Introduction
Lung cancer is the leading cause of cancer-related
mortality, among which non-small-cell lung cancer (NSCLC) accounts
for ~80% (1). Lung adenocarcinoma
(LUAD) and lung squamous cell carcinoma are the two major NSCLC
subtypes. The incidence of LUAD has increased in recent years, with
the 5-year survival rate value as low as 15% (2). A number of genes have been reported
to be dysregulated and to be involved in LUAD progression; however,
the mechanisms underlying the occurrence and development of LUAD
remain largely unknown (3–5).
The de novo purine biosynthetic pathway is an
energy-intensive pathway with a high conservation, in which
inositol monophosphate (IMP) is generated from phosphoribosyl
pyrophosphate (PRPP). Accumulating evidence has demonstrated that
de novo purine synthesis deregulation may be closely
implicated in carcinogenesis (6–8).
Cancer cells maintain a high level of de novo purine
synthesis, in order to maintain rapid growth (9). 5-Aminoimidazole-4-carboxamide
ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC, also
known as AICAR) is an enzyme that can catalyse the last two
reactions in the de novo purine biosynthetic pathway
(10). Recently, it has been
reported that ATIC is frequently upregulated and is important for
cancer progression and development. For instance, Li et al
(11) reported that ATIC was
evidently overexpressed in myeloma tissues. Additionally, the
aberrant upregulation of ATIC in hepatocellular carcinoma (HCC)
tissues, the association of an increased ATIC expression with a
poor prognosis, and the suppression of HCC cell viability and
migration by ATIC knockdown has been reported (12). However, the role of ATIC in LUAD
remains elusive.
Myc is a well-known master transcription factor and
modulates the expression of genes that are essential for cell
survival, growth and metastasis (13–15).
Myc, cyclin D1 (CCND1) and EGFR are the three more frequently
amplified oncogenes based on pan-cancer analysis (16). Previous studies have demonstrated
that ATIC can activate mTOR signalling, an upstream regulator of
Myc (12,17,18).
Thus, it was hypothesized that ATIC may increase the expression of
Myc by activating mTOR signalling.
The present study aimed to explore the role of ATIC
in the modulation of cell growth and migration in LUAD, and to
further elucidate the role of Myc in this process.
Materials and methods
Patient tissue samples
LUAD tissues and tumour-adjacent non-cancer tissues
were obtained prior to chemoradiotherapy from 56 patients with LUAD
post-pneumonectomy, between January, 2013 and January, 2015. The
inclusion criteria were as follows: i) Patients with primary LUAD;
ii) patients aged from 18 to 75 years; iii) patients or their
families signed the informed consent. The exclusion criteria were
the following: i) Patients with other types of malignant tumours;
ii) patients who received radiation or chemotherapy prior to
pneumonectomy. The present study received approval from the Ethics
Committee of National Cancer Center/National Clinical Research
Center for Cancer/Cancer Hospital and Shenzhen Hospital Chinese
Academy of Medical Sciences and Peking Union Medical College prior
to the initiation of the study (approval no. KYLX2012-107). Signed
written informed consent was acquired by all patients or their
families.
Bioinformatics analysis
The gene expression profiling interactive analysis
database (GEPIA; http://gepia.cancer-pku.cn/detail.php) was used to
analyse the expression of ATIC in LUAD tissues and normal lung
tissues. In order to evaluate ATIC expression in LUAD, ATIC was
searched for followed by GEPIA. Subsequently, ‘Expression DIY’ was
selected and the ‘Boxplot’ option was chosen. Finally, the ‘LUAD’
option was selected in ‘Dataset’ and the ‘Plot’ function was
implemented.
The StarBase database (http://starbase.sysu.edu.cn/) was used to analyse the
association between the expression levels of ATIC and Myc in the
LUAD samples. Firstly, the ‘Pan-Cancer’ option was selected, also
closing ‘RNA-RNA CoExpression’. Myc was then inserted in the ‘Query
Gene (lncRNA,mRNA,ncRNA,etc.)’ frame and ‘ATIC’ was inserted in the
‘Target Gene (lncRNA,mRNA,ncRNA,etc.)’ frame, concurrently
selecting ‘lung adenocarcinoma’ in the ‘Cancer’ frame, and the
‘search’ function was used to obtain the final result.
Cells and culture conditions
Three human LUAD cell lines, HCC827 (cat. no.
CRL-2868), NCI-H1435 (cat. no. CRL-5870) and HCC4006 (cat. no.
CRL-2871), and BEAS-2B cells (human normal lung epithelial cells;
cat. no. CRL-9609) were acquired from the American Type Culture
Collection (ATCC). HCC827 bears EGFR and TP53 gene mutations, and
was initially derived from a 39-year female patient with LUAD.
NCI-1435 bears APC, TERT and TP53 mutations, and was initially
derived from a 35-year female patient with LUAD. HCC4006 bears an
EGFR mutation, and was derived from the pleural effusion of a male
patient (aged >50 years) with LUAD. All cells were maintained in
RPMI-1640 medium with 10% foetal bovine serum (FBS) and 1% (v/v)
penicillin-streptomycin and incubated at 37°C in an atmosphere
containing 5% CO2. The cell culture medium, FBS and
penicillin-streptomycin reagents were purchased from Thermo Fisher
Scientific, Inc. Additionally, the cells were treated with 100
nmol/l rapamycin (Selleckchem) for 2 h prior to plasmid
transfection.
Lentivirus infection and plasmid
transfection
The shRNAs used to downregulate ATIC (sh-ATIC-1/2)
and Myc (sh-Myc-1/2) expression and the negative control vectors
(sh-NC) were obtained from Shanghai GenePharma Co., Ltd., using the
lentiviral interfering vector LV-2 (pGLVU6/Puro) (cat. no. C06002,
Shanghai GenePharma Co., Ltd.), containing an ATIC- or
Myc-targeting shRNA sequence. For lentiviral packaging, the
expression vector (pGLVU6/Puro, 20 mg) and packaging vectors (1
µg/ml; pHelper 1.0 and pHelper 2.0) were transfected into 293T
cells (cat. no. CRL-3216; ATCC) using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.), based on the manufacturer's
instructions. Following a 48-h culture, supernatants containing
sh-ATIC/Myc and sh-NC were harvested, respectively. Purification
was then performed using ultracentrifugation at 1,000 × g and 4°C
for 2 min (Himac CT15RE; Hitachi, Ltd.) and the lentiviral titre
was determined. For lentiviral infection, a total of
2×105 cells were seeded into each well of a 6-well plate
and cultured at 37°C overnight. The following day, the cells were
infected with the sh-ATIC-1/2, sh-Myc-1/2, or sh-NC lentiviruses
(Shanghai GenePharma Co., Ltd.) at multiplicity of infection (MOI)
of 5 with the help of polybrene (6 µg/ml). The 3rd generation
system was used in following experiments.
The ATIC overexpression vector (vector-ATIC; cat.
no. RC203490, OriGene Technologies, Inc.) established in
pCMV6-Entry vector was used to upregulate ATIC expression. LUAD
cells were transfected with vector-ATIC using Lipofectamine
3000® (Invitrogen; Thermo Fischer Scientific, Inc.),
according to the manufacturer's instructions. Vector-NC (cat. no.
PS100001, OriGene Technologies, Inc.) was used as a negative
control for vector-ATIC. Following 48 h of transfection, the cells
were harvested for transfection efficiency analysis. The shRNA
sequences are designed used using the website of Thermo Fisher
Scientific, Inc. (https://rnaidesigner.thermofisher.com/rnaiexpress/setOption.do?designOption=shrna&pid=3449414797065297167).
First, ‘shRNA’ was selected in ‘Target Design Options’, the
‘Accession number’ for ATIC (NM_004044.7) and Myc (NM_002467.6),
were then inserted, followed by the ‘RNAi Design’ function.
Subsequently, two target sequences with high Rank star rating we
selected and the function ‘Design shRNA Oligos’ was applied.
‘CTCGAGC’ was then inserted in the ‘Custom Loop Sequence’, for the
acquisition of the shRNA sequences. The shRNA sequences are listed
as follows: sh-ATIC-1 forward,
5′-CACCGGTTTGAATCTGGTCGCTTCCCTCGAGGGAAGCGACCAGATTCAAACC-3′ and
reverse,
5′-AAAAGGTTTGAATCTGGTCGCTTCCCTCGAGGGAAGCGACCAGATTCAAACC-3′;
sh-ATIC-2 forward,
5′-CACCGCGTATCTCAGATGCCCTTGACTCGAGTCAAGGGCATCTGAGATACGC-3′ and
reverse,
5′-AAAAGCGTATCTCAGATGCCCTTGACTCGAGTCAAGGGCATCTGAGATACGC-3′;
sh-Myc-1 forward,
5′-CACCGCTTCACCAACAGGAACTATGCTCGAGCATAGTTCCTGTTGGTGAAGC-3′ and
reverse,
5′-AAAAGCTTCACCAACAGGAACTATGCTCGAGCATAGTTCCTGTTGGTGAAGC-3′;
sh-Myc-2 forward,
5′-CACCGGAAACGACGAGAACAGTTGA/TCAACTGTTCTCGTCGTTTCC-3′ and reverse,
5′-AAAAGGAAACGACGAGAACAGTTGACTCGAGTCAACTGTTCTCGTCGTTTCC-3′.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the tissues and cells
(HCC827, NCI-H1435, HCC4006 and BEAS-2) using TRIzol®
reagent (Invitrogen; Thermo Fischer Scientific, Inc.) and then
subjected to cDNA synthesis using the PrimeScript RT Master Mix kit
(RR036A; Takara Bio, Inc.) for 15 min at 37°C and 5 sec at 85°C.
Subsequently, cDNA samples were used for PCR detection with 2X
SYBR-Green PCR Mastermix (Beijing Solarbio Science & Technology
Co., Ltd.) on a 7500 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The reactions were carried out as
follows: 95°C for 1 min, followed by 39 cycles of 95°C for 15 sec
and 60°C for 1 min. β-actin was used as the reference gene.
Relative mRNA expression was calculated based on the
2−∆∆Cq method (19).
The sequences of the primers used are presented in Table I.
 | Table I.Sequences of primers used for
RT-qPCR. |
Table I.
Sequences of primers used for
RT-qPCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| ATIC |
CGGCCAGCTCGCCTTATTTA |
ATTTGCTCCACAGCCTCCTC |
| Myc |
GCAATGCGTTGCTGGGTTAT |
CGCATCCTTGTCCTGTGAGT |
| β-actin |
CTCGCCTTTGCCGATCC |
TTCTCCATGTCGTCCCAGTT |
Western blot analysis
Total protein samples were obtained from tissues and
cells using RIPA lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.) supplemented with 1% protease inhibitor
(Beijing Solarbio Science & Technology Co., Ltd.). Following
centrifugation at 4°C for 30 min at a speed of at 12,000 × g, a
bicinchoninic acid protein assay kit (Thermo Fisher Scientific,
Inc.) was used to define the protein concentrations, according to
the manufacturer's specifications. A total of 20 µg of protein from
each sample were then loaded to a 10% SDS-polyacrylamide gel,
subjected to electrophoresis and transferred to polyvinylidene
difluoride membranes (PVDF; MilliporeSigma). The membranes were
then probed with primary antibodies overnight at 4°C, followed by
blocking with 5% non-fat milk for 1 h at room temperature. The
primary antibodies used included an anti-β-actin antibody (1:5,000
dilution; cat. no. ab8226; Abcam), an anti-ATIC antibody (1:3,000
dilution; cat. no. ab33520, Abcam), an anti-mTOR antibody (1:1,000
dilution; cat. no. 2972, Cell Signalling Technology, Inc.), an
anti-phosphorylated (p-)mTOR (p-mTOR) antibody (1:1,000 dilution;
cat. no. 5536, Cell Signalling Technology, Inc.) and an anti-Myc
antibody (1:2,500 dilution; cat. no. ab32072, Abcam). Subsequently,
all membranes were probed with HRP-conjugated goat anti-rabbit IgG
(cat. no. ab7090; Abcam) and goat anti-mouse IgG (cat. no. ab97040;
Abcam) second antibodies (1:10,000 dilution) at room temperature
for 1 h. Following incubation with ECL reagent (MilliporeSigma) for
30 sec at room temperature, the protein signals were measured using
ProfiBlot-48 (Tecan Group, Ltd.) and quantified using ImageJ
v2.1.4.7 (National Institutes of Health).
CCK-8 assay
LUAD cells (3,000) were placed into each well of a
96-well plate. Cell transfection was then carried out following
cell adherence. The cell culture medium was replaced with 10 µl
CCK-8 solution (Abcam) and 90 µl fresh culture medium after 1, 2,
3, 4 and 5 days of cell transfection. Following a 3-h incubation
with CCK-8 solution at 37°C, the OD values (450 nm) were examined
using a spectrophotometer (Thermo Fisher Scientific, Inc.).
Wound healing assay
LUAD cells at a density of 1×106
cells/well were seeded in 6-well plates and incubated at 37°C
overnight. Pipette tips were used for wound formation when the cell
confluency reached 100%. In addition, the culture medium was
replaced with FBS-free medium. The width was measured using
Image-Pro Plus software 6.0 (Media Cybernetics, Inc.) at 0 and 24 h
following wound formation. The cell migration rate was calculated
as follows: Cell migration (%) = (1-width24
h/width0 h) ×100%
Transwell chamber assay
Matrigel (25 µl Matrigel was diluted in 25 µl
serum-free medium and cultured at 37°C for 2 h)-coated Transwell
chambers (BD Biosciences) were used for cell invasion assessment. A
total of 1×105 LUAD cells resuspended in FBS-free medium
were placed into the top chamber, while 600 µl of culture medium
containing 15% FBS were added into the bottom chamber. Following
incubation at 37°C for 48 h, cells on the top of the filter were
removed using cotton swabs, and cells on the bottom were fixed in
4% paraformaldehyde and then stained with 1% crystal violet
(Beijing Solarbio Science & Technology Co., Ltd.) for 10 min at
room temperature. The invaded cells that were removed from the
bottom of the chambers were counted under an inverted microscope
(BX-42; Olympus Corporation).
Statistical analysis
Three independent experiments were performed for all
protocols used in the present study. Statistical analysis was
performed by using SPSS (version 23.0; IBM Corp.). The paired
Student's t-test was used to compare data between the cancer group
and para-carcinoma groups, while the unpaired Student's t-test was
used for other data comparisons between two groups. The one-way
ANOVA followed by Tukey's post hoc test were used for the data
analysis of two groups and multiple groups, respectively.
Kaplan-Meier curves with log-rank tests were used to analyse the
value of ATIC in predicting the overall survival rates of patients
with LUAD. An ATIC expression greater than the average mRNA
expression level from the RT-qPCR result was considered a high
expression, and a less than or average mRNA expression was
considered a low expression. Pearson's correlation analysis was
applied to examine the correlation between the expression levels of
ATIC and Myc in the LUAD cases. Fisher's analysis was used for data
comparisons in Table II. A value
of P<0.05 was considered to indicate a statistically significant
difference.
 | Table II.Association of ATIC expression with
the clinicopathological features of patients with LUAD. |
Table II.
Association of ATIC expression with
the clinicopathological features of patients with LUAD.
|
|
| ATIC
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Total no. of
patients (n=56) | Low (n=30) | High (n=26) | P-value |
|---|
| Age (years) |
|
|
| 0.423 |
|
≤60 | 32 | 18 | 14 |
|
|
>60 | 24 | 12 | 12 |
|
| Sex |
|
|
| 0.211 |
|
Male | 28 | 13 | 15 |
|
|
Female | 28 | 17 | 11 |
|
| TNM stage |
|
|
| 0.031 |
|
I–II | 42 | 26 | 16 |
|
|
III–IV | 14 | 4 | 10 |
|
| Tumour
differentiation |
|
|
| 0.035 |
|
Well | 34 | 22 | 12 |
|
|
Moderate-poor | 22 | 8 | 14 |
|
| Lymph node
metastasis |
|
|
| 0.030 |
| No | 28 | 19 | 9 |
|
|
Yes | 28 | 11 | 17 |
|
| Smoking |
|
|
| 0.274 |
|
Smokers | 46 | 26 | 20 |
|
|
Never | 10 | 4 | 6 |
|
Results
ATIC expression is elevated in
LUAD
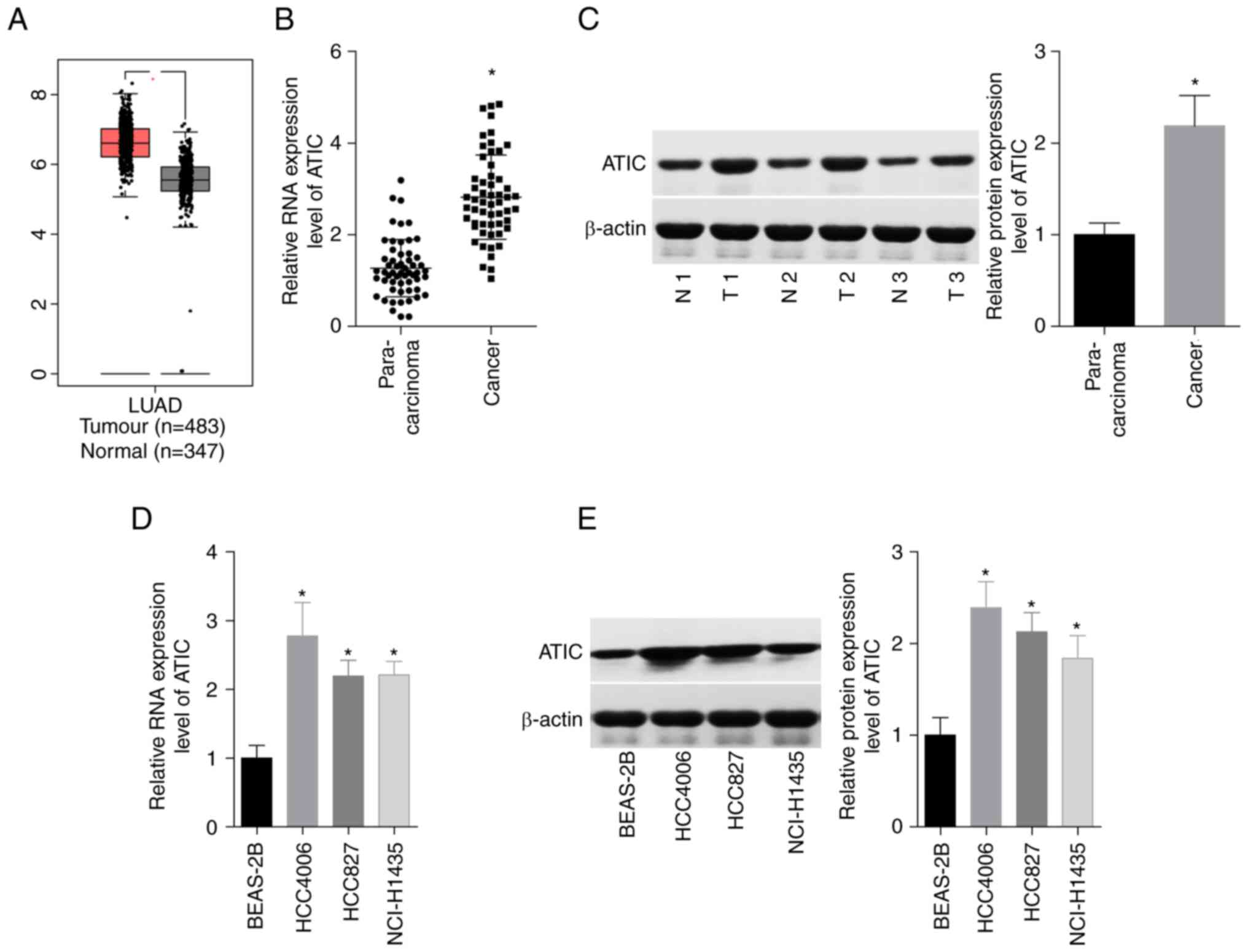
From the GEPIA database analysis, it was revealed
that the ATIC levels were upregulated in the LUAD tissues, as
compared with the normal lung tissues (Fig. 1A). Additionally, a 1-fold increase
in the expression of ATIC in the 56 paired LUAD tissues was
observed, as compared with the tumour-adjacent normal tissues using
RT-qPCR (Fig. 1B) and in 3 paired
LUAD tissues and the tumour-adjacent normal tissues using western
blot analysis (Fig. 1C). In
addition, the expression levels of ATIC in three human LUAD cell
lines (HCC827, NCI-H1435 and HCC4006) and one human normal lung
epithelial cell line (BEAS-2B) were compared. Consistently, the
results demonstrated that ATIC expression in the LUAD cells was ~
2–3-fold higher than that in the BEAS-2B cells (Fig. 1D and E). These results demonstrated
that ATIC expression was elevated in LUAD tissues and cell
lines.
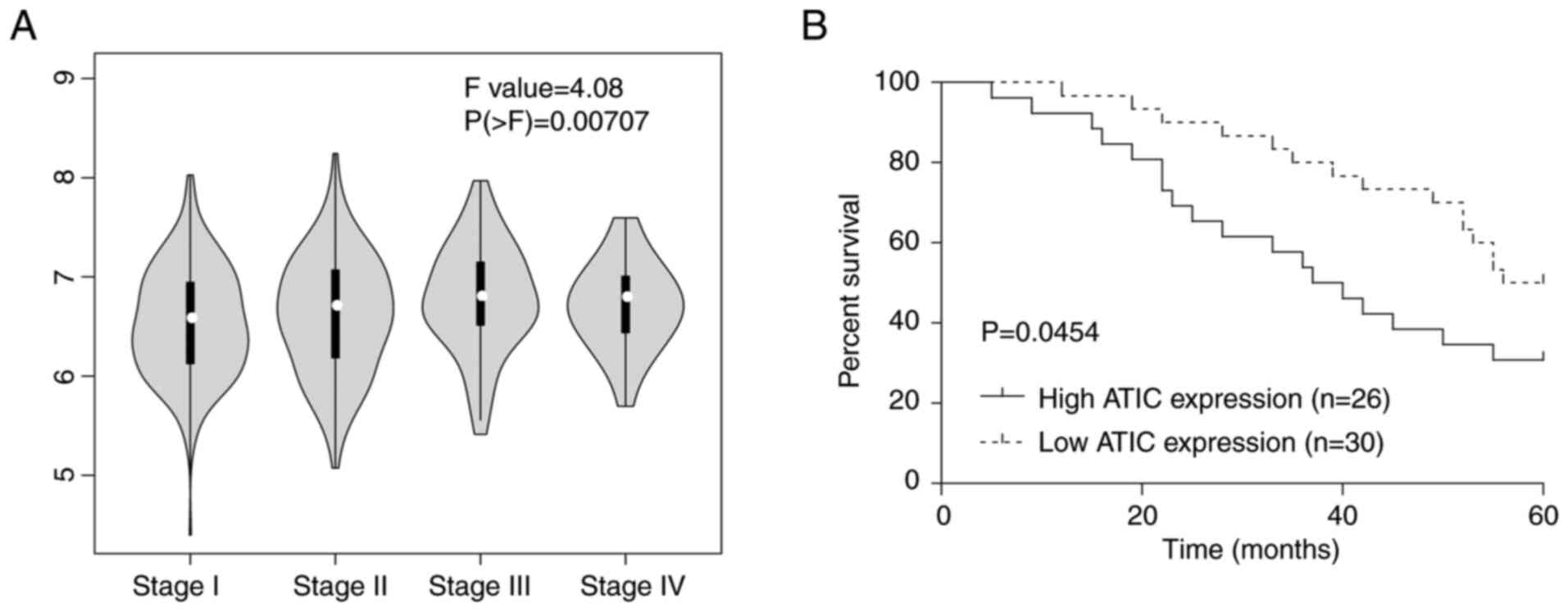
High expression of ATIC is a predictor
of an advanced tumour stage and lower survival rates in patients
with LUAD
Subsequently, the expression levels of ATIC in the
different stages of LUAD were assessed using the GEPIA database.
The results revealed that the ATIC level showed significant
difference between different stages of LUAD (Fig. 2A). In addition, the association
between the ATIC expression levels and the clinicopathological
features of patients with LUAD was evaluated. It was demonstrated
that ATIC expression was positively associated with the TNM stage
and lymph node metastasis, and that the high expression of ATIC was
a predictor of a poorer tissue differentiation (Table II). In addition, the 5-year
overall survival rate of patients with LUAD with a high ATIC
expression was lower than that of patients with a low ATIC
expression in the 56 clinical cases of LUAD examined (Fig. 2B). These results clearly
demonstrated that a high ATIC expression was associated with an
advanced stage and lower survival rates of patients with LUAD.
ATIC promotes cell growth and
migration in LUAD
Subsequently, both gain- and loss-of-function assays
were performed, in order to explore the role of ATIC in modulating
LUAD cell growth and migration. The HCC827 and NCI-H1435 cells
exhibited moderate ATIC expression levels in comparison with the
HCC4006 and BEAS-2B cells; thus, these two cell lines were used in
the following experiments. The expression of ATIC was observed to
be decreased by ~50–60% at the mRNA level and by 70–80% at the
protein level following transfection of the HCC827 and NCI-H1435
cells with sh-ATIC-1 and sh-ATIC-2 lentiviral vectors (Fig. S1A and B), whereas ATIC expression
was increased when the cells were transfected with vector-ATIC
(Fig. 1C and D). sh-ATIC-2 was
used in the following assay, due to its higher knockdown
efficiency, in comparison with sh-ATIC-1. The growth (Fig. 3A and B), migratory (Fig. 3C and D) and invasive (Fig. 3E) abilities of the HCC827 and
NCI-H1435 cells were enhanced by ~50–100% when ATIC was
overexpressed, whereas this effect was attenuated by ~50% when ATIC
was silenced (Fig. 3A and E).
These findings illustrated that ATIC promoted cell growth and
migratory abilities in LUAD.
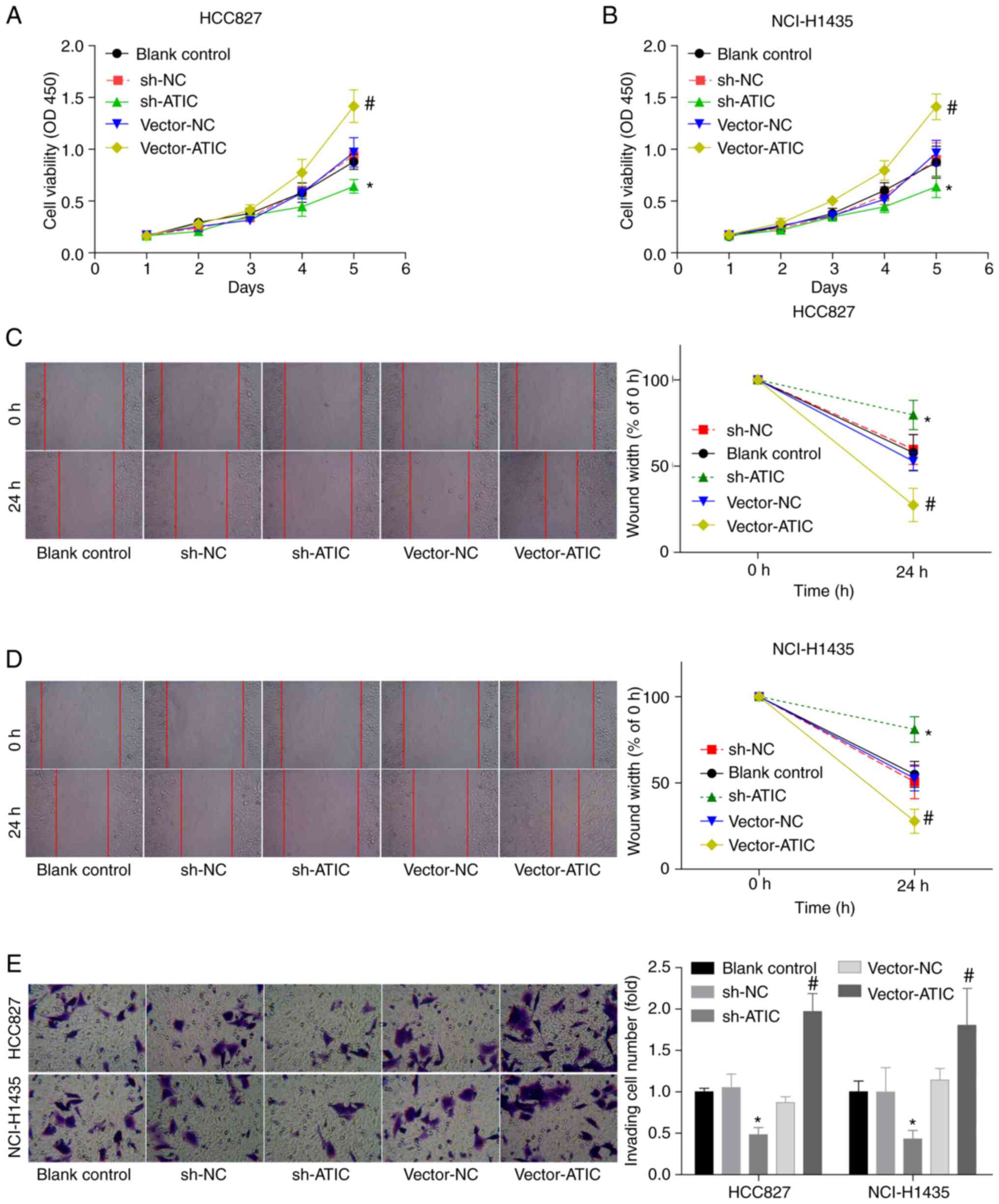 | Figure 3.ATIC promotes LUAD cell growth and
migration. (A and B) Cell growth ability in the blank control
(without treatment), sh-NC, sh-ATIC, vector-NC and vector-ATIC
groups was assessed using CCK-8 assay. (C and D) Cell migratory
ability in the blank control, sh-NC, sh-ATIC, vector-NC and
vector-ATIC groups was assessed using wound healing assay. (E) Cell
invasive ability in the blank control (without treatment), sh-NC,
sh-ATIC, vector-NC and vector-ATIC groups was assessed using
Transwell assay. *P<0.05, vs. sh-NC group;
#P<0.05, vs. vector-NC group. ATIC,
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP
cyclohydrolase; LUAD, lung adenocarcinoma. |
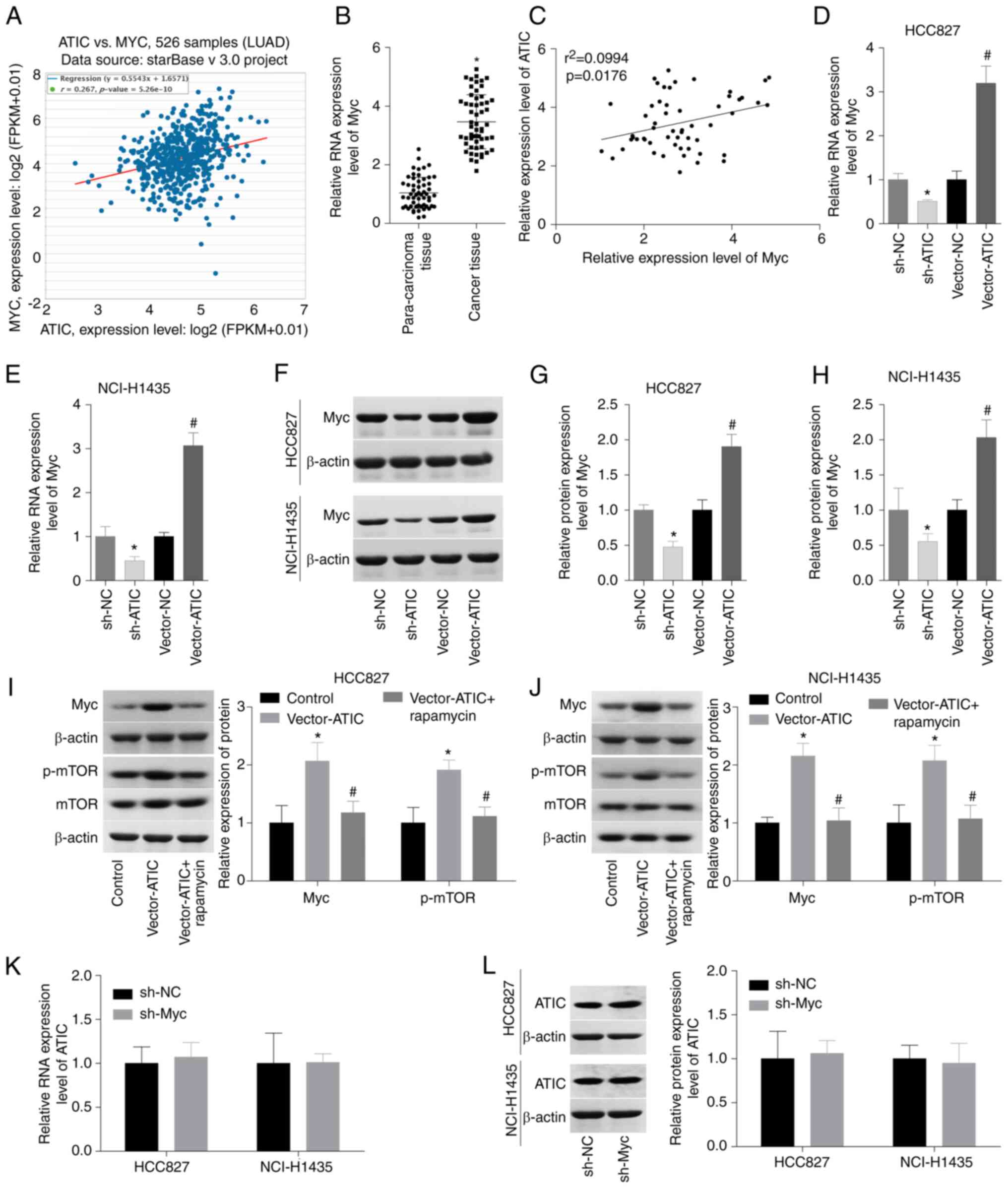
ATIC positively modulates Myc
expression in LUAD cells
In order to elucidate the mechanism through which
ATIC promotes LUAD progression, the association between ATIC and
Myc was subsequently evaluated in LUAD. According to the StarBase
analysis results, a weak association between the ATIC and Myc
expression levels in LUAD tissues was observed (Fig. 4A). Additionally, the Myc mRNA
expression levels were also analysed in the 56 paired LUAD tissues
and normal tissues. The results demonstrated that Myc expression
was elevated in LUAD, while a significant positive correlation was
observed with the ATIC levels in the LUAD cases (Fig. 4B and C). Subsequently, the effect
of ATIC on Myc expression in the HCC827 and NCI-H1435 cells was
evaluated. The silencing of ATIC led to a decrease in Myc
expression, while the overexpression of ATIC resulted in an
increase in Myc expression levels (Fig. 4D and H). It has been reported that
ATIC can activate the mTOR pathway which modulates Myc expression
(12,17,18);
thus, it was hypothesized that the increased expression of Myc
induced by ATIC may be related to mTOR activation. As was expected,
ATIC overexpression increased the expression of p-mTOR and Myc,
which was reversed by rapamycin, an mTOR inhibitor (Fig. 4I and J). Additionally, it was
assessed whether Myc modulates ATIC expression using qPCR and
western blot analysis. Transfection with sh-Myc-2 led to an 80%
decrease in the Myc mRNA levels and a 60% decrease in the Myc
protein levels in the HCC827 and NCI-H1435 cells (Fig. S1E and F). However, the ATIC mRNA
and protein levels did not exhibit an obvious change when Myc was
silenced in the HCC827 and NCI-H1435 cells (Fig. 4K and L). These results thus
demonstrated that ATIC positively modulated Myc expression in LUAD
cells.
ATIC promotes cell growth and
migration in a Myc-dependent manner
It was then examined whether Myc was involved in the
ATIC-mediated LUAD progression by using rescue experiments. The
growth, migratory and invasive abilities of the HCC827 and
NCI-H1435 cells were decreased ~0.5-fold when Myc was silenced, in
comparison with the control group. Additionally, the silencing of
Myc abolished the ability of ATIC to promote the growth (Fig. 5A and B), migration (Fig. 5C and D) and invasion (Fig. 5E) of the HCC827 and NCI-H1435
cells. These results confirmed that ATIC promoted LUAD cell growth
and migration in a Myc-dependent manner.
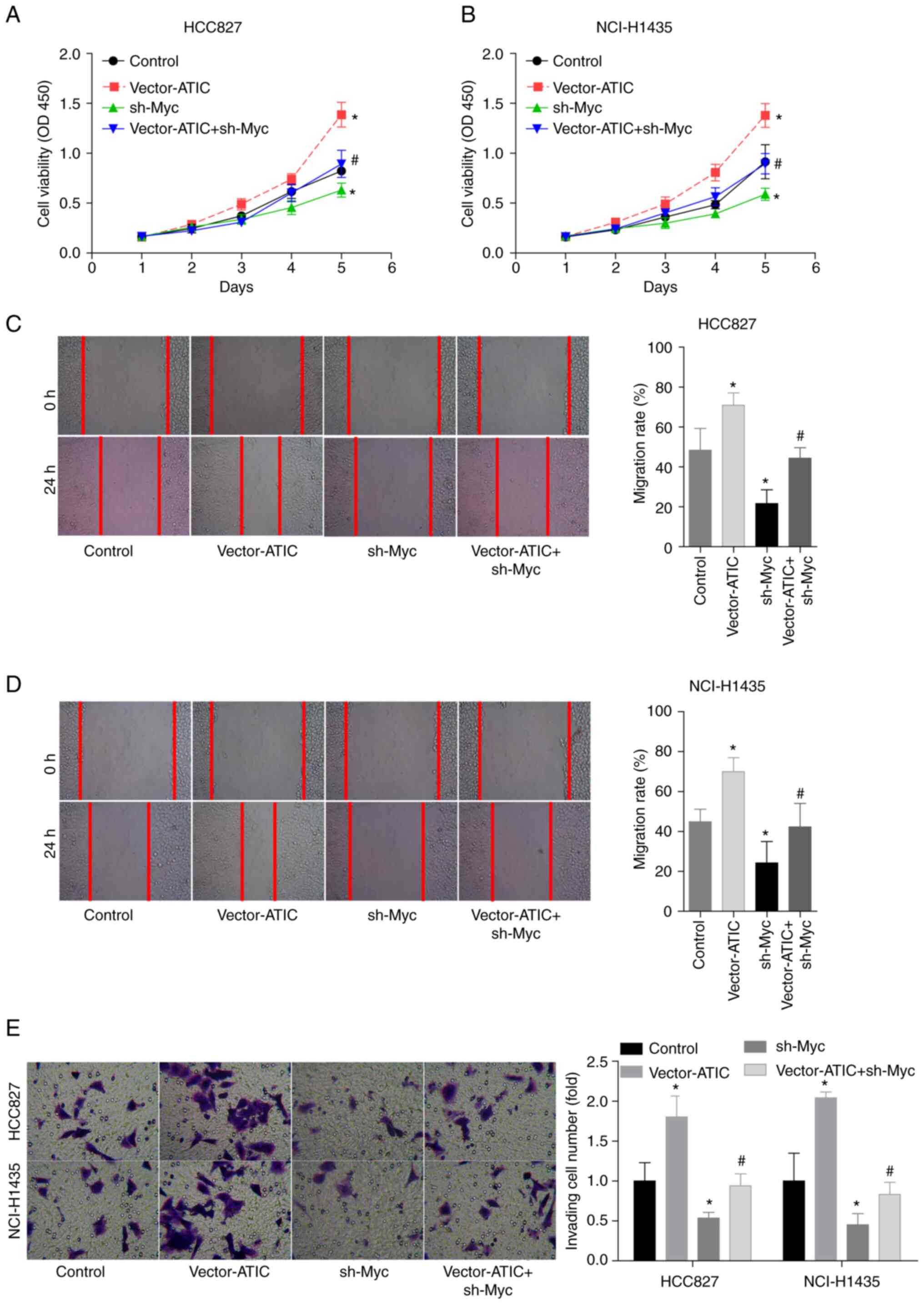 | Figure 5.ATIC promotes cell growth and
migration in a Myc-dependent manner. (A and B) Cell growth ability
in the control, vector-ATIC, sh-Myc and vector-ATIC + sh-Myc groups
was measured using CCK-8 assay. (C and D) Cell migration ability in
the control, vector-ATIC, sh-Myc and vector-ATIC + sh-Myc groups
was assessed using wound healing assay. (E) Cell invasive ability
in the control, vector-ATIC, sh-Myc and vector-ATIC + sh-Myc groups
was assessed using Transwell chambers. *P<0.05, vs. control
group; #P<0.05, vs. vector-ATIC group. ATIC,
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP
cyclohydrolase; LUAD, lung adenocarcinoma. |
Discussion
The main aim of the present study was to elucidate
the role of ATIC in LUAD progression. The results demonstrated that
ATIC expression was increased in LUAD tissues and cells, and that
the high expression of ATIC was associated with malignant clinical
phenotypes and lower survival rates of patients with LUAD.
Additionally, it was revealed that the overexpression of ATIC
significantly promoted cell growth viability, migration and
invasion by increasing Myc expression in LUAD.
ATIC is a 64 kDa bifunctional enzyme that modulates
the activities of two enzymes in the de novo purine
biosynthesis pathway, AICAR and IMP cyclohydrolase (20,21).
Previous studies have demonstrated that the de novo purine
biosynthesis pathway is closely implicated in the occurrence and
development of various types of cancer. For example, Lv et
al (7) found that de
novo nucleotide synthesis was increased in metastatic breast
cancer cells. In addition, blocking de novo synthesis with
phosphoribosyl pyrophosphate synthetase 2 (PRPS2) downregulation
has been shown to result in the marked inhibition of cell stemness
and lung metastasis (22). The
inhibition of AICAR activity can cooperate with pemetrexed to
suppress tumour growth (22). As
an enzyme that regulates the de novo purine biosynthesis
pathway, ATIC has also been reported to be involved in
carcinogenesis. Park and Shin (23) found that the polymorphism of ATIC
(347C >G) may be a factor affecting the response to methotrexate
in osteosarcoma. Li et al (12) demonstrated that ATIC was highly
expressed in HCC tissues and that the high expression of ATIC was
related to a poor prognosis; ATIC downregulation suppresses cell
proliferation, colony formation and migration by modulating the
adenosine monophosphate-activated protein kinase (AMPK)/mTOR
pathway. By using bioinformatics analysis, Zhu et al
(24) identified that ATIC was a
risk factor for LUAD that may present a high potential in
predicting the survival rates of patients with LUAD. Herein, it was
demonstrated for the first time, to the best of our knowledge, that
the expression of ATIC was markedly higher in LUAD tissues compared
with normal lung tissues. In addition, it was demonstrated that the
higher expression of ATIC was closely associated with advanced TNM
stages, higher lymph node metastasis rates, a poorer tissue
differentiation and lower survival rates. Moreover, ATIC expression
levels in three LUAD cell lines (HCC827, NCI-H1435 and HCC4006)
were assessed and it was observed that the ATIC levels were
increased in LUAD cells, in comparison with normal lung epithelial
BEAS-2B cells. The HCC827 and NCI-H1435 cells demonstrated moderate
ATIC expression levels in comparison with the HCC4006 and BEAS-2B
cells; thus, gain- and loss-of-function assays were performed using
these two cell lines, in order to assess the effect of ATIC on LUAD
progression in vitro. The results demonstrated that ATIC
overexpression led to the promotion of cell growth, migration and
invasion, while ATIC silencing inversely lead to the suppression of
cell growth, migration and invasion. The results of the present
study revealed that ATIC functioned as an oncogene in LUAD.
It was observed that ATIC expression positively
correlated with Myc expression in the LUAD cases. It has been
demonstrated that Myc is an oncogene in LUAD and that a high level
of Myc is closely linked to a decreased survival rate of patients
with LUAD (25,26). Through the modulation of Myc
expression, a number of genes have been reported to be involved in
the occurrence and development of LUAD. For instance,
lysophosphatidylcholine acyltransferase 1 has been reported to
promote the brain metastasis of LUAD by activating the PI3K/AKT/Myc
pathway (27). miR-1827 has been
reported to inhibit tumour growth by reducing Myc and Family with
sequence similarity 83 member F gene levels in LUAD (28). Additionally, Myc has been
identified to vitally contribute in facilitating nucleotide
biosynthesis by increasing the expression of the nucleotide
synthesis enzyme PRPS2 (29) and
phosphoribosylaminoimidazole carboxylase (30). In the present study, it was
revealed that ATIC overexpression increased Myc expression, whereas
Myc did not affect ATIC expression in LUAD cells. Since Myc is a
downstream factor of the mTOR pathway that can be activated by ATIC
(12,17,18),
it was hypothesized that the increased expression of Myc induced by
ATIC may be related to mTOR activation. This was verified by using
western blot analysis; rapamycin stimulation reversed the
ATIC-mediated increase in Myc expression.
In order to reveal whether Myc is involved in
ATIC-mediated LUAD progression, rescue experiments were also
performed. As was expected, the results demonstrated that the
silencing of Myc attenuated the promotion of cell growth, migration
and invasion induced by ATIC, suggesting that ATIC promoted LUAD
cell growth and migration by increasing Myc expression.
Several limitations should be underlined for the
present study. The roles of ATIC/Myc in tumour formation in
vivo and the drug resistance of LUAD were not explored. Thus,
the authors intend to investigate these parameters in future
studies. Furthermore, the mutation status of EGFR, anaplastic
lymphoma kinase and proto-oncogene tyrosine-protein kinase ROS in
clinical samples were not assessed, as well as their associations
with ATIC expression.
In conclusion, it was revealed that ATIC was highly
expressed in LUAD tissues and cells, and that the high expression
of ATIC was closely associated with lower survival rates and an
advanced clinical stage of patients with LUAD. The overexpression
of ATIC significantly promoted cell growth, migration and invasion
by increasing Myc expression in LUAD. The findings of the present
study may provide a possible novel diagnostic marker and treatment
target for LUAD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NN conceived the study performed the experiments and
revised the manuscript. JZ, XK and WZ performed western blotting
and RT-qPCR experiments and wrote the manuscript. CF, SL, JF and YY
analysed the data. All authors have read and approved the final
manuscript. NN and JZ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of National Cancer Center/National Clinical Research
Center for Cancer/Cancer Hospital and Shenzhen Hospital Chinese
Academy of Medical Sciences and Peking Union Medical College prior
to this study (approval no. KYLX2012-107). Signed written informed
consent was acquired by all patients or their families.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu Y, Jiang Q, Liu H, Xiao X, Yang D, Saw
PE and Luo B: DHX37 impacts prognosis of hepatocellular carcinoma
and lung adenocarcinoma through immune infiltration. J Immunol Res.
2020:88353932020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu Y, Liu Z, Li H, Feng S, Li Q, Li J and
Li S: VEGI downregulation is correlated with nodal metastasis and
poor prognosis in lung adenocarcinoma. Mol Clin Oncol. 14:252021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai J, Deng H, Luo L, You L, Liao H and
Zheng Y: Decreased expression of JAK1 associated with immune
infiltration and poor prognosis in lung adenocarcinoma. Aging
(Albany NY). 13:2073–2088. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robinson AD, Eich ML and Varambally S:
Dysregulation of de novo nucleotide biosynthetic pathway enzymes in
cancer and targeting opportunities. Cancer Lett. 470:134–140. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lv Y, Wang X, Li X, Xu G, Bai Y, Wu J,
Piao Y, Shi Y, Xiang R and Wang L: Nucleotide de novo synthesis
increases breast cancer stemness and metastasis via cGMP-PKG-MAPK
signaling pathway. PLoS Biol. 18:e30008722020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan TWM, Bruntz RC, Yang Y, Song H,
Chernyavskaya Y, Deng P, Zhang Y, Shah PP, Beverly LJ, Qi Z, et al:
De novo synthesis of serine and glycine fuels purine nucleotide
biosynthesis in human lung cancer tissues. J Biol Chem.
294:13464–13477. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamaoka T, Kondo M, Honda S, Iwahana H,
Moritani M, Ii S, Yoshimoto K and Itakura M:
Amidophosphoribosyltransferase limits the rate of cell
growth-linked de novo purine biosynthesis in the presence of
constant capacity of salvage purine biosynthesis. J Biol Chem.
272:17719–17725. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Verma P, Kar B, Varshney R, Roy P and
Sharma AK: Characterization of AICAR transformylase/IMP
cyclohydrolase (ATIC) from Staphylococcus lugdunensis. FEBS
J. 284:4233–4261. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li R, Chen G, Dang Y, He R, Liu A, Ma J
and Wang C: Upregulation of ATIC in multiple myeloma tissues based
on tissue microarray and gene microarrays. Int J Lab Hematol.
43:409–417. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li M, Jin C, Xu M, Zhou L, Li D and Yin Y:
Bifunctional enzyme ATIC promotes propagation of hepatocellular
carcinoma by regulating AMPK-mTOR-S6 K1 signaling. Cell
communication and signaling. Cell Commun Signal. 15:522017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hsieh AL, Walton ZE, Altman BJ, Stine ZE
and Dang CV: MYC and metabolism on the path to cancer. Semin Cell
Dev Biol. 43:11–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zack TI, Schumacher SE, Carter SL,
Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J,
Mermel CH, et al: Pan-cancer patterns of somatic copy number
alteration. Nat Genet. 45:1134–1140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen P, Reineke LC, Knutsen E, Chen M,
Pichler M, Ling H and Calin GA: Metformin blocks MYC protein
synthesis in colorectal cancer via mTOR-4EBP-eIF4E and
MNK1-eIF4G-eIF4E signaling. Mol Oncol. 12:1856–1870. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu D, Xie R, Xu Z, Zhao Z, Ding M, Chen W,
Zhang J, Mao E, Chen E, Chen Y, et al: mTOR-Myc axis drives
acinar-to-dendritic cell transition and the CD4+ T cell
immune response in acute pancreatitis. Cell Death Dis. 11:4162020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Greasley SE, Horton P, Ramcharan J,
Beardsley GP, Benkovic SJ and Wilson IA: Crystal structure of a
bifunctional transformylase and cyclohydrolase enzyme in purine
biosynthesis. Nat Struct Biol. 8:402–406. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vergis JM, Bulock KG, Fleming KG and
Beardsley GP: Human 5-aminoimidazole-4-carboxamide ribonucleotide
transformylase/inosine 5′-monophosphate cyclohydrolase. A
bifunctional protein requiring dimerization for transformylase
activity but not for cyclohydrolase activity. J Biol Chem.
276:7727–7733. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Racanelli AC, Rothbart SB, Heyer CL and
Moran RG: Therapeutics by cytotoxic metabolite accumulation:
Pemetrexed causes ZMP accumulation, AMPK activation, and mammalian
target of rapamycin inhibition. Cancer Res. 69:5467–5474. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park JA and Shin HY: ATIC gene
polymorphism and histologic response to chemotherapy in pediatric
osteosarcoma. J Pediatr Hematol Oncol. 39:e270–e274. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu J, Wang M and Hu D: Development of an
autophagy-related gene prognostic signature in lung adenocarcinoma
and lung squamous cell carcinoma. PeerJ. 8:e82882020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iwakawa R, Kohno T, Kato M, Shiraishi K,
Tsuta K, Noguchi M, Ogawa S and Yokota J: MYC amplification as a
prognostic marker of early-stage lung adenocarcinoma identified by
whole genome copy number analysis. Clin Cancer Res. 17:1481–1489.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ciribilli Y and Borlak J: Oncogenomics of
c-Myc transgenic mice reveal novel regulators of extracellular
signaling, angiogenesis and invasion with clinical significance for
human lung adenocarcinoma. Oncotarget. 8:101808–101831. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei C and Dong X, Lu H, Tong F, Chen L,
Zhang R, Dong J, Hu Y, Wu G and Dong X: LPCAT1 promotes brain
metastasis of lung adenocarcinoma by up-regulating PI3K/AKT/MYC
pathway. J Exp Clin Cancer Res. 38:952019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan G, Xu P and Tu P: miR-1827 functions
as a tumor suppressor in lung adenocarcinoma by targeting MYC and
FAM83F. J Cell Biochem. 121:1675–1689. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cunningham JT, Moreno MV, Lodi A, Ronen SM
and Ruggero D: Protein and nucleotide biosynthesis are coupled by a
single rate-limiting enzyme, PRPS2, to drive cancer. Cell.
157:1088–1103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barfeld SJ, Fazli L, Persson M, Marjavaara
L, Urbanucci A, Kaukoniemi KM, Rennie PS, Ceder Y, Chabes A,
Visakorpi T and Mills IG: Myc-dependent purine biosynthesis affects
nucleolar stress and therapy response in prostate cancer.
Oncotarget. 6:12587–12602. 2015. View Article : Google Scholar : PubMed/NCBI
|