Introduction
Breast cancer (BC) is the most common malignancy in
women, accounting for ~22% of all new cancer cases worldwide and
>1.05 million new cases annually (1). Globally, 450,000 patients succumb to
BC each year, which accounts for 3.7% of all cancer mortalities in
women (1,2). BC is a heterogeneous disease that has
a wide spectrum of clinical, pathological and prognostic subtypes.
BC occurs when normal epithelial cells become hyperplastic and
develop into carcinoma in situ, eventually transitioning to
an invasive carcinoma to facilitate metastasis (3,4).
Therefore, the molecular mechanism of BC tumorigenesis and
development requires further study to identify novel therapeutic
strategies for the treatment of this disease.
MicroRNAs (miRNAs/miRs) serve an important role in
cancer metastasis (5). Abnormal
expression profiles of miRNAs have been reported in numerous types
of BC (6). Moreover, the
dysregulation of miRNAs has been demonstrated to be involved in the
tumorigenesis and progression of BC, including miR-142 (7), miR-205 (8) and miR-92 (9). Previous studies have identified
various miRNAs that are aberrantly expressed in human malignancies
(10–12). miR-217-5p is downregulated in
gastric cancer (10), pancreatic
ductal adenocarcinoma (11) and
osteosarcoma (12). However, the
biological function of miR-217-5p in BC remains to be
elucidated.
The epithelial-mesenchymal transition (EMT) refers
to the biological process whereby cells of the epithelial phenotype
transform into those with a mesenchymal phenotype. This process
serves an important role in embryonic development, chronic
inflammation, tissue remodeling, cancer metastasis and various
fibrotic disease states (13). The
main characteristics of EMT are a reduction in the expression of
cell adhesion molecules, including E-cadherin, and the
transformation of the cytokeratin cytoskeleton (14). Vimentin-based cytoskeletons and
morphological features are typical characteristics of cells with
mesenchymal phenotypes (14).
Through the EMT, epithelial cells lose polarity and their
connection with the basement membrane, whereas other cells acquire
mesenchymal phenotypes, such as increased migration and invasion,
reduced apoptosis and the increased ability to degrade the
extracellular matrix (15). The
EMT is an important biological process that allows malignant tumor
cells derived from epithelial cells to acquire metastatic
capabilities (15).
Metadherin (MTDH) was first identified by Su et
al (16) in 2002. MTDH has a
functional role in several crucial aspects of tumor progression,
including transformation, proliferation, cell survival, evasion of
apoptosis, migration and invasion, metastasis, angiogenesis and
chemoresistance. MTDH is located on the 8q22 locus and has been
previously associated with the initiation and progression of
numerous types of cancer. MTDH is also known as astrocyte-elevated
gene-1 and is a type of oncogene (17). The upregulated expression of MTDH
has been observed in BC, ovarian, prostate, lung, esophageal and
colorectal cancer (17–19). Moreover, it promotes the
invasiveness of human BC cells by inducing the EMT (18). The NF-κB signaling pathway has been
reported to participate in the MTDH-induced EMT of BC cells in
vitro (18). Previous reports
suggest that MTDH activates NF-κB via the degradation of IκBα and
by directly binding to the p65 NF-κB subunit (19,20).
miR-217 possibly functions as a tumor suppressor in
pancreatic ductal adenocarcinoma by targeting KRAS (11,21).
Furthermore, miR-217 has been reported to induce endothelial cell
senescence via targeting of sirtuin 1 (22). Previously identified miR-217
targets include E2F transcription factor 3, enhancer of zeste
homolog 2 and runt-related transcription factor 2, the suppression
of which inhibits clear cell renal cell carcinoma, gastric cancer
and glioma, respectively (21,23,24).
However, miR-217 is also an oncogene that targets PTEN and
peroxisome proliferator-activated receptor γ co-activator 1-α to
enhance germinal center responses in mature B-cell lymphomagenesis
(25,26). Based on these previous reports, it
can be hypothesized that miR-217 can mediate context-dependent
functions during carcinogenesis. miR-217-5p is the main member of
the miR-217 family, and it is also the main research object of the
aforementioned studies.
The clinical significance of miR-217-5p expression
in BC remains controversial with previous studies reporting
conflicting results (27,28). The mechanism by which miR-217
targets MTDH to regulate BC cell function remains unclear.
Therefore, in the present study, miR-217-5p and MTDH expression
levels were examined in human BC tissue and cell lines. The
possible function of miR-217-5p and MTDH in BC was also
investigated.
Materials and methods
Clinical tissues
In total 35 pairs of BC tissues and matched adjacent
non-tumor tissues were obtained from patients (age, 28–65 years; 35
females) who were surgically treated at the Breast Center, The
Fourth Hospital of Hebei Medical University (Shijiazhuang, China)
between August 2020 and November 2021. These patients were all
pathologically diagnosed with breast cancer. None of the patients
received any treatment prior to surgery and written informed
consent was obtained from all patients. The clinicopathological
information of the patients is presented in Table I. Tissue samples were frozen in
liquid nitrogen and stored in a refrigerator at −80°C. The present
study was approved by the Ethics Committee of The Fourth Hospital
of Hebei Medical University (approval no. 2021KY056). The present
study was conducted in accordance with the principles described in
The Declaration of Helsinki.
 | Table I.Clinicopathological characteristics
of patients with breast cancer (n=35) in the present study. |
Table I.
Clinicopathological characteristics
of patients with breast cancer (n=35) in the present study.
|
Characteristics | Patients, n
(%) |
|---|
| Age, years |
|
|
≤50 | 13 (37.1) |
|
>50 | 22 (62.9) |
| Tumor size, cm |
|
| ≤3 | 28 (80) |
|
>3 | 7 (20) |
| Grade |
|
| II | 24 (68.6) |
|
III | 11 (31.4) |
| TNM stage |
|
| I | 14 (40) |
| II | 15 (42.9) |
|
III | 6 (17.1) |
| ER status |
|
|
Positive | 25 (71.4) |
|
Negative | 10 (28.6) |
| PR status |
|
|
Positive | 20 (57.1) |
|
Negative | 15 (42.9) |
| HER2 status |
|
|
Positive | 5 (14.3) |
|
Negative | 30 (85.7) |
| Ki-67, % |
|
|
≤20 | 13 (37.1) |
|
>20 | 22 (62.9) |
| Lymph node
status |
|
|
Positive | 13 (37.1) |
|
Negative | 22 (62.9) |
Cell culture and transfection
The human breast epithelial MCF10A cell line and BC
SK-BR3, MCF7 and BT549 cell lines were purchased from Procell Life
Science & Technology, Co., Ltd. Cells were cultured in
RPMI-1640 media (Invitrogen; Thermo Fisher Scientific, Inc.)
containing 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and
incubated with 5% CO2 at 37°C. miR-217-5p mimic,
miR-negative control (NC) mimic, miR-217-5p inhibitor and
miR-NC-inhibitor were synthesized by Shanghai GenePharma Co., Ltd.,
and transfected into SK-BR3 or BT549 cells at a final concentration
of 100 nM using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 6 h. Subsequent
experimentation was performed 48 h after transfection. The
following sequences were used: miR-217-5p mimic,
5′-UACUGCAUCAGGAACUGAUUGGA-3′; miR-NC mimic,
5′-UUGUCCGAACGUGUCACGU-3′; miR-217-5p inhibitor,
5′-UCCAAUCAGUUCCUGAUGCAGUA-3′; and miR-NC inhibitor,
5′-CAGCUGGUUGAAGGGGACCAAA-3′.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from BC
tissues and BC cell lines, including miRNA. RNA quality was
assessed using the A260/A280 ratio. This value was determined using
a microplate reader (Multiskan Sky; Thermo Fisher Scientific,
Inc.). RNA quantity was also assessed using a microplate reader.
Total RNA was reverse transcribed into complementary (c)DNA using a
Transcriptor First Strand cDNA Synthesis Kit [Roche Diagnostics
(Shanghai) Co., Ltd.] using the following temperature protocol
according to the manufacturer's instructions: 37°C for 15 min and
85°C for 5 sec. The volume of each RT reaction system was 20 µl,
including 1 µg of total RNA. DNase was used to remove genomic DNA
interference. Subsequently, qPCR was performed to examine gene
expression using a SYBR Green PCR Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The following thermocycling
conditions were used for qPCR: Initial denaturation at 95°C for 5
min; followed by 40 cycles of denaturation at 95°C for 5 sec, 60°C
for 30 sec and 72°C for 30 sec, with a final extension step at 72°C
for 2 min. The 2−ΔΔCq method was used to calculate the
relative expression levels using GAPDH or U6 as the internal
reference gene (29). The primer
sequences used for RT-qPCR are presented in Table II.
 | Table II.Primer sequences used for reverse
transcription-quantitative PCR. |
Table II.
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5′-3′) |
|---|
| miR-217-5p | F:
TACTGCATCAGGAACTGATTGGA |
|
| R:
CTCAACTGGTGTCGTGG |
| U6 | F:
CTGGTAGGGTGCTCGCTTCGGCAG |
|
| R: CAACTGGTGTCGTGG
AGTCGG |
| MTDH | F:
CTGTTGAAGTGGCTGAGGG |
|
| R:
GGTGGCTGCTTGCTGTTA |
| GAPDH | F:
GCACCGTCAAGGCTGAGAAC |
|
| R:
GGATCTCGCTCCTGGAAGATG |
| E-cadherin | F:
CGAGAGCTACACGTTCACGG |
|
| R:
GGGTGTCGAGGGAAAAATAGG |
| Vimentin | F:
GACGCCATCAACACCGAGTT |
|
| R:
CTTTGTCGTTGGTTAGCTGGT |
| slug | F:
TGTGACAAGGAATATGTGAGCC |
|
| R:
TGAGCCCTCAGATTTGACCTG |
| p65 | F:
ATGTGGAGATCATTGAGCAGC |
|
| R:
CCTGGTCCTGTGTAGCCATT |
| IκBα | F:
CTCCGAGACTTTCGAGGAAATAC |
|
| R:
GCCATTGTAGTTGGTAGCCTTCA |
Cell proliferation assay
Cell proliferation was analyzed using the Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay.
Briefly, transfected SK-BR3 or BT549 cells were seeded into 96-well
plates at a density of 2×103 cells/well. Following
incubation at 37°C for 24, 48 and 72 h, 10 µl CCK-8 solution was
added into each well and incubated at 37°C for 4 h. The absorbance
of each well at a wavelength of 450 nm was assessed using a
microplate reader (Multiskan Sky; Thermo Fisher Scientific,
Inc.).
Colony formation assay
Colony formation assays were performed to detect
cell proliferation. Transfected SK-BR3 and BT549 cells were seeded
at 1×103 cells/well in six-well plates and incubated
with 5% CO2 at 37°C. After 15 days, colonies were
visible to the naked eye. These colonies were fixed with 4%
paraformaldehyde for 15 min at ambient temperature and stained with
1% crystal violet (Beyotime Institute of Biotechnology) for 15 min
at ambient temperature. The number of colonies (containing >50
cells) was counted microscopically.
Wound healing assay
After 48 h of transfection, 2×105 SK-BR3
or BT549 cells were transferred into a 35-mm2 petri
dish, cultured at 37°C and grown to 90% confluence. A wound was
then created using a 200-µl pipette tip to scratch the monolayer
before any detached cells were washed away with PBS. The remaining
cells were cultured in serum-free culture media, and wound closure
was then observed. Images of the wounds were taken at 0 and 24 h
after the scratch was made using a light microscope (magnification,
×100; Olympus Corporation). Migratory ability was calculated as the
mobility ratio using the following formula: Mobility ratio=(scratch
width at 24 h-scratch width at 0 h)/scratch width at 0 h.
Western blotting
Total protein was extracted using RIPA lysis buffer
(Beyotime Institute of Biotechnology) from transfected SK-BR3 and
BT549 cell lines. Protein concentration was determined by BCA assay
(Thermo Fisher Scientific, Inc.). Next, the protein lysates (30 µg)
were separated using SDS-PAGE on a 10% gel and separated proteins
were then transferred onto PVDF membranes (MilliporeSigma).
Subsequently, the membranes were incubated with 5% skimmed milk at
room temperature for 2 h, before they were incubated with the
following primary antibodies against: MTDH (1:2,000; cat. no.
ab227981; Abcam), GAPDH (1:10,000; cat. no. 10494-1-Ig; ProteinTech
Group, Inc.), p65 (1:2,000; cat. no. 10745-1-Ig; ProteinTech Group,
Inc.), p-p65 (1:2,000; cat. no. 3033; Cell Signaling Technology),
IκBα (1:2,000; cat. no. 10268-1-Ig; ProteinTech Group, Inc.),
E-cadherin (1:2,000; cat. no. 20874-1-Ig; ProteinTech Group, Inc.),
Vimentin (1:20,000; cat. no. 10366-1-Ig; ProteinTech Group, Inc.)
and snail family transcriptional repressor 2 (slug; 1:2,000; cat.
no. 12129-1-AP; ProteinTech Group, Inc.), overnight at 4°C. After
washing with TBST (0.05% Tween 20), the membranes were incubated
with the HRP-conjugated goat polyclonal anti-rabbit IgG secondary
antibody (1:10,000; cat. no. SA00001-2; ProteinTech Group, Inc.)
for 1 h. Protein expression was determined by enhanced
chemiluminescence (MilliporeSigma).
Invasion assay
After transfection with miR-217-5p mimic, miR-NC
mimic, miR-217-5p inhibitor and miR-NC inhibitor for 48 h at 37°C,
a total of 5×105 SK-BR3 and BT549 cells were resuspended
in 200 µl serum-free medium and subsequently seeded into the upper
chamber of a Transwell insert (Corning, Inc.) precoated with 1
µg/µl Matrigel (BD Biosciences) overnight at 37°C. The lower
chamber was filled with 500 µl basal medium with 10% FBS to
stimulate cell invasion. Following 48 h of cell culture at 37°C,
the cells that did not cross the membrane were wiped with a cotton
swab, whereas the cells adhering to the lower surface of the
membrane were stained with 0.1% crystal violet solution at room
temperature for 10 min. The number of invading cells was counted in
five randomly selected fields using a light microscope
(magnification, ×200; Olympus Corporation).
Migration assay
A total of 5×105 transfected SK-BR3 and
BT549 cells were resuspended in 200 µl serum-free medium and were
subsequently seeded into the upper chamber of a Transwell insert
without Matrigel. All other procedures were performed as described
for the invasion assay.
Bioinformatics analysis
The TargetScan (http://www.targetscan.org/vert_70/) database was used
to predict the target genes of miR-217-5p.
Dual-luciferase reporter assay
The 3′-untranslated regions (UTR) of wild type or
mutant MTDH were inserted into the pmirGLO luciferase vector
(Shanghai GeneChem Co., Ltd.). These vectors and miR-217-5p mimic
or miR-NC mimic were co-transfected into BT549 cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C. The lysate was cleared and the
luciferase reporter activity was assayed at 48 h post-transfection
using the Dual-Luciferase Reporter Assay System (Promega
Corporation). The firefly luciferase activity was normalized using
Renilla luciferase activity.
Statistical analysis
Data were analyzed using SPSS 23.0 (IBM Corp.) and
GraphPad Prism 8 (GraphPad Software, Inc.). Data are presented as
the mean ± SD. All comparisons between two groups of cells were
performed using unpaired Student's t-tests. Paired Student's
t-tests were performed for comparisons between paired tumor and
adjacent non-tumor tissues. Two-way ANOVA followed by Bonferroni's
post hoc test were used to compare data from the dual-luciferase
reporter assay. The gene expression correlation between miR-217-5p
and MTDH was assessed using the Pearson's correlation coefficient
test. All experiments were performed at least three times.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-217-5p expression levels are
reduced in BC tissues and cell lines
miR-217-5p expression levels were demonstrated to be
markedly reduced in MCF7, BT549 and SK-BR3 cell lines compared with
MCF10A cells (Fig. 1A).
Furthermore, the expression levels of miR-217-5p were significantly
lower in BC tissues compared with those in the adjacent non-tumor
tissues (Fig. 1B). This indicated
that miR-217-5p expression levels were lower in both BC cell lines
and tissues.
Overexpression of miR-217-5p inhibits
cell proliferation, colony formation, migration and invasion of BC
cells
To investigate the specific function of miR-217-5p
in BC, miR-217-5p mimic was transfected into SK-BR3 cells, whereas
miR-217-5p inhibitor was transfected into BT549 cells (two cell
lines with highest transfection efficiency). Compared with the
negative control (NC), the results demonstrated that transfection
with the miR-217-5p mimic significantly increased miR-217-5p
expression levels, whereas miR-217-5p inhibitor transfection
significantly decreased miR-217-5p expression levels (Fig. 2A and C). The CCK-8 assay
demonstrated that compared with the NC, transfection with the
miR-217-5p mimic in SK-BR3 cells significantly inhibited cell
proliferation at 48 and 72 h, whereas transfection with the
miR-217-5p inhibitor into BT549 cells significantly promoted cell
proliferation at 48 and 72 h (Fig. 2B
and D). Furthermore, colony formation assays demonstrated that
miR-217-5p overexpression resulted in the significant inhibition of
BC cell proliferation, whereas miR-217-5p expression knockdown
significantly promoted colony formation compared with the NC group
(Fig. 2E and F). Wound healing
assays determined that transfection with the miR-217-5p inhibitor
into BT549 cells significantly promoted cell migration, whereas
miR-217-5p overexpression in SK-BR3 cells significantly suppressed
cell migration, compared with the NC group (Fig. 2G and H). Transwell assays
demonstrated that miR-217-5p knockdown significantly promoted cell
migration and invasion in BT549 cells, but miR-217-5p
overexpression significantly inhibited SK-BR3 cell migration and
invasion compared with the NC group (Fig. 2I and J). The aforementioned results
indicated that miR-217-5p exerted a tumor suppressive role in BC
cells.
 | Figure 2.Overexpression of miR-217-5p inhibits
cell proliferation, colony formation, migration and invasion of
breast cancer cells. (A) RT-qPCR analysis of miR-217-5p expression
levels in SK-BR3 cells transfected with miR-217-5p or miR-NC mimic.
(B) Cell proliferation in SK-BR3 cells transfected with miR-217-5p
mimic or miR-NC mimic. (C) RT-qPCR analysis of miR-217-5p
expression levels in BT549 cells. (D) Cell proliferation in SK-BR3
cells transfected with miR-217-5p inhibitor or the miR-NC
inhibitor. Colony formation by (E) SK-BR3 and (F) BT549 cells
transfected with miR-217-5p mimic or inhibitor, respectively, or
the corresponding NCs. Scale bar, 5 mm. Wound healing assays for
(G) SK-BR3 and (H) BT549 cells transfected with miR-217-5p mimic or
inhibitor, respectively, or the corresponding NCs. Scale bar, 500
µm. Magnification, ×100. Transwell assay of (I) SK-BR3 and (J)
BT549 cells transfected with miR-217-5p mimic or inhibitor,
respectively, or the corresponding NCs. Scale bar, 100 µm.
Magnification, ×200. **P<0.01 and ***P<0.005. miR, microRNA;
RT-qPCR, reverse transcription-quantitative PCR; NC, negative
control; OD, optical density. |
Overexpression of miR-217-5p prevents
the EMT in BC cells
E-cadherin, Vimentin and slug mRNA and protein
expression levels were analyzed using RT-qPCR and western blotting
to explore the effects of miR-217-5p on SK-BR3 and BT549 cells
further. Overexpression of miR-217-5p markedly enhanced E-cadherin
mRNA and protein expression levels, whereas Vimentin and slug
expression levels were markedly inhibited compared with the NC
group (Fig. 3A and C). However,
compared with the NC, miR-217-5p knockdown markedly decreased
E-cadherin mRNA and protein expression levels but markedly promoted
Vimentin and slug mRNA and protein expression levels (Fig. 3B and D). These results suggested
that miR-217-5p overexpression may suppress the EMT in BC.
Overexpression of miR-217-5p inhibits
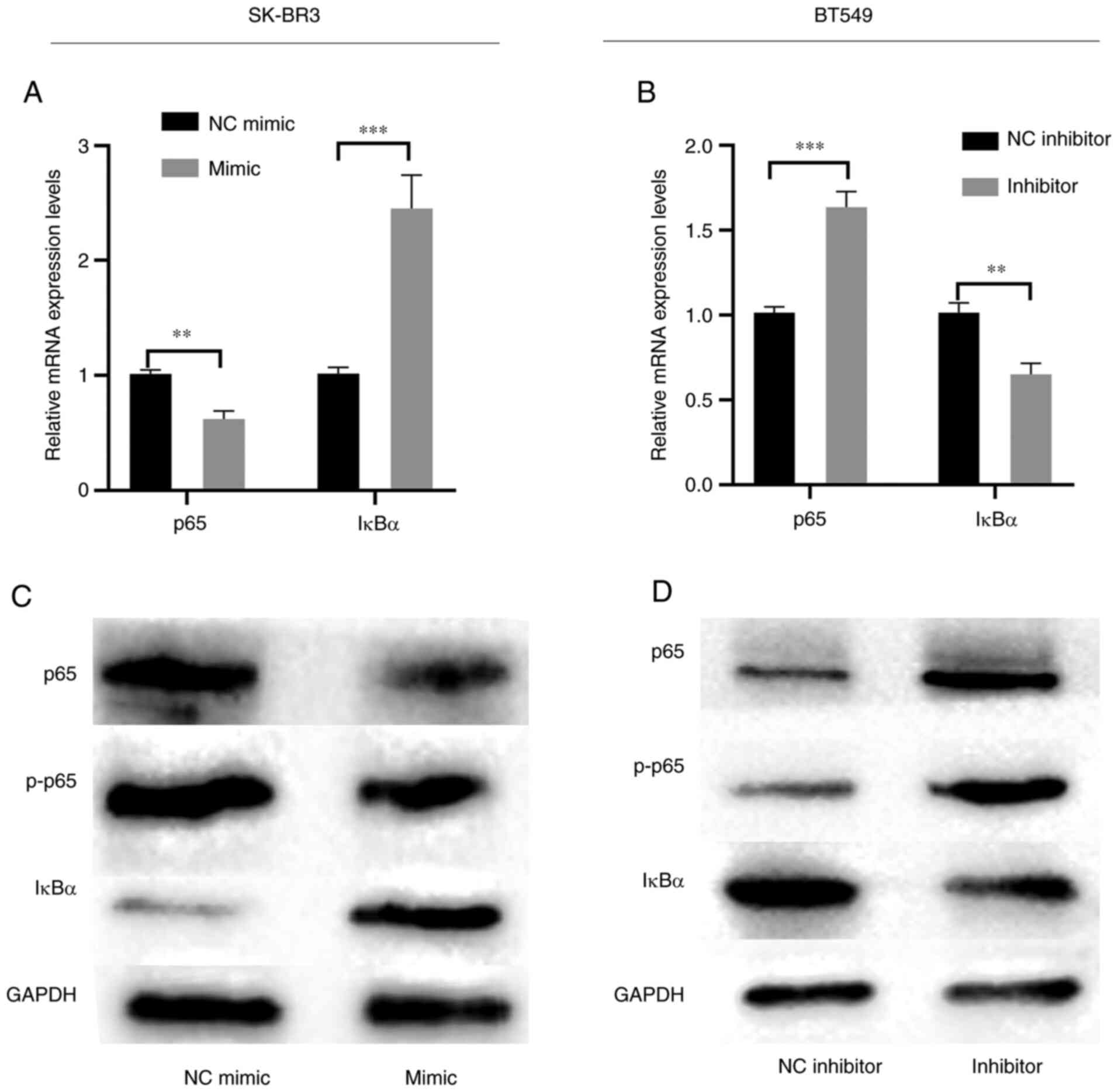
the NF-κB signaling pathway
The mRNA and protein expression levels of NF-κB
markers were subsequently assessed to determine the effects of
miR-217-5p on SK-BR3 and BT549 cells. RT-qPCR and western blotting
demonstrated that miR-217-5p overexpression markedly inhibited p65
phosphorylation but markedly promoted IκBα expression levels
compared with the NC (Fig. 4A and
C). However, miR-217-5p knockdown markedly suppressed IκBα mRNA
and protein expression levels but markedly promoted p65
phosphorylation compared with the NC (Fig. 4B and D). These results indicated
that the downregulation of miR-217-5p may activate the NF-κB
signaling pathway.
miR-217-5p directly targets MTDH in BC
cells
TargetScan (http://www.targetscan.org/vert_71/) was used to
predict miR-217-5p target genes. MTDH was determined to be a
candidate target for miR-217-5p. Therefore, dual-luciferase
reporter assays were performed to assess the potential binding
between MTDH mRNA and miR-217-5p (Fig.
5A). MTDH mRNA expression levels were demonstrated to be
markedly increased in MCF7, BT549 and SK-BR3 cell lines compared
with MCF10A cells (Fig. 5B).
Moreover, significantly higher MTDH mRNA expression levels were
demonstrated in BC tissues compared with adjacent non-tumor tissues
(Fig. 5C). Transfection with
miR-217-5p mimic significantly reduced the luciferase activity of
the wild-type MTDH 3′UTR compared with the miR-NC mimic. However,
there was no change in luciferase activity in the mutant-MTDH 3′UTR
group (Fig. 5D). Furthermore, MTDH
mRNA expression levels were demonstrated to be significantly
inversely correlated with those of miR-217-5p in BC tissues
(Fig. 5E). The results also
demonstrated that compared with the NC, miR-217-5p overexpression
markedly inhibited MTDH mRNA and protein expression levels
(Fig. 5F), whereas miR-217-5p
knockdown markedly promoted MTDH mRNA and protein expression levels
(Fig. 5G). These data therefore
suggested that miR-217-5p may directly target MTDH and that the
expression of miR-217-5p is inversely correlated with MTDH in BC
cells.
 | Figure 5.MTDH is a direct target of
miR-217-5p. (A) Schematic representation of MTDH 3′-UTR
demonstrating the putative miR-217-5p target site. RT-qPCR analysis
of MTDH mRNA expression levels in (B) BC cell lines and (C) BC
tissues. (D) Analysis of the relative luciferase activities of
MTDH-WT and MTDH-mut. (E) Pearson's correlation coefficient
analysis of miR-217-5p and MTDH expression levels in BC tissues.
RT-qPCR and western blotting analysis of MTDH expression levels in
(F) SK-BR3 and (G) BT549 cells transfected with miR-217-5p mimic or
inhibitor, respectively, or the corresponding negative control.
**P<0.01 and ***P<0.005. MTDH, metadherin; miR, microRNA;
UTR, untranslated region; RT-qPCR, reverse
transcription-quantitative PCR; BC, breast cancer; WT, wild-type;
mut, mutant; NC, negative control; luc, luciferase. |
Discussion
Previous studies have demonstrated that miRNAs serve
important roles in the growth and metastasis of malignant types of
cancer, including BC (7,30). miR-217 has been reported to
suppress tumors associated with gastric cancer (24), pancreatic ductal adenocarcinoma
(23), hepatocellular carcinoma
(31), osteosarcoma (12) and chronic myelogenous leukemia
(32). However, results from
previous studies on BC are controversial. Zhang et al
(27) reported that miR-217 is
frequently overexpressed in BC, which enhances tumor growth by
targeting Dachshund homolog 1 expression and promoting cell cycle
progression. However, Zhou et al (28) demonstrated that miR-217 suppresses
triple-negative BC cell proliferation, migration and invasion, at
least in part by downregulating Kruppel-like factor 5 expression.
miR-217-5p is an important member of the miR-217 family. In the
present study, miR-217-5p downregulation was detected in BC tissues
and BT549 and SK-BR3 cells. Therefore, miR-217-5p may serve
distinct roles in different BC cell lines.
The EMT is a process that describes the molecular
reprogramming and phenotypic changes during the transition of
polarized, immotile epithelial cells into motile mesenchymal cells,
which exhibit increased migratory and invasive capacities (13). The activation of the EMT is
characterized by the reduced expression of epithelial markers, such
as E-cadherin, coupled with the increased expression of mesenchymal
markers, such as Vimentin and slug. It has also been reported that
miRNAs can serve as regulators of malignant transformation and
metastasis (14). In the present
study, miR-217-5p was found to regulate the mRNA and protein
expression levels of E-cadherin, Vimentin and slug. Therefore,
these results indicated that miR-217-5p expression potentially
inhibits the EMT of BC cells.
Previous studies have demonstrated that the
activation of the NF-κB signaling pathway is associated with BC
development (33,34). IκB is a specific intracellular
inhibitor of NF-κB. In the majority of cell types, NF-κB exists in
an inactive form by forming a complex with IκB in the cytoplasm
(34). A previous study reported
that NF-κB is constitutively activated in hepatocellular carcinoma
cell lines (31). Results from the
present study demonstrated that the overexpression of miR-217-5p
significantly inhibited NF-κB signaling. These results therefore
indicated that miR-217-5p may prevent BC progression by potentially
regulating the NF-κB signaling pathway.
Given the potential importance of miR-217-5p in BC,
further experiments were performed to assess the consequences of
miR-217-5p activation. Using bioinformatics analysis combined with
the dual-luciferase reporter assay, MTDH was demonstrated to be a
direct target of miR-217-5p. A previous study reported that miR-217
can target MTDH to promote the progression of meningioma (35). MTDH has also been reported to be an
important oncogene in the carcinogenesis, progression and
metastasis of numerous malignancies, via regulating and PI3K/AKT,
ERK/MAPK, Wnt/β-catenin and aurora-A kinase signaling pathways
(36,37). Furthermore, the positive effects of
MTDH on cell migration, invasion and EMT have also been previously
determined in BC (36).
Furthermore, MTDH has been reported to regulate tumorigenesis by
interacting with several miRNAs, including miR-145 (38) and miR-630 (39). In the present study, miR-217-5p was
demonstrated to significantly negatively regulate MTDH expression
in BC. These results suggested that miR-217-5p may be involved in
BC progression via the inhibition of MTDH.
A previous study by Wang et al (40) reported that decreased miR-217
expression is associated with poor prognosis in patients with
colorectal cancer. In the present study, based on the available
data, none of the enrolled patients with BC experienced disease
progression or death. As the prognosis of breast cancer is
relatively good, it is usually difficult to observe differences in
survival after a short period of follow-up, such as several months.
The small sample size may also cause some bias. Therefore, in
future research, the sample size will be expanded and the follow-up
time extended, and related research on the impact of miR-217-5p on
the prognosis of breast cancer will continue to be conducted.
In conclusion, the data from the present study
suggested that miR-217-5p overexpression may inhibit cell
migration, invasion and the EMT via targeting MTDH expression in
BC. It can therefore be hypothesized that the miR-217-5p/MTDH axis
is likely to be involved in BC via the regulation of the NF-κB
signaling pathway. These findings indicated that miR-217-5p may
serve as an important potential target in BC therapy.
Acknowledgements
Not applicable.
Funding
Funding was provided by the Natural Science Foundation of Hebei
Province (grant nos. H2020206365 and H2021206071), the Special Fund
for Clinical Research of Wu Jieping Medical Foundation (grant no.
320.6750.2020-07-17) and the Beijing Xisike Clinical Oncology
Research Foundation (grant no. Y-SY201901-0021).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZS designed and conceptualized the present study.
LXY, BX and SL were responsible for data acquisition. LY, XK and MW
analyzed and interpretated the data. LXY drafted the manuscript. ZS
and XK revised the manuscript for critically important intellectual
content. LXY and ZS confirm the authenticity of all the raw data.
All authors contributed to the article and read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Fourth Hospital of Hebei Medical University
(approval no. 2021KY056). The present study was conducted in
accordance with the principles described in The Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
6:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Poortmans P, Marsiglia H, Heras MDL and
Algara M: Clinical and technological transition in breast cancer.
Rep Pract Oncol Radiother. 18:345–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rivenbark AG, O'Connor SM and Coleman WB:
Molecular and cellular heterogeneity in breast cancer: Challenges
for personalized medicine. Am J Pathol. 183:1113–1124. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel D: MicroRNAs: Genomics, biogenesis,
mechanism, and function. Cell. 116:281–297. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang L, Fu J, Wang S, Cen H, Zhang L,
Mandukhail SR, Du L, Wu Q, Zhang P and Yu X: MiR-142-3p enhances
chemosensitivity of breast cancer cells and inhibits autophagy by
targeting HMGB1. Acta Pharm Sin B. 10:1036–1046. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Greene SB, Herschkowitz JI and Rosen JM:
The ups and downs of miR-205: Identifying the roles of miR-205 in
mammary gland development and breast cancer. RNA Biol. 7:300–304.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Nakhle H, Burns PA, Cummings M, Hanby
AM, Hughes TA, Satheesha S, Shaaban AM, Smith L and Speirs V:
Estrogen receptor {beta}1 expression is regulated by miR-92 in
breast cancer. Cancer Res. 70:4778–4784. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu Y and Tang S: miR-217 targeted to MAPK1
for the inhibition of metastasis and invasion of gastric cancer
cell. Mod Med J Chin. 11:21–23. 2014.(In Chinese).
|
|
11
|
Zhao WG, Yu SN, Lu ZH, Ma YH and Chen J:
The miR-217 microRNA functions as a potential tumor suppressor in
pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis.
31:1726–1733. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei R, Deng Z and Su J: miR-217 targeting
Wnt5a in osteosarcoma functions as a potential tumor suppressor.
Biomed Pharmacother. 72:158–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Savary K, Termén S, Thuault S, Keshamouni
V and Moustakas A: Epithelial-mesenchymal transition as a mechanism
of metastasis. Lung Cancer Metastasis. Keshamouni V, Arenberg D and
Kalemkerian G: 1st edition. Springer; New York, NY: pp. 65–92.
2010
|
|
15
|
Abe-Yutori M, Chikazawa T, Shibasaki K and
Murakami S: Decreased expression of E-cadherin by porphyromonas
gingivalis-lipopolysaccharide attenuates epithelial barrier
function. J Periodontal Res. 52:42–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao
W, Volsky DJ and Fisher PB: Identification and cloning of human
astrocyte genes displaying elevated expression after infection with
HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid
subtraction hybridization, RaSH. Oncogene. 21:3592–3602. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin H, Shi X, Zhao Y, Peng M, Kong Y, Qin
D and Lv X: MicroRNA-30a mediates cell migration and invasion by
targeting metadherin in colorectal cancer. Technol Cancer Res
Treat. 17:1533033818758102018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo J, Xia B, Meng F and Lou G: MiR-137
suppresses cell growth in ovarian cancer by targeting AEG-1.
Biochem Biophys Res Commun. 441:357–363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gnosa S, Ticha I, Haapaniemi S and Sun XF:
MTDH genetic variants in colorectal cancer patients. Sci Rep.
6:231632016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gnosa S, Shen YM, Wang CJ, Zhang H,
Stratmann J, Arbman G and Sun XF: Expression of AEG-1 mRNA and
protein in colorectal cancer patients and colon cancer cell lines.
J Transl Med. 10:1092012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu Y, Zhao H, Li F and Xu S: MicroRNA-217
inhibits cell proliferation and invasion by targeting Runx2 in
human glioma. Am J Transl Res. 8:1482–1491. 2016.PubMed/NCBI
|
|
22
|
Menghini R, Casagrande V, Cardellini M,
Martelli E, Terrinoni A, Amati F, Vasa-Nicotera M, Ippoliti A,
Novelli G, Melino G, et al: MicroRNA 217 modulates endothelial cell
senescence via silent information regulator 1. Circulation.
120:1524–1532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li H, Zhao J, Zhang JW, Huang QY, Huang
JZ, Chi LS, Tang HJ, Liu GQ, Zhu DJ and Ma WM: MicroRNA-217,
down-regulated in clear cell renal cell carcinoma and associated
with lower survival, suppresses cell proliferation and migration.
Neoplasma. 60:511–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng
ZL, He MM, Wang FH, Li YH, ZhangH Z, Pelicano H, et al:
MicroRNA-217 inhibits tumor progression and metastasis by
downregulating EZH2 and predicts favorable prognosis in gastric
cancer. Oncotarget. 6:10868–10879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang S, Liu X, Liu J, Guo H, Xu H and
Zhang G: PGC-1 alpha interacts with microRNA-217 to functionally
regulate breast cancer cell proliferation. Biomed Pharmacother.
85:541–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yébenes VG, Bartolomé-Izquierdo N,
Nogales-Cadenas R, Pérez-Durán P, Mur SM, Martínez N, Lisio LD,
Robbiani DF, Pascual-Montano A, Cañamero M, et al: miR-217 is an
oncogene that enhances the germinal center reaction. Blood.
124:229–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Q, Yuan Y, Cui J, Xiao T and Jiang
D: MiR-217 promotes tumor proliferation in breast cancer via
targeting DACH1. J Cancer. 6:184–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou W, Song F, Wu Q, Rong L, Liu R, Wang
L, Liu C, Peng Y, Mao S, Feng J and Chen C: miR-217 inhibits
triple-negative breast cancer cell growth, migration, and invasion
through targeting KLF5. PLoS One. 12:e01763952017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Martello G, Rosato A, Ferrari F, Manfrin
A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T,
et al: A microRNA targeting dicer for metastasis control. Cell.
141:1195–1207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su J, Wang Q, Liu Y and Zhong M: miR-217
inhibits invasion of hepatocellular carcinoma cells through direct
suppression of E2F3. Mol Cell Biochem. 392:289–296. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nishioka C, Ikezoe T, Yang J, Nobumoto A,
Tsuda M and Yokoyama A: Downregulation of miR-217 correlates with
resistance of Ph (+) leukemia cells to ABL tyrosine kinase
inhibitors. Cancer Sci. 105:297–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
El-Ashmawy NE, El-Zamarany EA, Khedr EG
and Abo-Saif MA: Activation of EMT in colorectal cancer by
MTDH/NF-κB p65 pathway. Mol Cell Biochem. 457:83–91. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou J, Hao Z, Gu P, Bai J, Margolick JB
and Zhang Y: NF-kappaB pathway inhibitors preferentially inhibit
breast cancer stem-like cells. Breast Cancer Res Treat.
111:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ghafouri-Fard S, Abak A, Hussen BM, Taheri
M and Sharifi G: The emerging role of non-coding RNAs in pituitary
gland tumors and meningioma. Cancers (Basel). 13:59872021.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou CX, Wang CL, Yu AL, Wang QY, Zhan MN,
Tang J, Gong XF, Yin QQ, He M, He JR, et al: MiR-630 suppresses
breast cancer progression by targeting metadherin. Oncotarget.
7:1288–1299. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wan L and Kang K: Pleiotropic roles of
AEG-1/MTDH/LYRIC in breast cancer. Adv Cancer Res. 120:113–134.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meng X, Thiel KW and Leslie KK: Drug
resistance mediated by AEG-1/MTDH/LYRIC. Adv Cancer Res.
120:135–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu Q, Shan S, Li Y, Zhu D, Jin W and Ren
T: Long noncoding RNA SNHG1 promotes non-small cell lung cancer
progression by up-regulating MTDH via sponging miR-145-5p. FASEB J.
32:3957–3967. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang B, Shen ZL, Jiang KW, Zhao G, Wang
CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al: MicroRNA-217
functions as a prognosis predictor and inhibits colorectal cancer
cell proliferation and invasion via an AEG-1 dependent mechanism.
BMC Cancer. 15:4372015. View Article : Google Scholar : PubMed/NCBI
|