Introduction
Neuroblastoma (NB) is a tumor that originates from
precursor cells in the nervous system and is the most common
extra-cranial solid tumor in children (1,2). The
5-year survival rate in 2010 for children <1 year old was 95% in
the United States, and the understanding of this disease has
advanced tremendously in the past decade (2). Despite the ongoing comprehensive
research into this disease, NB still accounts for 6% of all
pediatric malignancies in the United States (3,4) and
11% of all pediatric cancer-related deaths in patients <15 years
old (2). Furthermore, the
long-term survival of high-risk patients with NB is <50% due to
the heterogeneous, aggressive and relapse-prone nature of these
tumors (5,6). Therefore, the identification of novel
prognostic markers for NB is required.
Targeting protein for Xenopus kinesin-like
protein 2 (TPX2; also known as C20orf1, C20orf2, DIL-2 and p100) is
required for microtubule formation and regulates cell movement
during key biological processes (7,8),
such as proliferation, apoptotic processes and cell division
(9–11). An oncogenic role for TPX2 has been
demonstrated in multiple malignancies, including gastric cancer,
colorectal carcinoma, hepatocellular carcinoma and bladder cancer
(12–15). In a lung cancer study, increased
TPX2 was observed during the establishment of drug tolerance and
was maintained into acquired resistance (16). A recent study suggested that TPX2
may be a prognostic marker to stratify high-risk NB (17). However, the function of TPX2 in NB
has not been completely elucidated.
The current prognostic assessment of NB is based on
the NB Risk Classification System, including The International NB
Staging System stage, tumor histology, age at diagnosis, chromosome
copy number alteration and MYCN proto-oncogene bHLH transcription
factor (MYCN) gene status (18–20).
Combined assessment of TPX2 expression and MYCN gene amplification
may be a prognostic marker to stratify high-risk NB patients. In
the present study, the prognostic significance of TPX2 combined
with MYCN gene amplification in patients with NB was evaluated and
the functional role of TPX2 in NB cell lines was elucidated.
Materials and methods
Patients and sample collection
NB tissue samples and data of 43 patients with NB
who underwent surgical resection between March 1980 and February
2010 at Mie University Hospital (Tsu, Japan) were obtained
retrospectively from the Department of Pathology and from patient
records. All patients were managed according to the Japan NB Study
Group protocol (21). The median
patient age was 8 months (ranging from 1 month to 8 years) and
23.3% of the patients were female. The clinicopathological
characteristics of the patients are listed in Table I. The amplification status of MYCN
from 39 of 43 patients was obtained from the Department of
Pathology, Mie University Hospital, as assessed by fluorescence
in situ hybridization. The protocol for the present research
project was approved (approval no. 1464) by the Institutional
Review Board of Mie University Hospital. Informed consent was
obtained from the parents/guardians of the patients using the
opt-out scheme.
 | Table I.Clinicopathological characteristics
of patients (n=43). |
Table I.
Clinicopathological characteristics
of patients (n=43).
| Clinicopathological
characteristic | Value |
|---|
| Median age of
onset, months | 8 (0–96) |
| Sex, n (%) |
|
|
Male | 33 (76.7) |
|
Female | 10 (23.3) |
| Primary tumor site,
n (%) |
|
|
Mediastinum | 11 (25.6) |
| Adrenal
gland | 19 (44.2) |
|
Abdomen | 12 (27.9) |
|
Neck | 1 (2.3) |
| Tumor size, n
(%) |
|
| >5
cm | 23 (53.5) |
| ≤5
cm | 20 (46.5) |
| Ipsilateral lymph
node metastasis, n (%) |
|
|
Present | 23 (53.5) |
|
Absent | 20 (46.5) |
| Contralateral lymph
node metastasis, n (%) |
|
|
Present | 7 (16.3) |
|
Absent | 36 (83.7) |
| Distant metastasis,
n (%)a |
|
|
Present | 15 (34.9) |
|
Absent | 28 (65.1) |
| Distant metastasis
site, n |
|
|
Liver | 7 |
|
Bone | 6 |
|
Marrow | 7 |
| Lymph
node | 6 |
|
Skin | 1 |
| Complete resection,
n (%) |
|
|
Yes | 24 (55.8) |
| No | 19 (44.2) |
| INSS stage, n
(%) |
|
| 1 | 7 (16.3) |
| 2A | 7 (16.3) |
| 2B | 6 (14.0) |
| 3 | 8 (18.6) |
| 4 | 11 (25.6) |
| 4s | 4 (9.3) |
| MYCN amplification,
n (%) |
|
|
Present | 7 (16.3) |
|
Absent | 32 (74.4) |
|
Unknown | 4 (9.3) |
| Pre-operative
chemotherapy, n (%) |
|
|
Present | 12 (27.9) |
|
Absent | 31 (72.1) |
| Radiation, n
(%) |
|
|
Present | 10 (23.3) |
|
Absent | 33 (76.7) |
| PBSCT, n (%) |
|
|
Present | 3 (7.0) |
|
Absent | 40 (93.0) |
| BMT, n (%) |
|
|
Present | 4 (9.3) |
|
Absent | 39 (90.7) |
| Prognosis, n
(%) |
|
|
Alive | 32 (74.4) |
|
Dead | 11 (25.6) |
Survival information and gene expression data for
247 NB samples were also obtained from the Therapeutically
Applicable Research to Generate Effective Treatments (TARGET)
project (https://ocg.cancer.gov/programs/TARGET/data-matrix)
conducted in November 2018. The TARGET-NB dataset was used for
survival analysis and association between TPX2 expression and other
gene expression. The clinicopathological characteristics of the
TARGET-NB data set are listed in Table SI.
Cell lines and culture
The MYCN-amplified IMR-5 cell line and the
MYCN-non-amplified KP-N-SI(FA) cell line were acquired from the
Cell Resource Center of Biomedical Research, Institute of
Development, Aging and Cancer (Tohoku University). Both cell lines
were maintained in RPMI-1640 medium (cat. no. 30264-56; Nacalai
Tesque, Inc.) supplemented with 10% fetal bovine serum (cat. no.
s1580-500; Biowest), glutamine (2 mmol/l), penicillin (100,000
U/l), streptomycin (100 mg/l) and gentamicin (40 mg/l) at 37°C in a
5% CO2 atmosphere. The cell lines were tested and
authenticated.
Total RNA extraction and cDNA
synthesis
Total RNA was extracted from NB cells and tissues
using the RNeasy mini kit (cat. no. 217004) and RNeasy FFPE kit
(cat. no. 217504; both from Qiagen GmbH) in accordance with the
manufacturer's instructions. RNA quality and concentration were
determined using a Denovix® DS-11+ spectrophotometer
(DeNovix, Inc.). cDNA was synthesized from 5 µg of total RNA with
random hexamer primers, dNTPs, 5X buffer and
SuperScript® III Reverse Transcriptase (cat. no.
8080-044; Invitrogen; Thermo Fisher Scientific, Inc.) as previously
described (22).
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR analyses of NB cells and tissues were
conducted using the Power SYBR Green PCR Master Mix (cat. no.
4367659; Applied Biosystems; Thermo Fisher Scientific, Inc.) and
the Applied Biosystems 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's instructions and as previously described (22). The qPCR cycling conditions were as
follows: 95°C for 10 min, followed by 40 cycles of 15 sec at 95°C
and 60 sec at 60°C. Primers for TPX2 and GAPDH mRNAs were as
follows: TPX2 forward, 5′-TGAGGCAGCCATATCAAGAA-3′ and reverse,
5′-TGGCACATCTCTTGGCTTTC-3′; and GAPDH forward,
5′-GGAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-AATGAAGGGGTCATTCATGG-3′.
Relative expression levels of TPX2 mRNA were calculated by
normalization to the levels of endogenous GAPDH mRNA using the
2−∆∆Cq method (23).
RT-qPCR assays were performed in triplicate for each sample and the
mean value was calculated.
TPX2 RNA interference
TPX2-specific small interfering RNA (siRNA) (cat.
no. 4392429; Silencer® Predesigned siRNA) and negative
control siRNA (cat. no. 4390844; Silencer Negative Control siRNA)
were purchased from Ambion®; Thermo Fisher Scientific,
Inc. The sequences of TPX2 siRNA were as follows: Sense,
5′-GGAAAGUGAACUUUACAUCUtt-3′ and antisense,
5′-AGAUGUAAAGUUCACUUCCtt-3′. Cells were seeded in six-well culture
plates at 2×105 cells per well in 2 ml RPMI-1640. Cells
were cultured for 24 h and then incubated with siRNA
oligonucleotides using Lipofectamine® RNAiMAX Reagent
(cat. no. 13778-150) and OptiMEM® I (cat. no. 31985-062)
(both Invitrogen; Thermo Fisher Scientific, Inc.) for 20 min at
room temperature in accordance with the manufacturer's
instructions. The final siRNA oligonucleotide concentration was 50
nM. After 48 h, transfected cells were examined by RT-qPCR as
aforementioned or used for further experiments.
Immunohistochemical staining
Immunohistochemical staining of NB tissues was
performed using an anti-human TPX2 antibody (1:1,000; cat. no.
ab32795; Abcam) as previously described (22).
Western blotting
At 48 h after transfection, cells were lysed in RIPA
buffer (cat. no. F015; BioDynamics Laboratory Inc.) with protease
inhibitors, and lysates were centrifuged for 5 min at 12,000 × g at
4°C. The protein concentration was measured using the BCA protein
assay kit (cat. no. 23227; Thermo Fisher Scientific, Inc.). Total
protein samples (10 µg) were separated on 5-20% gradient
polyacrylamide gels (cat. no. c520L; ATTO Corporation) and then
transferred onto a polyvinylidene difluoride filter (cat. no.
WSE4050; ATTO Corporation). The filter was first blocked with 5%
skimmed milk for 1 h at room temperature and then incubated with
primary antibodies against TPX2 (1:5,000) and β-actin (1:20,000;
cat. no. 691001) (both MP Biomedicals, LLC) for 15 min at room
temperature. Following the primary incubation, membranes were
incubated with horseradish peroxidase-conjugated anti-mouse IgG
antibody (1:10,000 for TPX2 and 1:40,000 for β-actin) (cat. no.
W402B; Promega Corporation) for 30 min at room temperature. The
protein bands were visualized by a WSE-6100 LuminoGraph imaging
system (ATTO Corporation).
Immunofluorescence staining
For double immunofluorescence of cell lines and NB
tissues, sections were incubated with primary antibodies against
TPX2 (1:500) and N-Myc (1:200; cat. no. 51705; Cell signaling
Technology, Inc.) overnight at 4°C. Alexa Fluor® 546
goat anti-mouse IgG (1:500; cat. no. A-11030) and Alexa
Fluor® 488 goat anti-rabbit IgG (1:200; cat. no.
A-11008) (both Invitrogen; Thermo Fisher Scientific, Inc.) were
used as secondary antibodies for 1 h at room temperature. Nuclear
counterstaining was performed using ProLong® Gold
Antifade Reagent with DAPI (cat. no. 727434; Invitrogen; Thermo
Fisher Scientific, Inc.). Confocal images were acquired using a
BX53 inverted microscope with a DP74 digital camera system
(Olympus). Further details are provided in Appendix 1.
Cell proliferation assay
Cell proliferation of TPX2 siRNA- and control
siRNA-transfected NB cells was evaluated with a Cell Counting Kit-8
(cat. no. CK04; Dojindo Laboratories, Inc.) in accordance with the
manufacturer's instructions. Additional experimental details on the
assay are provided in Appendix
1.
Cell cycle analysis and apoptosis
assay
The DNA content of TPX2 siRNA- and control
siRNA-transfected NB cells was evaluated using the Muse™ Cell Cycle
Assay kit (cat. no. MCH100106) and Muse Annexin V & Dead Cell
Kit (cat. no. MCH100105) using the Muse Cell Analyzer (all
MilliporeSigma) in accordance with the manufacturer's instructions.
Additional experimental details on these assays are provided in
Appendix 1.
Statistical analysis
Statistical analysis was performed using JMP
software version 10 (SAS Institute, Inc.) and MedCalc Statistical
Software version 19.1.2 (MedCalc Software bvba). Differences
between groups were estimated by one-way ANOVA followed by Tukey's
post hoc test, and Wilcoxon rank-sum test was used when
appropriate. Two-way repeated measures ANOVA was used for the
comparisons of repeated measurements. Pearson's correlation
coefficient assay was used to analyze expression correlation.
Shapiro-Wilk tests were performed to evaluate the normality of
distribution and Levene's tests were conducted to assess the
equality of variance for comparable groups. For RT-qPCR data, an
unpaired two-tailed Student's t-test was used to calculate
significant ΔCq differences between two groups. For time-to-event
analyses, survival estimates were calculated using the Kaplan-Meier
method and groups were compared using the log-rank test. Receiver
operating characteristic (ROC) curves with Youden's index were
established to determine the cut-off values for analyzing
prognosis. All P-values were calculated using the two-tailed test
and P<0.05 was considered to indicate a statistically
significant difference.
Results
Association between patient
characteristics and TPX2 expression
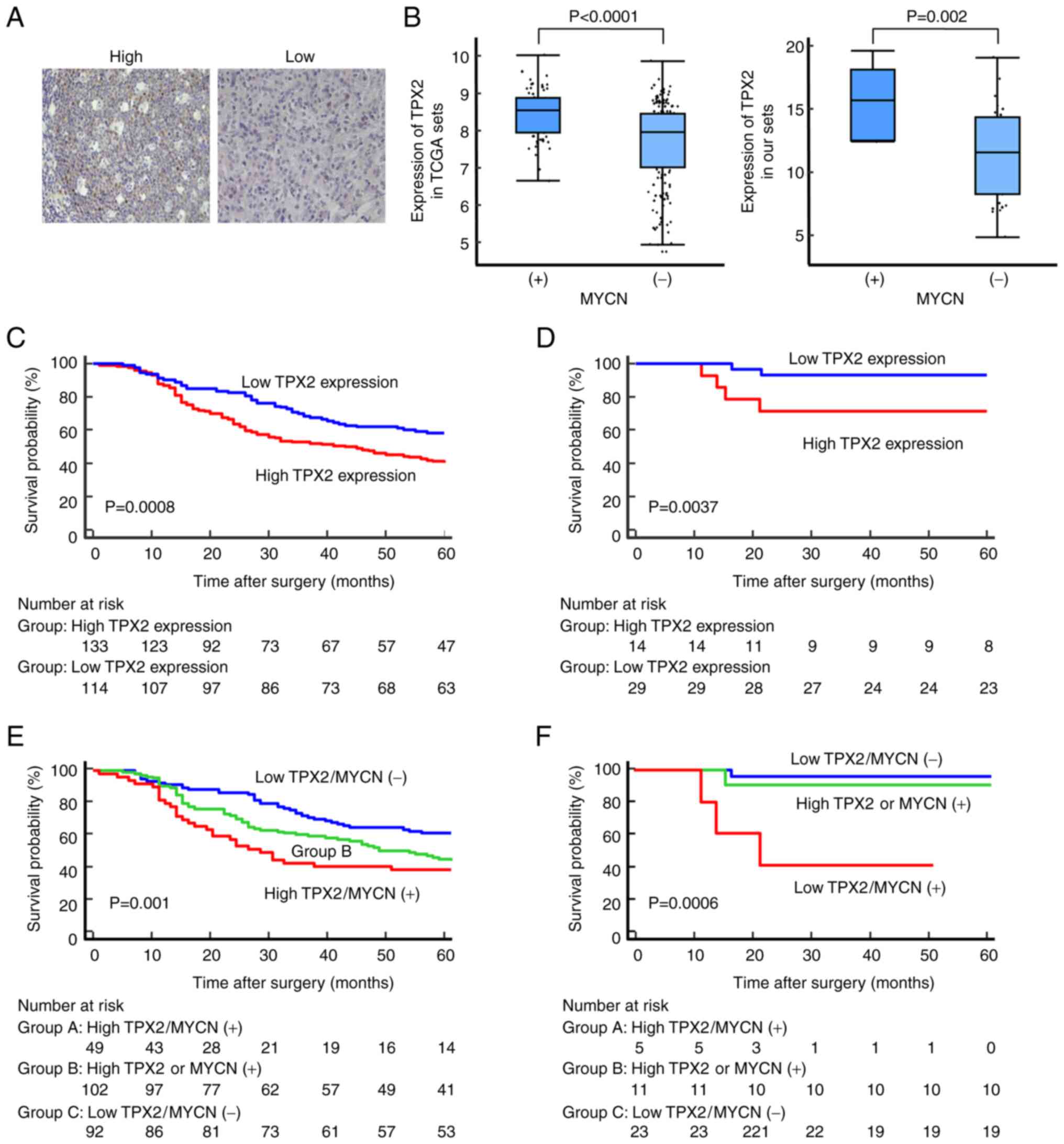
Representative immunostaining images of NB tissues
from validation set are revealed in Fig. 1A. The median was used as the
cut-off point for the definition of high and low TPX2 expression
TPX2 mRNA expression was significantly higher in NB with MYCN gene
amplification compared with that in patients with a single copy of
MYCN in both the TARGET-NB set (P<0.0001) and the validation set
(P=0.002) (Fig. 1B; Table II). In clinical trials of NB,
Aurora kinase inhibitor has been shown to exhibit preclinical
activity (24); therefore, the
correlation between Aurora kinase A (AURKA) and TPX2 (activator of
AURKA) was examined. A significant linear correlation was observed
between TPX2 expression and AURKA expression in the TARGET-NB set
(r2=0.78, P<0.0001; Fig. S1A). In the TARGET-NB set, TPX2
expression was significantly associated with age at diagnosis,
stage, MYCN status and tumor histology (Table SII).
 | Table II.Association between patient
characteristics and TPX2 expression in the validation set. |
Table II.
Association between patient
characteristics and TPX2 expression in the validation set.
| Clinicopathological
characteristic | TPX2 expression
(mean ± SD) | P-value |
|---|
| Age of onset,
months |
| 0.39 |
|
≥12 | 11.3±3.4 |
|
|
<12 | 12.3±4.0 |
|
| Sex |
| 0.68 |
|
Male | 11.5±3.6 |
|
|
Female | 12.1±3.9 |
|
| Primary tumor
site |
| 0.59 |
|
Abdomen | 11.2±3.4 |
|
|
Other | 11.9±3.8 |
|
| Primary tumor size,
cm |
| 0.58 |
|
>5 | 12.0±4.1 |
|
| ≤5 | 11.4±3.3 |
|
| Distant
metastasis |
| 0.45 |
|
Present | 12.3±3.9 |
|
|
Absent | 11.4±3.5 |
|
| Complete
resection |
| 0.55 |
|
Yes | 12.0±3.7 |
|
| No | 11.3±3.6 |
|
| INSS stage |
| 0.36 |
|
1/2/3/4s | 11.4±3.3 |
|
| 4 | 12.6±4.5 |
|
| MYCN
amplification |
| 0.002 |
|
Present | 15.4±2.6 |
|
|
Absent | 11.0±3.4 |
|
| INPC stage |
| 0.96 |
|
Favorable | 11.6±3.3 |
|
|
Unfavorable | 11.7±4.1 |
|
High TPX2 expression is significantly
associated with poor prognosis in patients with NB
To evaluate the association between TPX2 expression
and the outcome of NB patients, data of NB tumor samples from
TARGET (containing complete clinical data and survival information)
were analyzed. The cut-off value was defined according to the best
predictive value as calculated by ROC analysis (Fig. S1B and C). The cut-off values for
TARGET-NB set and the validation set of the present study were set
at 8.10 and 13.96, respectively. It was found that high expression
of TPX2 in patients with NB was significantly associated with
decreased overall survival (OS) (P=0.0008, log-rank test; Fig. 1C). Data from the validation set
revealed the same result (P=0.0037, log-rank test) (Fig. 1D). These results showed that high
expression of TPX2 in patients with NB was significantly correlated
with a poor prognosis for OS. However, in the multivariate analysis
of the TARGET-NB set, only the INSS stage was a strong independent
prognostic factor (Table
SIII).
Combined assessment of TPX2 expression
and MYCN gene amplification
Considering the prognostic role of MYCN gene
amplification in NB (25), it was
hypothesized that the combined assessment of TPX2 expression
(high/low expression) and MYCN status (MYCN amplification +/-) may
result in an increased predictive effect. The patients were divided
into three groups on the basis of the expression levels of TPX2 and
MYCN. It was found that NB patients with high expression of TPX2
and MYCN gene amplification had the poorest prognosis in both the
TARGET-NB set and the present validation set (Fig. 1E and F). The colocalization of TPX2
with N-Myc in NB cells was also observed using double fluorescence
immunohistochemistry. Similar results were observed in NB tissues
(Fig. 2A).
Inhibition of TPX2 suppresses cell
proliferation in NB cells
To explore the functional role of TPX2 in NB cells,
proliferation was evaluated in the MYCN-amplified cell line (IMR5)
and MYCN-non-amplified cell line [KP-N-SI(FA)] with TPX2 knockdown
mediated by siRNA. At 48 h post-transfection of TPX2 siRNA, TPX2
expression was significantly decreased in both the IMR5 and
KP-N-SI(FA) cells (Fig. S1D). It
was found that NB cell proliferation was significantly decreased
following TPX2 knockdown compared with that in controls in both
cell lines (Fig. 2B).
Knockdown of TPX2 expression induces
G2/M arrest in NB cells
Next, the influence of TPX2 on the cell cycle
distribution of NB cells was evaluated. After TPX2 knockdown in the
MYCN-amplified cell line and MYCN-non-amplified cell line, the
percentage of G2/M cells was increased compared with that in the
controls (Fig. 2C).
Inhibition of TPX2 promotes early
apoptosis in NB cells
To determine whether increased apoptosis induction
may explain the observed phenotype in TPX2 knockdown NB cells, cell
death and apoptosis were determined using an Annexin V & Dead
Cell kit. Apoptosis assays revealed that TPX2 knockdown
significantly decreased NB cell growth and increased the percentage
of early apoptotic cells (Fig.
2D).
Discussion
In the present study, the prognostic role of TPX2 in
NB and the association between TPX2 expression and MYCN gene
amplification in NB were evaluated using the TARGET-NB data set and
a validation set. It was found that high expression of TPX2 was
significantly associated with the poor OS of patients with NB in
both data sets. The expression of TPX2 was higher in patients with
MYCN gene amplification, and patients with high TPX2 expression and
MYCN amplification had the poorest OS compared with patients with
either low TPX2 expression or a single copy of MYCN. The
colocalization of TPX2 with N-Myc in NB cells and tissue was
observed. Knockdown of TPX2 in NB cell lines suppressed the
proliferation of NB cells and blocked the cell cycle progression.
Further analysis indicated that early apoptosis was significantly
increased in NB cells after TPX2 knockdown.
TPX2 is a microtubule-associated protein and a
critical factor for mitosis and spindle assembly (8). TPX2 recruits and activates AURKA
during mitosis (26,27). TPX2 binding and phosphorylation of
the Thr288 site of AURKA affect the conformation of AURKA, inducing
AURKA activity (28). In addition,
TPX2 enters the nucleus in response to DNA double-strand breaks
induced by ionizing radiation and regulates γ-histone 2AX (γ-H2AX)
levels in DNA repair (29). TPX2
and its partner AURKA are frequently upregulated in human tumors
(8,30). Knockdown of TPX2 or AURKA leads to
elevated γ-H2AX (17,31), which is associated with increased
apoptosis (32). van Gijn et
al (33) reported TPX2/AURKA
signaling as a potential therapeutic target in genomically unstable
cancer cells. However, the most current treatment strategy
targeting TPX2/AURKA signaling is only focusing on Aurora kinase
inhibitors (34).
Several studies have reported the prognostic role of
TPX2 expression in various types of human malignancies, including
gastric, colon, hepatic, pancreatic, lung, salivary gland and
cervical cancer (13,15,35–39).
In colorectal cancer, TPX2 promotes the progression of colorectal
adenoma to carcinoma (15).
Ognibene et al (17)
reported that high TPX2 expression in NB was associated with
unfavorable OS and relapse-free survival using Versteeg (n=88) and
SEQC (n=498) datasets. TPX2 expression in MYCN gene-amplified
patients with NB was significantly higher compared with expression
in MYCN single copy patients (17). Ooi et al (40) found that spindle assembly genes,
including TPX2 and AURKA genes, are upregulation and are predictive
of a poor outcome in NB bearing MYCN amplification in high-risk NB
patients with MYCN amplification. Another study showed that colon
cancer patients with high expression of MYC and AURKA/TPX2 have the
poorest OS (41). The present
study results were in line with the aforementioned findings. In the
present study, colocalization of TPX2 with N-Myc in the nucleus and
cytoplasm of NB cells was observed. The findings of the present
study also revealed that high TPX2 expression leads to a worse
prognosis in NB patients with MYCN amplification. AURKA inhibitor
alters the structure of AURKA and results in structural
destabilization of the MYC-AURKA complex (42), preventing phospho-MYC-AURKA protein
complex formation and leading to N-Myc degradation and cell death
in cancer (43). These findings
and mechanisms could explain the association of high TPX2 with poor
outcome and the increase of TPX2, an activator of AURKA, in MYCN
gene-amplified patients with NB.
Knockdown of TPX2 expression using siRNA in multiple
cancer cell lines has been shown to significantly inhibit cell
proliferation and viability (39,44,45).
The results of the present study revealed that proliferation was
significantly decreased in both a MYCN-amplified cell line and a
MYCN-non-amplified cell line with TPX2 knockdown. Further apoptosis
analysis demonstrated that TPX2 may play an anti-apoptotic role in
NB cells. Knockdown of TPX2 promotes cell death by decreasing the
resistance to apoptosis. The expression of TPX2 is strictly
controlled by the cell cycle. During the S/G2 phases,
TPX2 localizes to mitotic spindle poles and then is degraded after
completion of cytokinesis (8). Chu
et al (46) found that the
Aurora-A/TPX2 molecular axis regulated the cell cycle progression
of breast cancer cells. The present results identified that
knockdown of TPX2 led to a cell cycle arrest at the G2/M
phase.
The present study has several limitations. First, it
is a single-institution study and the expression of TPX2 was
assessed in only 43 NB tissues. Second, due to the long-term span
of this retrospective study, it was not possible to collect enough
paraffin-embedded tissue sections to assess the expression of TPX2
protein in NB specimens by immunohistochemical staining. Third, the
role of TPX2, such as that in DNA repair and chemoresistance, under
NB-relevant drugs was not evaluated. Finally, the study did not
provide evidence showing interaction between TPX2 and MYCN. Further
experimental evidence is necessary to prove the mechanism
underlying the effect between TPX2 and MYCN.
In conclusion, the present study provided evidence
for the clinical role and biological function of TPX2 in NB. The
present findings suggested that clinical assessment of TPX2
expression in primary tumors may provide prognostic information for
children with NB, particularly for NB patients with MYCN
amplification. Considering the high incidence of NB in childhood
tumors and the poor treatment effect, targeting TPX2 may be a novel
therapy for NB.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YK, CY, YT and YOku conceptualized and designed the
study. YK, YS, YN, KM, KA, KO, MI, KU and MH provided the samples
and clinical information, and helped analyze results. YK, CY, YT,
YOki, YOku, AY, TK, TS, MK and MO analyzed and interpreted the
data. YK, CY, YT and YOku performed statistical analysis. CY, YK,
YT and YOku drafted the manuscript. All authors revised the
manuscript for important intellectual content. YK and CY confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The protocol for this research project was approved
(approval no. 1464) by the Institutional Review Board of Mie
University Hospital (Tsu, Japan). The study was conducted in
accordance with The Declaration of Helsinki. Informed consent was
obtained from the parents/guardians of the patients using the
opt-out scheme.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TPX2
|
targeting protein for Xenopus
kinesin-like protein 2
|
|
NB
|
neuroblastoma
|
|
TARGET
|
Therapeutically Applicable Research to
Generate Effective Treatments
|
|
siRNA
|
small interfering RNA
|
|
ROC
|
receiver operating characteristic
|
|
OS
|
overall survival
|
|
AURKA/Aurora-A
|
aurora kinase A
|
References
|
1
|
Pizzo PA PD: Principles and Practice of
Pediatric Oncology. Sixth edition. Wolters Kluwer Health/Lippincott
Williams and Wilkins; Philadelphia: 2010
|
|
2
|
Smith MA, Altekruse SF, Adamson PC, Reaman
GH and Seibel NL: Declining childhood and adolescent cancer
mortality. Cancer. 120:2497–2506. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
American Cancer Society, . Key Statistics
About Neuroblastoma. https://www.cancer.org/cancer/neuroblastoma/about/key-statistics.htmlDecember
17–2021
|
|
4
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer Statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park JR, Kreissman SG, London WB, Naranjo
A, Cohn SL, Hogarty MD, Tenney SC, Haas-Kogan D, Shaw PJ, Kraveka
JM, et al: Effect of tandem autologous stem cell transplant vs
single transplant on event-free survival in patients with high-risk
neuroblastoma: A randomized clinical trial. JAMA. 322:746–755.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matthay KK, Maris JM, Schleiermacher G,
Nakagawara A, Mackall CL, Diller L and Weiss WA: Neuroblastoma. Nat
Rev Dis Primers. 2:160782016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wieczorek M, Bechstedt S, Chaaban S and
Brouhard GJ: Microtubule-associated proteins control the kinetics
of microtubule nucleation. Nat Cell Biol. 17:907–916. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Neumayer G, Belzil C, Gruss OJ and Nguyen
MD: TPX2: Of spindle assembly, DNA damage response, and cancer.
Cell Mol Life Sci. 71:3027–3047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heidebrecht HJ, Buck F, Steinmann J,
Sprenger R, Wacker HH and Parwaresch R: p100: A novel
proliferation-associated nuclear protein specifically restricted to
cell cycle phases S, G2, and M. Blood. 90:226–233. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garrido G and Vernos I: Non-centrosomal
TPX2-dependent regulation of the Aurora a kinase: Functional
implications for healthy and pathological cell division. Front
Oncol. 6:882016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moss DK, Wilde A and Lane JD: Dynamic
release of nuclear RanGTP triggers TPX2-dependent microtubule
assembly during the apoptotic execution phase. J Cell Sci 122(Pt
5). 644–655. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan L, Li Q, Yang J and Qiao B:
TPX2-p53-GLIPR1 regulatory circuitry in cell proliferation,
invasion, and tumor growth of bladder cancer. J Cell Biochem.
119:1791–1803. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tomii C, Inokuchi M, Takagi Y, Ishikawa T,
Otsuki S, Uetake H, Kojima K and Kawano T: TPX2 expression is
associated with poor survival in gastric cancer. World J Surg
Oncol. 15:142017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu H, Liu J, Feng J, Zhang Q, Bian T, Li
X, Sun H, Zhang J and Liu Y: Overexpression of TPX2 predicts poor
clinical outcome and is associated with immune infiltration in
hepatic cell cancer. Medicine (Baltimore). 99:e235542020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sillars-Hardebol AH, Carvalho B, Tijssen
M, Beliën JA, de Wit M, Delis-van Diemen PM, Pontén F, van de Wiel
MA, Fijneman RJ and Meijer GA: TPX2 and AURKA promote 20q
amplicon-driven colorectal adenoma to carcinoma progression. Gut.
61:1568–1575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shah KN, Bhatt R, Rotow J, Rohrberg J,
Olivas V, Wang VE, Hemmati G, Martins MM, Maynard A, Kuhn J, et al:
Aurora kinase A drives the evolution of resistance to
third-generation EGFR inhibitors in lung cancer. Nat Med.
25:111–118. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ognibene M, Podestà M, Garaventa A and
Pezzolo A: Role of GOLPH3 and TPX2 in neuroblastoma DNA damage
response and cell resistance to chemotherapy. Int J Mol Sci.
20:47642019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Irwin MS, Naranjo A, Zhang FF, Cohn SL,
London WB, Gastier-Foster JM, Ramirez NC, Pfau R, Reshmi S, Wagner
E, et al: Revised neuroblastoma risk classification system: A
report from the children's oncology group. J Clin Oncol.
39:3229–3241. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bosse KR and Maris JM: Advances in the
translational genomics of neuroblastoma: From improving risk
stratification and revealing novel biology to identifying
actionable genomic alterations. Cancer. 122:20–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cohn SL, Pearson AD, London WB, Monclair
T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et
al: The International neuroblastoma risk group (INRG)
classification system: An INRG Task Force report. J Clin Oncol.
27:289–297. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakagawara A, Li Y, Izumi H, Muramori K,
Inada H and Nishi M: Neuroblastoma. Jpn J Clin Oncol. 48:214–241.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matsushita K, Uchida K, Saigusa S, Ide S,
Hashimoto K, Koike Y, Otake K, Inoue M, Tanaka K and Kusunoki M:
Glycolysis inhibitors as a potential therapeutic option to treat
aggressive neuroblastoma expressing GLUT1. J Pediatr Surg.
47:1323–1330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mossé YP, Fox E, Teachey DT, Reid JM,
Safgren SL, Carol H, Lock RB, Houghton PJ, Smith MA, Hall D, et al:
A phase II study of alisertib in children with recurrent/refractory
solid tumors or leukemia: Children's oncology group phase I and
pilot consortium (ADVL0921). Clin Cancer Res. 25:3229–3238. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tonini GP, Boni L, Pession A, Rogers D,
Iolascon A, Basso G, Cordero di Montezemolo L, Casale F, Pession A,
Perri P, et al: MYCN oncogene amplification in neuroblastoma is
associated with worse prognosis, except in stage 4s: The Italian
experience with 295 children. J Clin Oncol. 15:85–93. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bayliss R, Sardon T, Vernos I and Conti E:
Structural basis of Aurora-A activation by TPX2 at the mitotic
spindle. Mol Cell. 12:851–862. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Asteriti IA, Rensen WM, Lindon C, Lavia P
and Guarguaglini G: The Aurora-A/TPX2 complex: A novel oncogenic
holoenzyme? Biochim Biophys Acta. 1806:230–239. 2010.PubMed/NCBI
|
|
28
|
Lake EW, Muretta JM, Thompson AR,
Rasmussen DM, Majumdar A, Faber EB, Ruff EF, Thomas DD and Levinson
NM: Quantitative conformational profiling of kinase inhibitors
reveals origins of selectivity for Aurora kinase activation states.
Proc Natl Acad Sci USA. 115:e11894–e11903. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Neumayer G, Helfricht A, Shim SY, Le HT,
Lundin C, Belzil C, Chansard M, Yu Y, Lees-Miller SP, Gruss OJ, et
al: Targeting protein for xenopus kinesin-like protein 2 (TPX2)
regulates γ-histone 2AX (γ-H2AX) levels upon ionizing radiation. J
Biol Chem. 287:42206–42222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pérez de Castro I and Malumbres M: Mitotic
stress and chromosomal instability in cancer: The case for TPX2.
Genes Cancer. 3:721–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Do TV, Hirst J, Hyter S, Roby KF and
Godwin AK: Aurora A kinase regulates non-homologous end-joining and
poly(ADP-ribose) polymerase function in ovarian carcinoma cells.
Oncotarget. 8:50376–50392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Parry JA, Chin A, Duensing S and
Duensing A: Soluble histone H2AX is induced by DNA replication
stress and sensitizes cells to undergo apoptosis. Mol Cancer.
7:612008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
van Gijn SE, Wierenga E, van den Tempel N,
Kok YP, Heijink AM, Spierings DCJ, Foijer F, van Vugt MATM and
Fehrmann RSN: TPX2/Aurora kinase A signaling as a potential
therapeutic target in genomically unstable cancer cells. Oncogene.
38:852–867. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Niu H, Manfredi M and Ecsedy JA:
Scientific rationale supporting the clinical development strategy
for the investigational Aurora a kinase inhibitor alisertib in
cancer. Front Oncol. 5:1892015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma Y, Lin D, Sun W, Xiao T, Yuan J, Han N,
Guo S, Feng X, Su K, Mao Y, et al: Expression of targeting protein
for xklp2 associated with both malignant transformation of
respiratory epithelium and progression of squamous cell lung
cancer. Clin Cancer Res. 12:1121–1127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang B, Jia C, Huang Y, He H, Li J, Liao
H, Liu X, Liu X, Bai X and Yang D: TPX2 level correlates with
hepatocellular carcinoma cell proliferation, apoptosis, and EMT.
Dig Dis Sci. 60:2360–2372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Warner SL, Stephens BJ, Nwokenkwo S,
Hostetter G, Sugeng A, Hidalgo M, Trent JM, Han H and Von Hoff DD:
Validation of TPX2 as a potential therapeutic target in pancreatic
cancer cells. Clin Cancer Res. 15:6519–6528. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shigeishi H, Ohta K, Hiraoka M, Fujimoto
S, Minami M, Higashikawa K and Kamata N: Expression of TPX2 in
salivary gland carcinomas. Oncol Rep. 21:341–344. 2009.PubMed/NCBI
|
|
39
|
Chang H, Wang J, Tian Y, Xu J, Gou X and
Cheng J: The TPX2 gene is a promising diagnostic and therapeutic
target for cervical cancer. Oncol Rep. 27:1353–1359.
2012.PubMed/NCBI
|
|
40
|
Ooi WF, Re A, Sidarovich V, Canella V,
Arseni N, Adami V, Guarguaglini G, Giubettini M, Scaruffi P,
Stigliani S, et al: Segmental chromosome aberrations converge on
overexpression of mitotic spindle regulatory genes in high-risk
neuroblastoma. Genes Chromosomes Cancer. 51:545–556. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Takahashi Y, Sheridan P, Niida A, Sawada
G, Uchi R, Mizuno H, Kurashige J, Sugimachi K, Sasaki S, Shimada Y,
et al: The AURKA/TPX2 axis drives colon tumorigenesis cooperatively
with MYC. Ann Oncol. 26:935–942. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Richards MW, Burgess SG, Poon E,
Carstensen A, Eilers M, Chesler L and Bayliss R: Structural basis
of N-Myc binding by Aurora-A and its destabilization by kinase
inhibitors. Proc Natl Acad Sci USA. 113:13726–13731. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dauch D, Rudalska R, Cossa G, Nault JC,
Kang TW, Wuestefeld T, Hohmeyer A, Imbeaud S, Yevsa T, Hoenicke L,
et al: A MYC-Aurora kinase A protein complex represents an
actionable drug target in p53-altered liver cancer. Nat Med.
22:744–753. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wei P, Zhang N, Xu Y, Li X, Shi D, Wang Y,
Li D and Cai S: TPX2 is a novel prognostic marker for the growth
and metastasis of colon cancer. J Transl Med. 11:3132013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang Y, Guo W and Kan H: TPX2 is a
prognostic marker and contributes to growth and metastasis of human
hepatocellular carcinoma. Int J Mol Sci. 15:18148–18161. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chu TLH, Connell M, Zhou L, He Z, Won J,
Chen H, Rahavi SMR, Mohan P, Nemirovsky O, Fotovati A, et al: Cell
cycle-dependent tumor engraftment and migration are enabled by
Aurora-A. Mol Cancer Res. 16:16–31. 2018. View Article : Google Scholar : PubMed/NCBI
|