Introduction
As one of the deadliest malignant tumors, pancreatic
cancer (PC) currently ranks tenth among the most frequent types of
cancer in men and the ninth in women in USA (1). In addition, PC is the third highest
cause of cancer-related mortality, accounting for ~227,000 deaths
annually worldwide (2). Due to its
atypical symptoms, PC is difficult to diagnose, thus resulting in
increased incidence and mortality rates (3). The most common non-specific symptoms
of PC include abdominal pain and weight loss, while the 5-year
survival rate of patients remains <5% (4,5).
Although significant progress has been made in the diagnosis and
management of PC, its causes remain poorly understood (6). Therefore, the present study aimed to
uncover the biological mechanisms underlying the development of PC
and identify relatively effective therapeutic approaches.
Neuronal pentraxin 1 (NPTX1), also known as
neuropilin-1 is a member of the long pentraxin family of proteins.
It is mainly expressed in central neurons and displays promotive
effects on neurite growth and regulates cellular properties
(7,8). NPTX1 was first identified as an
epigenetic target in PC by applying genome scanning technology,
which provides a global DNA methylation analysis (9). Notably, emerging evidence has
suggested that NPTX1 is involved in the development of different
types of cancer. For example, NPTX1 is shown to exhibit
antiproliferative effects on colon cancer, while NPTX1
overexpression is found to downregulate cyclin A2 and CDK2 in colon
cancer cells (10). In addition,
Zhou et al (11) report
that NPTX1 is a novel epigenetic regulation gene in lung cancer and
NPTX1 overexpression can attenuate lung cancer progression.
Furthermore, NPTX1 is found to be involved in the progression of PC
(12,13). In the current study, the mechanism
of NPTX1 in PC was further investigated.
Gemcitabine (GEM), the most important cytidine
analogue, exhibits antitumor activity in several tumor models
(14). It has been also suggested
that cisplatin (DDP) has great potential in treating various solid
tumors (15). To date, several
studies have investigated the effects of GEM and DDP on PC. For
example, Heinemann (16) reported
that treatment with single-agent gemcitabine achieved clinical
benefit and symptoms improvement in 20–30% of patients with a
higher 1-year survival and a median survival. Another study
indicated that the combination of gemcitabine and cisplatin
significantly improves the quality of life in patients with locally
advanced or metastatic PC, prolonging the survival time with
tolerable toxicity (17). The
aforementioned studies supported the antitumor activity of both GEM
and DDP in PC. Therefore, the present study also investigated the
effects of GEM and DDP on PC.
RNA-binding protein 10 (RBM10), a member of the RBP
family, is located on chromosome Xp11.23 (18). Previous studies demonstrated that
RBM10 could cell promote apoptosis and inhibit cell proliferation
(19,20). Xiao et al (21) demonstrate that RBM10 is
downregulated in PC, while its expression is associated with the
prognosis of PC. Other studies reveal that AKT, which is possibly
inhibited by RBM10, could be involved in the progression of various
types of cancer via regulating NPTX1 (22,23).
Therefore, the present study aimed to investigate the regulatory
effect of RBM10 on NPTX1 expression and to uncover their potential
interaction.
Materials and methods
Cell culture, treatment and
transfection
The normal human pancreatic ductal epithelial cell
line HPDE6-C7 and the PC cell lines PANC-1, CAPAN-1, SW1990 and
BxPC-3 were obtained from ATCC. The cells were cultured in DMEM
supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified incubator with 5% CO2. Subsequently, cells
were treated with 100 µM GEM for 72 h or 1 µM DDP for 24 h. To
overexpress NPTX1 and RBM10, PANC-1 and BxPC-3 cells were
transfected with NPTX1 and RBM10 overexpression plasmids (ov-NPTX1
and ov-RBM10; Hunan Fenghui Biotechnology Co., Ltd.) at 37°C for 48
h using Lipofectamine® 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Transfected cells were used for
subsequent experiments 48 h later.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from PANC-1 and BxPC-3 cells
(5×106 cells/ml) using TRIzol® reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Subsequently, the extracted RNA was reverse
transcribed into cDNA using the PrimeScript RT Reagent kit (Takara
Bio, Inc.) according to the manufacturer's instructions. To analyze
gene expression, qPCR was performed on the ABI PRISM 7000 Sequence
Detection System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The following
thermocycling conditions were used: Initial denaturation at 95°C
for 7 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30
sec, and then a final extension at 72°C for 30 sec. The primer
sequences for PCR are presented as below: NPTX1:
5′-ACCGAGGAGAGGGTCAAGAT-3′ (forward) and 5′-GTGGGAATGTGAGCTGGAAC-3′
(reverse); RBM10: 5′-AGGGCAAGCATGACTATGA-3′ (forward) and
5′-GTGGAGAGCTGGATGAAGG-3′ (reverse); GAPDH:
5′-GGGAAACTGTGGCGTGAT-3′ (forward) and 5′-GAGTGGGTGTCGCTGTTGA-3′
(reverse). The 2−ΔΔCq method (24) was employed to determine the
relative gene expression levels, which normalized to GAPDH level.
The experiments were repeated at least 3 times.
Western blot analysis
Total proteins were extracted from PANC-1 and BxPC-3
cells using RIPA lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.). The protein concentration was measured using
a BCA protein assay kit (Beyotime Institute of Biotechnology). The
protein samples (30 µg/lane) were separated by 10% SDS-PAGE and
were then transferred onto PVDF membranes. Following blocking with
5% non-fat milk at room temperature for 2 h, the membranes were
incubated with the appropriate primary antibodies against NPTX1
(catalog no. bs-4893R; 1:500; Bioss), Bcl-2 (catalog no. ab32124;
1:1,000), Bax (catalog no. ab32503; 1:1,000), cleaved PARP (catalog
no. ab32064; 1:1,000), MMP12 (catalog no. ab52897; 1:1,000), ZEB1
(catalog no. ab203829; 1:500), RBM10 (catalog no. ab72423; 1:2,000)
and GAPDH (catalog no. ab8245; 1:500) (all Abcam) at 4°C overnight.
Subsequently, the membranes were incubated with the corresponding
HRP-conjugated Goat Anti-Rabbit IgG secondary antibodies (catalog
no. ab205718; 1:2,000; Abcam) for 2 h at room temperature. Finally,
the protein bands were visualized by using an ECL reagent (Merck
KGaA). The protein intensity was quantified with ImageJ software
v1.8.0 (National Institutes of Health).
Cell counting kit-8 (CCK-8) assay
PANC-1 and BxPC-3 cells were seeded into 96-well
plates for 24 h. Subsequently, 10 µl CCK-8 reagent (Beyotime
Institute of Biotechnology) was added into each well and the cells
were incubated for 24, 48 and 72 h at 37°C in a humidified
incubator with 5% CO2. The absorbance of each well was
detected at a wavelength of 450 nm using a microplate reader
(Thermo Fisher Scientific, Inc.).
TUNEL assay
The apoptosis rate of PANC-1 and BxPC-3 cells was
determined using a TUNEL assay kit (Beyotime Institute of
Biotechnology). Briefly, PANC-1 and BxPC-3 cells were fixed with 4%
paraformaldehyde for 15 min and were then permeabilized with 0.25%
Triton-X 100 for 20 min at room temperature. Subsequently, cells
were incubated with TUNEL reaction solution at 37°C for 1 h
followed by staining with DAPI for 30 min at room temperature.
Finally, images of the TUNEL-positive cells randomly selected from
5 fields of view were captured under a fluorescence microscope
(magnification, ×200).
Wound healing assay
PANC-1 and BxPC-3 cells were inoculated into 6-well
plates at a density of 6×104 cells/well and cultured in
serum-free DMEM until reaching 90–100% confluence. Subsequently, a
straight linear wound was created across the cell monolayer using a
200-µl pipette tip. The cell debris was removed by washing with PBS
three times and the cells were then incubated at 37°C with 5%
CO2. Wound closure was measured using randomly selected
cells from 3 fields of view at 0 and 24 h post-treatment with a
light microscope (magnification, ×200). Finally, ImageJ software
v1.8.0 was used to determine cell migration.
Transwell assay
The invasive ability of PANC-1 and BxPC-3 cells was
evaluated by using a 24-well Transwell (CLS3396; Corning, Inc.)
assay. Briefly, PANC-1 and BxPC-3 cells (1×105
cells/well) were added into the upper chamber of the Transwell
inserts, while the lower compartment of the Transwell chamber was
filled with medium supplemented with 10% FBS. Following incubation
at 37°C for 24 h, cells were fixed with 4% paraformaldehyde for 30
min and stained with 0.1% crystal violet solution for 30 min at
37°C. Finally, images of the invading cells randomly selected from
5 fields of view were captured under a light microscope
(magnification, ×400).
RNA binding protein
immunoprecipitation (RIP) assay
Cells were scraped from culture dishes and incubated
with glycine after fixing by formaldehyde (0.3%) at room
temperature for 10 min. Then the cells were transferred into 1.5-ml
tubes and lysed with RIP buffer. Subsequently, the cells were
incubated with the anti-RBM10 antibody (catalog no. ab72423; 1:20;
Abcam) at 37°C overnight. Precipitated RNA was extracted using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and
analyzed by PCR amplification.
Bioinformatics analysis
StarBase (http://starbase.sysu.edu.cn/) predicted the
relationship between RBM10 and NPTX1. The RNA-Protein Interaction
Prediction (RPISeq; http://pridb.gdcb.iastate.edu/RPISeq) database
predicted the interaction probabilities of RBM10 and NPTX1.
Statistical analysis
The data are expressed as mean ± standard deviation.
All data were analyzed by using SPSS software (version 20.0; IBM
Corp.). Differences between two groups were compared using
Student's t-test, while those among multiple groups using ANOVA and
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
NPTX1 is downregulated in PC
cells
To determine the expression levels of NPTX1 in PC
cell lines, RT-qPCR and western blot analyses were performed. As
shown in Fig. 1A and B, the
relative mRNA and protein expression levels of NPTX1 were
significantly decreased in PC cell lines, particularly in PANC-1
and BxPC-3 cells. Therefore, PANC-1 and BxPC-3 cells were used for
the subsequent experiments.
NPTX1 overexpression inhibits
proliferation and promotes apoptosis in PC cells
As shown in Fig.
2A-D, NPTX1 was notably upregulated in PANC-1 and BxPC-3 cells
following cell transfection with ov-NPTX1. CCK-8 assays revealed
that PANC-1 and BxPC-3 cell viability was significantly reduced in
NPTX1-overexpressing cells, suggesting that NPTX1 overexpression
exerted antiproliferative effects on PC cells (Fig. 2E and F). Furthermore, the apoptosis
rate was significantly increased in PANC-1 and BxPC-3 cells
overexpressing NPTX1 (Fig. 2G and
H). Additionally, the protein level of Bcl-2 was decreased,
while the contents of Bax and cleaved poly ADP-ribose polymerase
(PARP) were significantly increased in PANC-1 and BxPC-3 cells
after transfection with ov-NPTX1 (Fig.
2I and J).
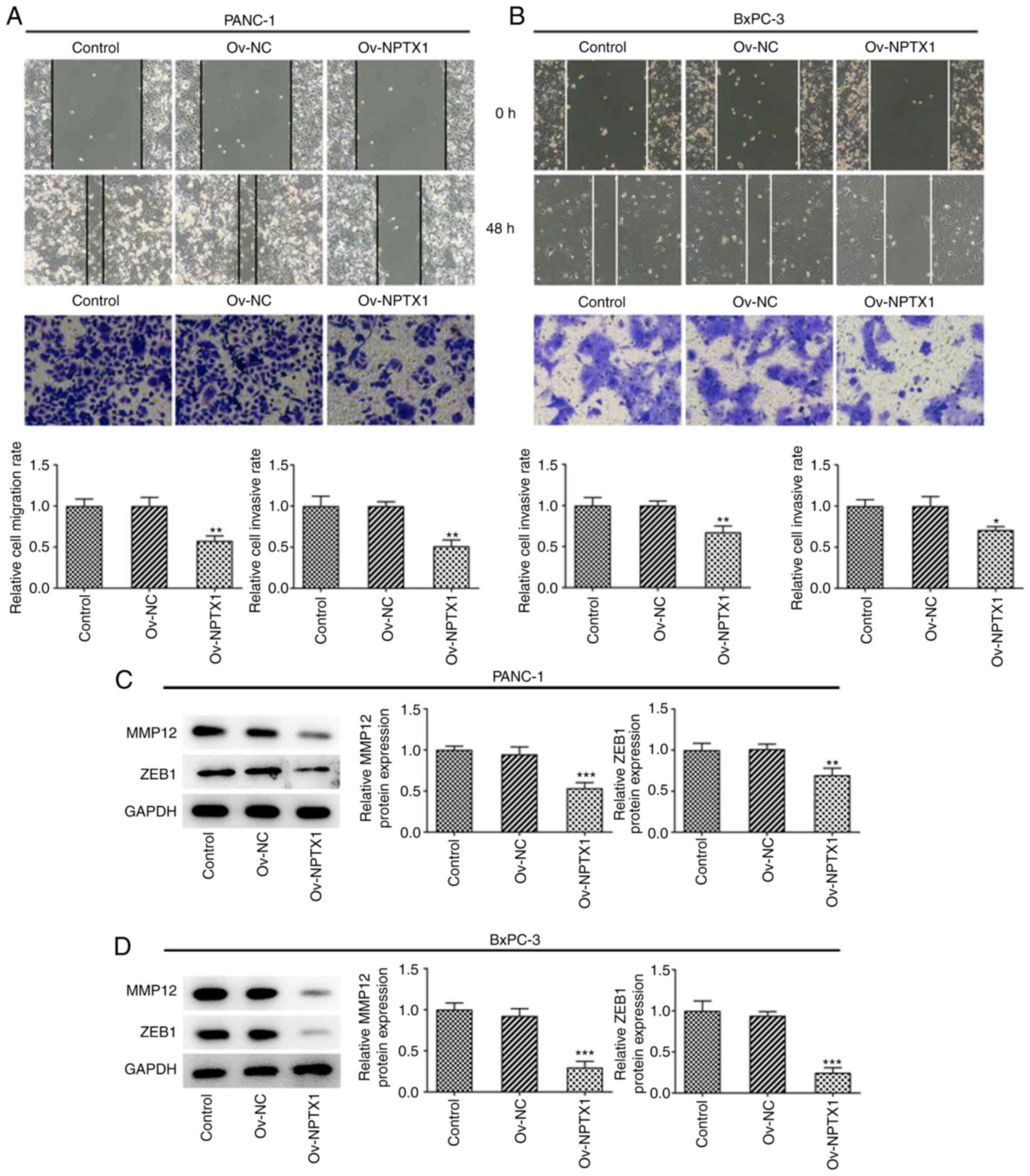
NPTX1 overexpression attenuates the
migration and invasion ability of PC cells
The results showed that NPTX1 overexpression reduced
the migration and invasion abilities of PANC-1 cells compared with
the untransfected cells (Fig. 3A).
Accordingly, NPTX1 overexpression attenuated the migration and
invasion abilities of BxPC-3 cells (Fig. 3B). As shown in Fig. 3C and D, MMP12 and zinc finger
E-box-binding homeobox 1 were significantly downregulated in PC
cells overexpressing NPTX1. The aforementioned findings indicated
that NPTX1 overexpression exerted an inhibitory effect on PC cell
migration and invasion.
NPTX1 overexpression enhances the
sensitivity of PANC-1 and BxPC-3 cells to GEM and DDP
CCK-8 assays revealed that the viability of PANC-1
and BxPC-3 cells treated with GEM was decreased in a dose-dependent
manner (Fig. 4A and B). In
addition, NPTX1 overexpression further decreased the viability of
PANC-1 and BxPC-3 cells treated with GEM compared with cells
treated with negative control overexpression plasmid (ov-NC). This
finding supported that NPTX1 overexpression could enhance the
sensitivity of PC cells to GEM. Furthermore, NPTX1 overexpression
notably decreased the viability of DDP-treated PANC-1 and BxPC-3
cells compared with the ov-NC group. Similarly, NPTX1
overexpression exerted the same effect on the sensitivity of PC
cells to DDP (Fig. 4C and D).
RBM10 overexpression stabilizes the
mRNA and protein expression levels of NPTX1
Bioinformatics analysis using the StarBase
(http://starbase.sysu.edu.cn/) and
RNA-Protein Interaction Prediction (RPISeq; http://pridb.gdcb.iastate.edu/RPISeq) databases
predicted that RBP could interact with NPTX1. In addition, the mRNA
expression and protein level of RBM10 in PC cells were
significantly downregulated compared with normal pancreatic ductal
epithelial cell line HPDE6-C7 cells (Fig. 5A and B). RT-qPCR and western blot
analyses demonstrated that the expression of RBM10 was markedly
increased in PANC-1 and BxPC-3 cells following cell transfection
with ov-RBM10 (Fig. 5C-F). As
shown in Fig. 5G-J, NPTX1 was
significantly upregulated in PANC-1 and BxPC-3 cells overexpressing
RBM10. Furthermore, RIP assays showed that RBM10 could bind with
NPTX1 mRNA (Fig. 5K). To further
verify that the expression of RBM10 could stabilize the expression
of NPTX1, a mRNA stability assay was performed using actinomycin D.
As shown in Fig. 5L, RBM10
overexpression enhanced the expression of NPTX1 in PANC-1 and
BxPC-3 cells.
Discussion
PC, an aggressive type of cancer of the digestive
system, has become a severe health problem globally (25). The lack of diagnostic and
prognostic biomarkers allowing the early diagnosis and prognosis of
patients with PC have contributed to the poor survival rate in
these patients (26). Therefore,
PC screening and treatment have come to represent a major challenge
in the clinical setting (27). The
present study demonstrated that NPTX1 was downregulated in PC cell
lines. NPTX1 overexpression suppressed the cell proliferation of
PANC-1 and BxPC-3 cells by CCK-8 assay and promoted cell apoptosis
by Tunel assay and the detection of levels of Bcl2, Bax and cleaved
PARP. In addition, NPTX1 overexpression also was found to inhibit
the invasion and migration of PANC-1 and BxPC-3 cells with
decreased levels of MMP12 and zinc finger E-box-binding homeobox 1.
Moreover, upregulated NPTX1 enhanced the sensitivity of 0–1 µM of
GEM and DDP in PANC-1 and BxPC-3 cells. Mechanistic investigations
showed that NPTX1 was combined with RBM10 and overexpression of
RBM10 increased the stability of the mRNA and protein levels of
NPTX1.
It has been reported that NPTX1, which was first
identified in the central nervous system, serves a key role in
regulating neural lineage specification (28). In addition, it has been suggested
that NPTX1 is involved in cancer progression. For example, a
previous study demonstrated that NPTX1 silencing promoted cell
proliferation, migration and EMT process in head and neck squamous
cell (29). In addition, Zhao
et al (22) found that
NPTX1 suppresses the growth ability of HCC cells and contributes to
mitochondria-related apoptosis by an AKT-mediated signaling
mechanism.
As a pyrimidine nucleoside analog and anticancer
drug, GEM shows high efficacy against several types of solid tumors
(30). DDP is considered as one of
the most effective anticancer drugs, owing to its ability to
activate or silence different genes to activate the cellular
self-defense system (31). In
addition, GEM and DDP are used to treat several types of cancer,
such as lung, ovarian, bladder and breast cancer (32–35).
The results of the present study demonstrated that treatment of
PANC-1 and BxPC-3 cells with GEM or DDP could decrease cell
viability in a dose-dependent manner.
In the present study, the role of NPTX1 in PC was
investigated. The results demonstrated that NPTX1 was downregulated
in PC cells, while NPTX1 overexpression attenuated the
proliferation, migration, invasion and expression of
apoptosis-related proteins in these cells. Notably, NPTX1
overexpression could promote cell apoptosis and enhance the
sensitivity of PC cells to GEM and DDP.
RBM10 is involved in the repair of damaged tissue as
well as in different cellular processes (36,37).
Loiselle and Sutherland (37) and
Rodor et al (38) revealed
that RBM10 is involved in cell proliferation and tissue
infiltration, thus accelerating the progression of different
diseases. Through the StarBase database, it was predicted that
RBM10 RBP can be combined with NPTX1. The RNA-Protein Interaction
Prediction (RPISeq) database (http://pridb.gdcb.iastate.edu/RPISeq) also predicted
that the interaction probability of RBM10 with NPTX1 was 0.85
(>0.5 means combined). The results of the present study showed
that RBM10 could interact with NPTX1. Furthermore, RBM10 was
downregulated in PC cells, while its overexpression notably
upregulated NPTX1, thus suggesting that RBM10 overexpression could
regulate the expression of NPTX1. This finding was further verified
by mRNA stability assays using actinomycin D. However, there were
certain limitations to the present study. For example, the effect
of GEM and DDP co-administration on the aforementioned processes
was not investigated. Therefore, further studies will be conducted
to fully uncover the role of combination use of GEM and DDP in PC.
In addition, it has been recently reported that interferon
signaling pathways are involved in the occurrence of PC and chronic
pancreatitis (39,40). It is therefore hypothesized that
interferon signaling pathways are important for the occurrence of
PC and chronic pancreatitis and the relationship between NPTX1 and
the interferon system in PC will be explored in a study.
Overall, the present study demonstrated that RBM10
could interact with NPTX1, while NPTX1 overexpression could enhance
the proliferation, migration, invasion and apoptosis of PC cells
via targeting RBM10. Additionally, NPTX1 overexpression could
enhance the sensitivity of PC cells to GEM and DDP chemotherapy,
which provides a novel biological marker for GEM and DDP-resistant
PC patients.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JW and GL designed the study, drafted and revised
the manuscript. KA and LS analyzed the data and searched the
literature. JW, GL and KA performed the experiments. JW and GL
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peto R: The fraction of cancer
attributable to lifestyle and environmental factors in the UK in
2010. Br J Cancer. 105 (Suppl 2):S12011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, Yang G, You L, Yang J, Feng M, Qiu
J, Zhao F, Liu Y, Cao Z, Zheng L, et al: Role of the microbiome in
occurrence, development and treatment of pancreatic cancer. Mol
Cancer. 18:1732019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chu LC, Goggins MG and Fishman EK:
Diagnosis and detection of pancreatic cancer. Cancer J. 23:333–342.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Quaresma M, Coleman MP and Rachet B:
40-year trends in an index of survival for all cancers combined and
survival adjusted for age and sex for each cancer in England and
Wales, 1971-2011: A population-based study. Lancet. 385:1206–1218.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raimondi S, Maisonneuve P and Lowenfels
AB: Epidemiology of pancreatic cancer: An overview. Nat Rev
Gastroenterol Hepatol. 6:699–708. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsui CC, Copeland NG, Gilbert DJ, Jenkins
NA, Barnes C and Worley PF: Narp, a novel member of the pentraxin
family, promotes neurite outgrowth and is dynamically regulated by
neuronal activity. J Neurosci. 16:2463–2478. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schlimgen AK, Helms JA, Vogel H and Perin
MS: Neuronal pentraxin, a secreted protein with homology to acute
phase proteins of the immune system. Neuron. 14:519–526. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hagihara A, Miyamoto K, Furuta J, Hiraoka
N, Wakazono K, Seki S, Fukushima S, Tsao MS, Sugimura T and
Ushijima T: Identification of 27 5′ CpG islands aberrantly
methylated and 13 genes silenced in human pancreatic cancers.
Oncogene. 23:8705–8710. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng X, Pan K, Zhao W, Zhang J, Yuan S,
Wen X, Zhou W and Yu Z: NPTX1 inhibits colon cancer cell
proliferation through down-regulating cyclin A2 and CDK2
expression. Cell Biol Int. 42:589–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou C, Qin Y, Xie Z, Zhang J, Yang M, Li
S and Chen R: NPTX1 is a novel epigenetic regulation gene and
associated with prognosis in lung cancer. Biochem Biophys Res
Commun. 458:381–386. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yue W, Wang T, Zachariah E, Lin Y, Yang
CS, Xu Q, DiPaola RS and Tan XL: Transcriptomic analysis of
pancreatic cancer cells in response to metformin and aspirin: An
implication of synergy. Sci Rep. 5:133902015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yue W, Yang CS, DiPaola RS and Tan XL:
Repurposing of metformin and aspirin by targeting AMPK-mTOR and
inflammation for pancreatic cancer prevention and treatment. Cancer
Prev Res (Phila). 7:388–397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mini E, Nobili S, Caciagli B, Landini I
and Mazzei T: Cellular pharmacology of gemcitabine. Ann Oncol. 17
(Suppl 5):v7–v12. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ghosh S: Cisplatin: The first metal based
anticancer drug. Bioorg Chem. 88:1029252019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heinemann V: Gemcitabine: Progress in the
treatment of pancreatic cancer. Oncology. 60:8–18. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Ni Q, Jin M, Li Z, Wu Y, Zhao Y
and Feng F: Gemcitabine or gemcitabine plus cisplatin for in 42
patients with locally advanced or metastatic pancreatic cancer.
Zhonghua Zhong Liu Za Zhi. 24:404–407. 2002.PubMed/NCBI
|
|
18
|
Thiselton DL, McDowall J, Brandau O,
Ramser J, d'Esposito F, Bhattacharya SS, Ross MT, Hardcastle AJ and
Meindl A: An integrated, functionally annotated gene map of the
DXS8026-ELK1 interval on human Xp11.3-Xp11.23: Potential hotspot
for neurogenetic disorders. Genomics. 79:560–572. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao J, Sun Y, Huang Y, Song F, Huang Z,
Bao Y, Zuo J, Saffen D, Shao Z, Liu W and Wang Y: Functional
analysis reveals that RBM10 mutations contribute to lung
adenocarcinoma pathogenesis by deregulating splicing. Sci Rep.
7:404882017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang K, Bacon ML, Tessier JJ, Rintala-Maki
ND, Tang V and Sutherland LC: RBM10 modulates apoptosis and
influences TNF-α gene expression. J Cell Death. 5:1–19. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao W, Chen X, Li X, Deng K, Liu H, Ma J,
Wang Z, Hu Y and Hou J: RBM10 regulates human TERT gene splicing
and inhibits pancreatic cancer progression. Am J Cancer Res.
11:157–170. 2021.PubMed/NCBI
|
|
22
|
Zhao Y, Yu Y, Zhao W, You S, Feng M, Xie
C, Chi X, Zhang Y and Wang X: As a downstream target of the AKT
pathway, NPTX1 inhibits proliferation and promotes apoptosis in
hepatocellular carcinoma. Biosci Rep. 39:BSR201816622019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin X, Di X, Wang R, Ma H, Tian C, Zhao M,
Cong S, Liu J, Li R and Wang K: RBM10 inhibits cell proliferation
of lung adenocarcinoma via RAP1/AKT/CREB signalling pathway. J Cell
Mol Med. 23:3897–3904. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Previdi MC, Carotenuto P, Zito D, Pandolfo
R and Braconi C: Noncoding RNAs as novel biomarkers in pancreatic
cancer: What do we know? Future Oncol. 13:443–453. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gedge K: Pancreatic cancer: A symptomless
killer. J Perioper Pract. 27:158–161. 2017.PubMed/NCBI
|
|
28
|
Boles NC, Hirsch SE, Le S, Corneo B, Najm
F, Minotti AP, Wang Q, Lotz S, Tesar PJ and Fasano CA: NPTX1
regulates neural lineage specification from human pluripotent stem
cells. Cell Rep. 6:724–736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu S, Zhou C, Zou B, Zhang H and Feng M:
MiR-4295 facilitates cell proliferation and metastasis in head and
neck squamous cell carcinoma by targeting NPTX1. Genes Immun.
21:4–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miao H, Chen X and Luan Y: Small molecular
gemcitabine prodrugs for cancer therapy. Curr Med Chem.
27:5562–5582. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shen DW, Pouliot LM, Hall MD and Gottesman
MM: Cisplatin resistance: A cellular self-defense mechanism
resulting from multiple epigenetic and genetic changes. Pharmacol
Rev. 64:706–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abrams TJ, Lee LB, Murray LJ, Pryer NK and
Cherrington JM: SU11248 inhibits KIT and platelet-derived growth
factor receptor beta in preclinical models of human small cell lung
cancer. Mol Cancer Ther. 2:471–478. 2003.PubMed/NCBI
|
|
33
|
Koch M, Krieger ML, Stölting D, Brenner N,
Beier M, Jaehde U, Wiese M, Royer HD and Bendas G: Overcoming
chemotherapy resistance of ovarian cancer cells by liposomal
cisplatin: Molecular mechanisms unveiled by gene expression
profiling. Biochem Pharmacol. 85:1077–1090. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Steinberg RL, Thomas LJ, Brooks N, Mott
SL, Vitale A, Crump T, Rao MY, Daniels MJ, Wang J, Nagaraju S, et
al: Multi-institution evaluation of sequential gemcitabine and
docetaxel as rescue therapy for nonmuscle invasive bladder cancer.
J Urol. 203:902–909. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seidman AD: Gemcitabine as single-agent
therapy in the management of advanced breast cancer. Oncology
(Williston Park). 15 (2 Suppl 3):S11–S14. 2001.
|
|
36
|
Jackson TC, Du L, Janesko-Feldman K, Vagni
VA, Dezfulian C, Poloyac SM, Jackson EK, Clark RS and Kochanek PM:
The nuclear splicing factor RNA binding motif 5 promotes caspase
activation in human neuronal cells, and increases after traumatic
brain injury in mice. J Cereb Blood Flow Metab. 35:655–666. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Loiselle JJ and Sutherland LC:
Differential downregulation of Rbm5 and Rbm10 during skeletal and
cardiac differentiation. In Vitro Cell Dev Biol Anim. 50:331–339.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rodor J, FitzPatrick DR, Eyras E and
Cáceres JF: The RNA-binding landscape of RBM10 and its role in
alternative splicing regulation in models of mouse early
development. RNA Biol. 14:45–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Minaga K, Watanabe T, Hara A, Kamata K,
Omoto S, Nakai A, Otsuka Y, Sekai I, Yoshikawa T, Yamao K, et al:
Identification of serum IFN-α and IL-33 as novel biomarkers for
type 1 autoimmune pancreatitis and IgG4-related disease. Sci Rep.
10:148792020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fujisawa M, Kanda T, Shibata T, Sasaki R,
Masuzaki R, Matsumoto N, Nirei K, Imazu H, Kuroda K, Sugitani M, et
al: Involvement of the interferon signaling pathways in pancreatic
cancer cells. Anticancer Res. 40:4445–4455. 2020. View Article : Google Scholar : PubMed/NCBI
|