, Fan
X, Xu P and Mo Y: Genetic variants in MIR17HG affect the
susceptibility and prognosis of glioma in a Chinese Han population.
BMC Cancer. 20:9762020. View Article : Google Scholar : PubMed/NCBI
|