Introduction
Head and neck carcinoma is the seventh most common
cancer by worldwide incidence (1).
Cancers of this type develop in the craniocervical region,
including the oral cavity, nasal cavity, pharynx, and larynx.
Approximately 90% of lesions are histopathologically diagnosed as
head and neck squamous cell carcinoma (HNSCC) (1,2). In
recent decades, HNSCC treatments such as surgical and
radiotherapeutic techniques, along with combined modality
therapies, have improved remarkably, and disease-free survival of
patients has been dramatically extended. However, the five-year
survival rate of patients with HNSCC has remained 50–60% with no
significant change, in part because of tumor cell invasion followed
by metastasis (3,4). The largest studies have reported that
the incidence of distant metastases in HNSCC patients is
approximately 3% at initial presentation (5,6).
However, distant metastasis during the course of the disease varies
between 9 and 38% (7–9). Autopsy studies have revealed an even
higher incidence of distant metastasis (10,11).
Overall survival at 24 months after diagnosis of distant metastases
is only 4–26.2% (12,13), and the median time from distant
metastasis to death is only 3.3-10 months (12–14).
Thus, epidemiological data indicate that the control of invasion
and metastasis is an urgent issue for therapy of HNSCC.
Rho-family GTPases are small guanine
nucleotide-binding proteins (G-proteins) that serve as critical
regulators of cell adhesion, migration, and spreading, exerting
their effects by modulating cytoskeletal dynamics through
downstream effector proteins (15). The activity of small G-proteins is
switched by guanine-nucleotide binding. The GTP-bound form
activates effector proteins. However, when GTP is cleaved, the
protein transitions to a GDP-bound form that cannot activate
effectors. Guanine nucleotide exchange factors (GEFs) facilitate
the exchange of GDP to GTP on small G-proteins, while
GTPase-activating proteins (GAPs) and GDP dissociation inhibitors
(GDIs) negatively regulate the activity of small G-proteins.
Another factor affecting the activity of Rho-family GTPases is
phosphorylation. Phosphorylation at Ser-188 on RhoA by
cAMP/cGMP-dependent kinase negatively regulates RhoA activity by
enhancing interaction with GDI (16). The correlation between expression
or activation of Rho-family GTPases and tumor progression is
context dependent. However, RhoA is overexpressed in 80% of HNSCCs
compared with adjacent non-neoplastic epithelial tissues (17). In addition, cortactin-dependent
expression and activation of RhoA has been shown to promote cell
cycle progression in the hypopharyngeal squamous cell carcinoma
cell line FaDu (18). Similarly,
activation and serine-phosphorylation of RhoA are associated with
cell motility and invasion under the regulation of protein kinase C
(PKC) ε in UMSCC11A and UMSCC36 cell lines (19). Epidermal growth factor receptor
(EGFR) and the hyaluronan receptor CD44 are known to form a complex
that promotes growth and cell migration via leukemia-associated
RhoGEF (LARG), a RhoA specific GEF, in the HNSCC cell line HSC-3
(20). Vav guanine nucleotide
exchange factor 2 (VAV2) is another Rho-specific GEF that is
overexpressed in HNSCCs and has been shown to associate with
regenerative proliferation in a RhoA-dependent manner (21). Considered together, these findings
support the hypothesis that activation of RhoA promotes HNSCC
progression.
Crumbs (Crb)-family single-transmembrane proteins
are expressed in normal epithelial tissues, where Crb molecules
contribute to the establishment of apicobasal cell polarity. The
mammalian crumbs3 (Crb3) protein is a homolog of the
Drosophila Crb protein, and Crb3 seems to be
expressed in epithelial tissues (22). Crb3-knockout (KO) mice show
defects in the formation of glandular epithelium in lung, kidney,
and colon tissues (23,24). The expression and function of Crb3
in normal squamous epithelium (including skin, esophagus, and oral
tissues) have not (to our knowledge) been examined. Our recent
study demonstrated that Crb3 is strongly expressed in colon
adenocarcinomas, especially in metastatic foci, in comparison with
non-neoplastic colon epithelium, where the protein promotes
invasion and metastasis by activating fibroblast growth factor
receptor (FGFR) signaling (25).
However, the expression and function of Crb3 in other carcinomas
remains poorly elucidated. In the present study, we revealed that
Crb3 is endogenously expressed in HNSCC tissues. HNSCC consists of
heterogeneous subgroups that arise from the oral cavity, larynx,
oropharynx, and hypopharynx. Oral squamous cell carcinoma (OSCC) is
the largest subgroup that constitutes approximately 40% of all
HNSCC. Functional analyses of Crb3 were performed by either
knock-down or knock-out of Crb3 in OSCC cell lines. Our
results suggested that Crb3 is expressed in OSCC cell lines and
promotes cell migration. To investigate how Crb3 regulates the
motility of OSCC cells, the activity of RhoA was evaluated. A RhoA
activation assay revealed that the GTP-bound form of RhoA is
significantly depleted in Crb3-KO OSCC cell lines. This
result represents the first evidence that Crb3 promotes cell
migration and RhoA activation in squamous cell carcinomas.
Materials and methods
Cell lines
Human OSCC cell lines Ca9-22, HSC-2, and HSC-3 were
obtained from the JCRB (Japanese Collection of Research
Bioresources) Cell Bank. The primary normal human dermal fibroblast
(NHDF) cell was obtained from ATCC (#PCS-201-012, American Type
Culture Collection). Crb3-KO derivatives of OSCC cell lines
were created by targeting a genomic sequence (CCGTTCCTGCTGGCCCGCTG)
in the Crb3 allele using CRISPR-Cas9-based technology
(25). The isolation of
single-cell clones of Crb3-KO cells was performed by serial
dilution. Deficiency of Crb3 protein was confirmed by
immunoblotting. All cell lines were cultured in RPMI-1640 medium
(#189-02025, FUJIFILM Wako Chemicals) supplemented with 10~15%
fetal bovine serum (FBS) (Hyclone, #SH30071, Cytiva) and
Penicillin-Streptomycin Solution (#168-23191, FUJIFILM Wako
Chemicals) at 37°C in a humidified 5% CO2 incubator. All
the OSCC cell lines used in this study were authenticated by STR
analysis employing the GenePrint 10 System (Promega) as shown in a
previous project (26) The
patterns of STR markers of Ca9-22, HSC-2, and HSC-3 used in this
study were 100% identical with JCRB0625 (Ca9-22), JCRB0622 (HSC-2),
and JCRB0623 (HSC-3), respectively.
Immunohistochemistry and absorption
test
Immunohistological experiments using tumor tissues
surgically obtained from patients with HNSCC were approved by the
Research Ethics Committee of Niigata University (approval no.
2019-0101). The patients >20 years old who were diagnosed with
HNSCC by two pathologists and underwent radical surgical treatment
at the Niigata University Hospital from April 1, 2017 to March 31,
2019 were included. All participants provided informed consent to
participate in this study. The patient who refused to participate
in the study were excluded. A total of fourteen cases of primary
OSCC tissue (5 cases from tongue, 3 cases from oral floor, 3 cases
from pharynx, 2 cases from larynx, and 1 case from gingiva) and 3
cases of paired cervical lymph node metastatic lesions from
consecutive patients were analyzed. TNM staging was determined
according to the 8th edition of the Union for International Cancer
Control TNM classification. For the immunohistochemistry,
paraffin-embedded sections were deparaffinized, and antigen
retrieval was performed in 10 mM sodium citrate buffer (pH 6.0)
using a Pascal Pressure Chamber. Antigen-retrieved sections were
incubated overnight at 4°C with anti-human Crb3 monoclonal antibody
in phosphate-buffered saline (PBS) containing 1% bovine serum
albumin (BSA), then stained using a Histofine DAB
(diaminobenzidine) substrate kit (#425011, Nichirei Bioscience).
The anti-human Crb3 monoclonal antibody (anti-Crb3 antibody) was
obtained as described previously (25). The DAB-stained sections were
counterstained with Mayer's hematoxylin (#30004, Mutoh Pure
Chemical Industries). For the absorption test, 2.0 µg of anti-Crb3
antibody and 0.75 µg of the epitope peptide
(NH2-VGARVPPTPNLKLPPEERLI, 1:25 molar ratio) were mixed in 1.2 ml
of PBS containing 1% BSA and incubated for 15 min at room
temperature before initiating the primary antibody reaction.
Immunoblotting
Commercially obtained antibodies employed in this
study were as follows: anti-β-tubulin (1/2,000 dilution; #PM054-7,
Medical & Biological Laboratories), anti-RhoA (1/250 dilution;
#ARH05, Cytoskeleton, Inc.), anti-SNAIL2 (1/1,000 dilution;
#12129-1-AP, Proteintech), and anti-E-cadherin (1/1,000 dilution;
#610181, BD Biosciences). Antibody dilutions and reactions were
performed using Can Get Signal Solution 1 (#NKB-201, Toyobo).
Freshly cultured cells were washed in PBS and lysed by adding 1X
Laemmli sample buffer. After incubation at 95°C for 3 min, protein
samples were separated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) using Tris-glycine buffer. For
immunoblotting using Crb3 antibody, blocking of protein-bound PVDF
membranes was performed using 0.2% BSA (#017-25771, FUJIFILM Wako
Chemicals). For immunoblotting using anti-RhoA antibody, blocking
of membranes was performed using TBST (10 mM Tris, pH 7.2, 0.05%
Tween-20) containing 1% skimmed milk. For immunoblotting with all
other primary antibodies, blocking of membranes was performed with
PBST (PBS, 0.05% Tween-20) containing 1% skimmed milk. All blocking
reactions consisted of a 30-min incubation. The reactions with
horseradish peroxidase (HRP)-conjugated secondary antibody and
membrane washing were conducted in PBST or TBST. Immunoreactivity
was visualized using ImmunoStar® Zeta (#291-72401,
FUJIFILM Wako Chemicals) or ImmunoStar® LD (#296-69901,
FUJIFILM Wako Chemicals) and detected using the ChemiDoc Touch MP
Imaging System (#17001402JA, Bio-Rad).
Immunofluorescence
Freshly cultured cells were fixed in 4%
paraformaldehyde for 10 min at room temperature. Fixed cells were
washed twice in PBS and permeabilized in PBST for 10 min at room
temperature. Permeabilized cells were blocked in PBST containing 1%
BSA for 30 min at room temperature. Immunoreaction with anti-Crb3
antibody or anti-RhoA antibody (1:1,000) diluted in PBS was carried
out overnight at 4°C on a reciprocal shaker. Immunoreaction of
fluorescent dye-conjugated secondary antibody was performed for 30
min at room temperature. Counterstaining with Hoechst 33342
(#H-3570, Thermo Fisher Scientific) was performed simultaneously
with the secondary antibody reaction. Fluorescent images were
acquired using an inverted fluorescence microscope (IX71,
Olympus).
Transfection of siRNA
Crb3 gene silencing in OSCC cells was
performed using Silencer Select pre-designed siRNAs (Thermo Fisher
Scientific). The product number and target sequence of siRNAs used
to knock-down Crb3 were as follows: siCrb3−1
(#s40936, CAGGGAAGAAGGUACUUCA), siCrb3−2 (#s195567,
AGUGCUUAAUAGCAGGGAA). Silencer select Negative Control No. 1 siRNA
(#4390843, Thermo Fisher Scientific) was used as a control. siRNAs
(10 nM final concentration) were transfected using ScreenFect™
siRNA (#299-75001, FUJIFILM Wako Chemicals) and a forward
transfection protocol.
MTT assay
Cells (1×104 per well) were seeded into
24-well plates in RPMI-1640 medium supplemented with 10% FBS (10%
FBS/RPMI-1640); plates were incubated at 37°C in a 5%
CO2 incubator. Cell growth was assessed using MTT
reagent (Cell Count Reagent SF, #07553-44, Nacalai Tesque) starting
24 h after seeding, and analysis was repeated every 24 h thereafter
by measuring the absorbance at 450 nm using an iMark microplate
reader (#1681135JA, Bio-Rad). To assess the effect of RhoA
inhibitors, culture medium was replaced with fresh medium
containing 15 µM Y16 (#Y-12649, MedChemExpress, NJ, USA) or 20 µM
Rhosin (#555460, Merck, NJ, USA) at 24 h after seeding the
cells.
Transwell migration assay
To evaluate cell migration ability, 1×105
OSCC cells suspended in 200 µl Opti-MEM were loaded into the upper
compartment of a Transwell chamber (#3422, Corning, Inc.). The
lower chamber was loaded with 500 µl of Opti-MEM supplemented with
10% FBS. At 24–48 h after seeding, the cells in the upper chamber
were removed with a cotton swab, and the remaining cells (those
that had migrated through the Transwell membrane) were stained with
Hoechst 33342. Fluorescent images were captured using an inverted
fluorescence microscope (IX71). The number of nuclei was counted in
three different fields using ImageJ software (version 1.52a,
http://imagej.net/). To investigate whether RhoA
inhibitors affect cell migration, the cells were pretreated with
10% FBS/RPMI-1640 containing 15 µM Y16 or 20 µM Rhosin for 24 h.
Y16 or Rhosin was added to the growth medium in both the upper and
lower chambers to ensure consistent exposure.
Xenograft model of OSCC lung
metastases
All procedures were in accordance with the protocols
approved by the Animal Care and Use Committee of Niigata University
School of Medicine (approval number: SA00875). Six immunodeficient
mice (SHO-Prkdcscid Hrhr) were
obtained from Charles River Laboratories International, Inc.. The
animals were maintained at 22–24°C and 40–60% humidity under a
light-dark (12–12 h) cycle of ad libitum feeding in a
specific pathogen-free environment. Suspensions of wild-type or
Crb3-KO HSC-2 cells (1×106/200 µl PBS) were
injected into tail veins of 10-week-old female mice using 29-gauge
insulin syringes. The average weight was 28.5 gram/mouse at the
start of the experiment. Wild-type (n=3) and Crb3-KO (n=3)
cell-injected mice were sacrificed concurrently between 61–75 days
after injection. 20% of body weight reduction from baseline or the
defined period of 75 days were used to determine the endpoint of
the experiment. Cervical dislocation was conducted for euthanasia
of mice. The animals were anesthetized by intraperitoneal injection
of combination anesthetic (0.3 mg/kg of medetomidine, 4.0 mg/kg of
midazolam, and 5.0 mg/kg of butorphanol) before euthanasia.
Isolated lung specimens were fixed with 10% buffered formalin for
48 h and embedded in paraffin.
Detection of serum-induced activation
of RhoA
Cells (1×106) were seeded in a 35-mm dish
and incubated for 24 h. Cells then were washed twice with PBS and
serum-starved for 2 h in Opti-MEM at 37°C in 5% CO2.
Next, cells were stimulated for 30 min in RPMI-1640 containing 10%
FBS at 37°C in 5% CO2. Isolation of activated RhoA
protein from the cell lysate was performed using the Rho Activation
Assay Biochem Kit (# BK036-S, Cytoskeleton) according to the
manufacturer's instructions. The protein samples were subjected to
15% SDS-PAGE followed by immunoblot analysis using
anti-RhoA-specific antibody.
Statistical analysis
The data from MTT and Transwell assays are presented
as the mean ± SD of triplicate experiments. Statistical
significance was determined using a one-way ANOVA followed by
Bonferroni's post hoc test. BellCurve for Excel (version 3.23), a
statistical add in software was purchased (https://bellcurve.jp/ex/, Social Survey Research
Information Co., Ltd.) and used by adding into Microsoft Excel 2013
to conduct statistical analyses. P<0.05 was considered to
indicate a statistically significant difference.
Results
Crb3 is expressed in HNSCC patient
tissues
Immunohistochemistry was performed on formalin-fixed
paraffin-embedded tissue sections from fourteen patients with HNSCC
using a monoclonal anti-Crb3 antibody (Figs. 1, S1 and S2; Table
SI). Absorption tests were performed in parallel to evaluate
the specificity of the antibody and the level of background
staining (Figs. S1 and S2). All of the primary HNSCC tissues and
metastatic lesions in cervical lymph nodes showed positive staining
for Crb3; the absorption tests did not detect background staining,
confirming the specificity of the antibody (Figs. 1, S1 and S2). HNSCCs appeared to display stronger
Crb3 staining than did adjacent non-neoplastic squamous epithelial
tissues (Fig. S1). Most
Crb3-positive cells were observed in the prickle cell layer, with
Crb3 protein exhibiting apically distributed localization in
juxtanuclear cytoplasm in non-neoplastic tissues (Fig. S1).
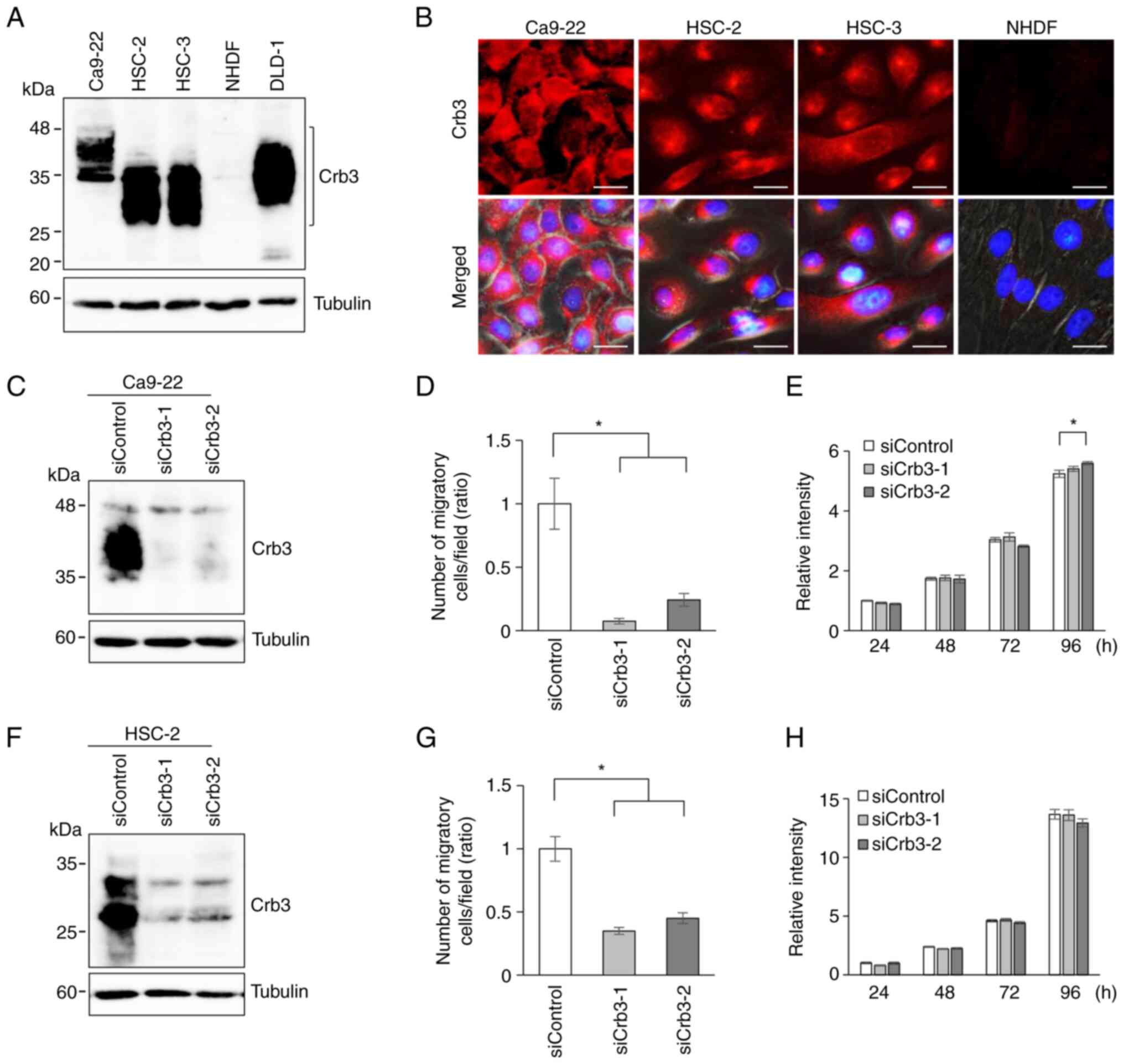
Crb3 is expressed in OSCC cell
lines
Next, the endogenous expression of Crb3 protein in
OSCC cell lines Ca9-22, HSC-2, and HSC-3 was evaluated by
immunoblotting using a monoclonal anti-Crb3 antibody. Cell lysates
from the colon cancer cell line DLD-1 and normal human dermal
fibroblasts (NHDFs) were employed as positive and negative
controls, respectively. Expression of Crb3 was detected in Ca9-22,
HSC-2, and HSC-3. Although Crb3 is predicted to have a molecular
mass of approximately 13 kDa, immunoblotting detected the protein
as multiple smeary bands in the 25- to 45-kDa range (Fig. 2A). These data suggested that Crb3
protein is modified by N-glycosylation in OSCC cells, as had been
reported in adenocarcinoma cells and MDCK cells (27,28).
In addition, immunofluorescent staining of OSCC cell lines was
performed. Crb3 localized primarily in cytoplasmic granules in
HSC-2 and HSC-3 cells, while Ca9-22 cells showed dispersed
localization of Crb3 in the cytoplasm and partly in the plasma
membrane (Fig. 2B). These
subcellular localization patterns of Crb3 were commonly observed in
OSCC cells in patient tissues (Figs.
1, S1 and S2).
OSCC cells with Crb3 knock-down
exhibit decreased cell motility
Because Crb3 is involved in colon adenocarcinoma
cell migration and metastasis, Crb3 function in OSCC cell migration
was investigated by siRNA-based knock-down (KD) of Crb3.
Crb3 expression in Ca9-22 and HSC-2 cells was knocked down
using two siRNAs (siCrb3-1 and siCrb3-2) that target different
sites on the Crb3 transcript. The efficiency of knock-down
by siRNAs was evaluated by detecting endogenously expressed Crb3
protein using immunoblotting (Fig. 2C
and F). Ca9-22 cells treated with siCrb3-1 or siCrb3-2
displayed 92 and 75% reduction in cell motility, respectively,
compared to control siRNA-treated cells (Figs. 2D and S3A). Similarly, HSC-2 cells treated with
siRNAs showed 65% and 55% reduction compared to controls (Figs. 2G and S3B). Although the small difference in
proliferation of siCrb3-2 treated Ca9-22 cells was observed at 96 h
(Fig. 2E), siCrb3-1 treated cells
did not show such difference. The proliferation was not
significantly affected by knocking down Crb3 in HSC-2 cells
(Fig. 2H).
Knockout of Crb3 affects both cell
motility and proliferation in OSCC cells
To assess whether the siRNA experimental results
were due to off-target effects, Crb3-KO cell clones were
established using the CRISPR-Cas9 system. Crb3-KO clones
derived from Ca9-22 or HSC-2 were isolated by serial dilution. The
deficiency of Crb3 protein in Crb3-KO clones of Ca9-22
(Fig. 3A) or HSC-2 (Fig. 3E) was confirmed by immunoblotting.
The motility of parent and Crb3-KO clones of OSCC cell lines
was examined by a Transwell cell migration assay. Compared to
parent Ca9-22 cells, Crb3-KO clones (CKO#1 and CKO#2)
exhibited 60 and 97% reduction in cell migration (Figs. 3B and S3C). Similarly, Crb3-KO clones
(HKO#1 and HKO#2) showed more than 90% reduction in migration
compared to parent HSC-2 cells (Figs.
3F and S3D). However, unlike
Crb3-KD cells, the proliferation of Crb3-KO OSCC
clones was slightly suppressed (CKO#1: 29%, CKO#2: 36%, HKO#1: 28%,
HKO#2: 28%) compared to that of the respective parent cells
(Fig. 3C and G). To investigate
whether Crb3 promotes migration via the epithelial-mesenchymal
transition (EMT) mechanism in OSCC cells, the expression of EMT
markers was evaluated. Immunoblotting revealed that E-cadherin was
expressed in both parent and Crb3-KO cells at different
expression levels. In addition, the expression of EMT inducer snail
family transcriptional repressor-2 (SNAI2) was not dramatically
altered in Crb3-KO cells compared to parent cells for either
cell line (Fig. 3D and H).
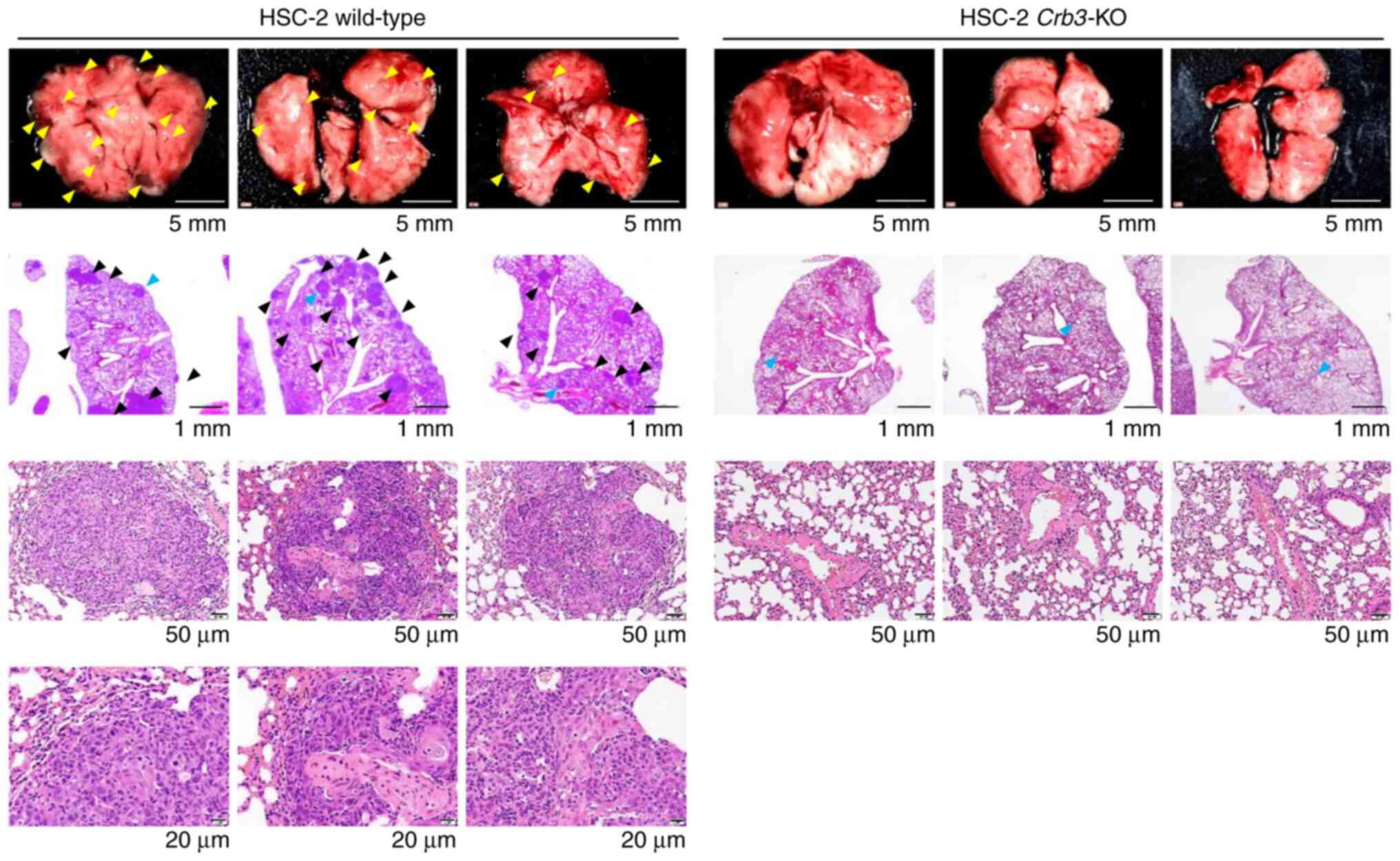
Crb3-KO OSCC cells show a significant
reduction of lung metastases
The metastatic potentials of parent and
Crb3-KO cells were evaluated using a xenograft model of
hematogenous lung metastases (Fig.
4). Parent or Crb3-KO HSC-2 cells were injected into the
tail veins of SCID Hairless Outbred (SHO-Prkdcscid
Hrhr) mice. As a result, no metastases were observed
in Crb3-KO HSC-2 cell-injected mice during the study period
(n=3), whereas parent cell-injected mice developed multiple
metastases in the lungs (n=3). These results indicate that Crb3
plays a key role in OSCC metastasis to the lung.
RhoA activation contributes to OSCC
cell migration and proliferation
Rhosin is a small molecule inhibitor of RhoA
activation. Rhosin contacts RhoA within the GEF-binding pocket, in
proximity to tryptophan 58 of RhoA, to block the interaction of
general Rho-GEFs with RhoA (29).
To investigate whether the RhoA pathway is involved in the
malignant behavior of OSCC, cell proliferation was analyzed by MTT
assay using unmodified Ca9-22 or HSC-2 cells. Cell proliferation of
both cell lines was partially inhibited in the presence of higher
concentrations of Rhosin (Fig. 5A and
E). Y16, another inhibitor of RhoA activation, specifically
blocks the interaction between RhoA and regulator
G-protein-signaling RhoGEFs (RGS-RhoGEFs) (30). As with Rhosin, the addition of 25
µM Y16 inhibited the proliferation of Ca9-22 and HSC-2 (Fig. 5B and F). In addition, Transwell
chamber assays revealed that the cell migration of Ca9-22 (Figs. 5C and S3E) and HSC-2 (Figs. 5G and S3F) was significantly inhibited in the
presence of 20 µM Rhosin or 15 µM Y16 without affecting
proliferation.
RhoA activation is abrogated in
Crb3-KO cells
Rho-family small GTPases are major regulators of the
cytoskeleton and are related to biological processes including cell
migration. To address whether Crb3 affects the RhoA signaling
pathway, the amount of activated RhoA in parent and Crb3-KO
OSCC clones was examined. Endogenously expressed GTP-bound
Rho-family GTPases were captured biochemically from lysates of
serum-stimulated OSCC cells using rhotekin Rho-binding domain
(RBD)-immobilized beads. Total cell lysates and RBD-captured
samples were analyzed by immunoblotting using an anti-RhoA-specific
antibody. Immunoblot analysis showed that GTP-bound RhoA is
significantly depleted in Crb3-KO clones in both Ca9-22 and
HSC-2 (Fig. 5D and H), suggesting
that Crb3 functions as an upstream regulator of the RhoA pathway in
OSCC cell migration.
Discussion
Despite advances in multimodal therapy, the
long-term survival of patients with HNSCC has not been meaningfully
improved. Distant metastasis is one of the major factors adversely
affecting the prognosis of such patients. Finding practical
diagnostic markers and developing molecular targets against tumor
metastasis based on biological analysis of HNSCC are urgent issues.
Our study sought to assess a novel regulatory molecule involved in
malignant behaviors in OSCC, a cell type that constitutes the
largest subgroup of HNSCCs (1).
Crb originally was discovered as an essential
fly gene employed by Drosophila for ectodermal
embryogenesis. Three Crb paralogs have been identified in
the human genome, and Crb3 likely is expressed in all human
epithelial cells. In a previous study, we showed that a novel
monoclonal antibody raised against a C-terminal peptide unique to
isoform A of human Crb3 successfully detects endogenously expressed
Crb3 in colon adenocarcinomas and adjacent non-neoplastic tissues
(25). However, the expression and
function of Crb3 in human tissues remain poorly understood.
Immunostaining revealed that Crb3 is expressed in
OSCC cell lines and tissues from patients with HNSCC. Subcellular
localization of Crb3 was observed predominantly as cytoplasmic
granules in most HNSCC tissues, but a diffuse staining pattern
typically was observed in the cells of neighboring tissues
(Fig. 1). Staining of HSC-2 and
HSC-3 showed a cytoplasmic granule pattern, whereas a more diffuse
pattern was observed in Ca9-22 cells (Fig. 2B). These differences in subcellular
localization of Crb3 may reflect differences in the character of
the originating tissues. However, tumor-adjacent non-neoplastic
tissues displayed much weaker staining compared to OSCC tissues
(Fig. S1). Crb3-KO mice
exhibit severe defects in ductal epithelia in the intestine,
kidney, and lung without gross anatomical defects at the body
surface (23,24). These results may indicate that Crb3
is not essential for the development of the normal squamous
epithelium. In contrast, the results of the present study indicated
that OSCC cell migration was inhibited either by the knock-down or
knock-out of Crb3, implying that Crb3-dependent cell
migration occurs only in cancer cells in squamous epithelial
tissues. EMT markers were not significantly altered by a deficiency
of Crb3, suggesting that EMT is not a major downstream process by
which Crb3 enhances the motility of OSCC cells. Crb3-KO OSCC
clones were viable, albeit while displaying impaired cell growth;
in contrast, knock-down of Crb3 did not appear to affect
proliferation in OSCCs. Crb3 heterozygous mutant mice do not
show apparent defects (23,24),
suggesting that low-level expression of Crb3 is sufficient to drive
proliferation of Crb3-KD cells.
The Rho family of GTPases comprises 20 members,
which are involved in divergent cellular processes including cell
migration and proliferation; these effects are mediated by the
regulation of downstream effector molecules. The knock-down or
knock-out of RhoA triggers compensatory changes in the
expression of other Rho family genes; for instance, the
induction of RhoB and RhoC has been reported under such conditions
(31,32). To exclude the effect of
compensation, analysis of the RhoA function in OSCC cell migration
was demonstrated using inhibitors rather than knock-down or
knock-out of RhoA. Specifically, 20 µM Rhosin and 15 µM Y16
were employed in assays demonstrating that the inhibition of RhoA
activation affects cell migration without affecting proliferation.
However, we note that exposure of cells to 50 µM Rhosin or 25 µM
Y16 inhibits cell proliferation (Fig.
5A, B, E and F), suggesting that RhoA plays a bifunctional role
in OSCC cells in an activity-dependent manner.
To assess whether Crb3 affects RhoA signaling, the
activation of RhoA was assayed using parent cells and
Crb3-KO clones. Crb3-KO clones of Ca9-22 and HSC-2
demonstrated decreased serum-induced activation of RhoA compared to
that in the respective parent cells. However, RhoA activation was
not decreased by knock-down of Crb3 using siRNAs. Similarly,
the proliferation of Ca9-22 and HSC-2 was not decreased
significantly by knock-down of Crb3 (Fig. 2E and H), whereas Crb3-KO
clones exhibited impaired proliferation. These results may indicate
that depletion of Crb3 by siRNA treatment is not sufficient for
inactivation of RhoA and reduction of proliferation, or it may
indicate that Crb3-dependent activation of RhoA is coupled to
proliferation rather than cell migration in OSCC cells.
Members of the RGS-RhoGEF family of proteins, which
includes LARG, PDZ-RhoGEF, and p115-RhoGEF, are regulated by the
Gα12/13 subunits of heterotrimeric G-proteins (33). Transwell cell migration assays
indicated that the migration of OSCC cells was reduced
significantly not only by the general RhoA inhibitor Rhosin but
also by the RGS-RhoGEF-specific inhibitor Y16, suggesting that Crb3
affects RhoA activation in a RGS-RhoGEF-dependent manner.
There are two limitations of this study. First, the
detailed molecular mechanism of RhoA activation was not clarified.
The intracellular domain of Crb3 protein contains the PDZ-domain
binding motif (PBM) and the FERM-domain binding motif (FBM).
Intriguingly, LARG and PDZ-RhoGEF contain the PDZ-domain in their
N-terminal regions. Therefore, the molecular interaction between
the PBM of Crb3 and RhoGEFs should be investigated. The other
limitation is that the number of samples analyzed by
immunohistochemistry was still small. Although The Cancer Genome
Atlas dataset of HNSCC cohorts displays no apparent correlation
between Crb3 mRNA expression and tumor malignancy or patient
prognosis (data not shown), analyses of protein expression may be
more appropriate to assess the clinical significance of Crb3 in
HNSCC. Accordingly, further pathological investigations along with
immunohistochemistry should be performed using larger tissue
samples.
This study showed, for the first time (to our
knowledge), that Crb3 is expressed in HNSCC patient tissues and
OSCC cell lines. Additionally, functional analyses of Crb3, using
knock-down or knock-out cells, demonstrated that Crb3 is involved
in cell migration and proliferation, and that these effects are
mediated by changes in RhoA activity in OSCC cells. These findings
reveal novel aspects of Crb3 function, an insight that has
potential value for the identification of molecular targets with
activity against OSCC metastasis.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Japan Society for the
Promotion of Science (JSPS) Grant-in-Aid for Scientific Research
(C) (project number 20K07368), and Yamaguchi Educational and
Scholarship Foundation to HI.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HI, AH and EK designed the project. YY and HI
carried out the main experiments and data acquisition. YY and AH
prepared tissue samples. EK performed pathological analyses. HI
wrote the paper. HI, AH and EK supervised. YY and HI confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All experiments using HNSCC tissues surgically
obtained from patients were approved by the Research Ethics
Committee of Niigata University (approval no. #2019-0101; Niigata,
Japan). Informed consent was obtained from all participants by
providing an opportunity to opt-out through the website of Niigata
University School of Medicine. All study procedures adhered to the
principles of the Declaration of Helsinki. The animal experiments
conducted in this study were approved by the Animal Care and Use
Committee of Niigata University School of Medicine (approval no.
SA00875; Niigata, Japan). Ethical approval was not sought for the
use of primary NHDF cells in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Crb3
|
crumbs3
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
OSCC
|
oral squamous cell carcinoma
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnson DE, Burtness B, Leemans CR, Lui
VWY, Bauman JE and Grandis JR: Head and neck squamous cell
carcinoma. Nat Rev Dis Primers. 6:922020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Argiris A, Karamouzis MV, Raben D and
Ferris RL: Head and neck cancer. Lancet. 371:1695–1709. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Michiels S, Le Maître A, Buyse M,
Burzykowski T, Maillard E, Bogaerts J, Vermorken JB, Budach W,
PajakT F, Ang KK, et al: Surrogate endpoints for overall survival
in locally advanced head and neck cancer: Meta-analyses of
individual patient data. Lancet Oncol. 10:341–350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuperman DI, Auethavekiat V, Adkins DR,
Nussenbaum B, Collins S, Boonchalermvichian C, Trinkaus K, Chen L
and Morgensztern D: Squamous cell cancer of the head and neck with
distant metastasis at presentation. Head Neck. 33:714–718. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu JC, Bhayani M, Kuchta K, Galloway T
and Fundakowski C: Patterns of distant metastasis in head and neck
cancer at presentation: Implications for initial evaluation. Oral
Oncol. 88:131–136. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Leibel SA, Scott CB, Mohiuddin M, Marcial
VA, Coia LR, Davis LW and Fuks Z: The effect of local-regional
control on distant metastatic dissemination in carcinoma of the
head and neck: Results of an analysis from the RTOG head and neck
database. Int J Radiat Oncol Biol Phys. 21:549–556. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garavello W, Ciardo A, Spreafico R and
Gaini RM: Risk factors for distant metastases in head and neck
squamous cell carcinoma. Arch Otolaryngol Neck Surg. 132:7622006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alvi A and Johnson JT: Development of
distant metastasis after treatment of advanced-stage head and neck
cancer. Head Neck. 19:500–505. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kotwall C, Sako K, Razack MS, Rao U,
Bakamjian V and Shedd DP: Metastatic patterns in squamous cell
cancer of the head and neck. Am J Surg. 154:439–442. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nishijima W, Takooda S, Tokita N, Takayama
S and Sakura M: Analyses of distant metastases in squamous cell
carcinoma of the head and neck and lesions above the clavicle at
autopsy. Arch Otolaryngol Head Neck Surg. 119:65–68. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duprez F, Berwouts D, De Neve W, Bonte K,
Boterberg T, Deron P, Huvenne W, Rottey S and Mareel M: Distant
metastases in head and neck cancer. Head Neck. 39:1733–1743. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiegand S, Zimmermann A, Wilhelm T and
Werner JA: Survival after distant metastasis in head and neck
cancer. Anticancer Res. 35:5499–5502. 2015.PubMed/NCBI
|
|
14
|
Pisani P, Airoldi M, Allais A, Valletti
PA, Battista M, Benazzo M, Briatore R, Cacciola S, Cocuzza S,
Colombo A, et al: Metastatic disease in head & neck oncology.
Acta Otorhinolaryngol Ital. 40 (Suppl 1):S1–S86. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vega FM and Ridley AJ: Rho GTPases in
cancer cell biology. FEBS Lett. 582:2093–2101. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ellerbroek SM, Wennerberg K and Burridge
K: Serine phosphorylation negatively regulates RhoA in vivo. J Biol
Chem. 278:19023–19031. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adnane J, Muro-Cacho C, Mathews L, Sebti
SM and Muñoz-Antonia T: Suppression of rho B expression in invasive
carcinoma from head and neck cancer patients. Clin Cancer Res.
8:2225–2232. 2002.PubMed/NCBI
|
|
18
|
Croucher DR, Rickwood D, Tactacan CM,
Musgrove EA and Daly RJ: Cortactin modulates RhoA activation and
expression of Cip/Kip cyclin-dependent kinase inhibitors to promote
cell cycle progression in 11q13-amplified head and neck squamous
cell carcinoma cells. Mol Cell Biol. 30:5057–5070. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pan Q, Bao LW, Teknos TN and Merajver SD:
Targeted disruption of protein kinase Cε reduces cell invasion and
motility through inactivation of RhoA and RhoC GTPases in head and
neck squamous cell carcinoma. Cancer Res. 66:9379–9384. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bourguignon LYW, Gilad E, Brightman A,
Diedrich F and Singleton P: Hyaluronan-CD44 interaction with
leukemia-associated RhoGEF and epidermal growth factor receptor
promotes Rho/Ras Co-activation, phospholipase Cε-Ca2+
signaling, and cytoskeleton modification in head and neck squamous
cell carcinoma cells. J Biol Chem. 281:14026–14040. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lorenzo-Martín LF, Fernández-Parejo N,
Menacho-Márquez M, Rodríguez-Fdez S, Robles-Valero J, Zumalave S,
Fabbiano S, Pascual G, García-Pedrero JM, Abad A, et al: VAV2
signaling promotes regenerative proliferation in both cutaneous and
head and neck squamous cell carcinoma. Nat Commun. 11:47882020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tepass U, Theres C and Knust E: Crumbs
encodes an EGF-like protein expressed on apical membranes of
Drosophila epithelial cells and required for organization of
epithelia. Cell. 61:787–799. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Whiteman EL, Fan S, Harder JL, Walton KD,
Liu CJ, Soofi A, Fogg VC, Hershenson MB, Dressler GR, Deutsch GH,
et al: Crumbs3 is essential for proper epithelial development and
viability. Mol Cell Biol. 34:43–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Charrier LE, Loie E and Laprise P: Mouse
Crumbs3 sustains epithelial tissue morphogenesis in vivo. Sci Rep.
5:176992016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iioka H, Saito K, Sakaguchi M, Tachibana
T, Homma K and Kondo E: Crumbs3 is a critical factor that regulates
invasion and metastasis of colon adenocarcinoma via the specific
interaction with FGFR1. Int J Cancer. 1–15. 2019.
|
|
26
|
Saito K, Mitsui A, Sumardika IW, Yokoyama
Y, Sakaguchi M and Kondo E: PLOD2-driven IL-6/STAT3 signaling
promotes the invasion and metastasis of oral squamous cell
carcinoma via activation of integrin β1. Int J Oncol. 58:292021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Makarova O, Roh MH, Liu CJ, Laurinec S and
Margolis B: Mammalian Crumbs3 is a small transmembrane protein
linked to protein associated with Lin-7 (Pals1). Gene. 302:21–29.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iioka H, Saito K and Kondo E: Crumbs3
regulates the expression of glycosphingolipids on the plasma
membrane to promote colon cancer cell migration. Biochem Biophys
Res Commun. 519:287–293. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shang X, Marchioni F, Sipes N, Evelyn CR,
Jerabek-Willemsen M, Duhr S, Seibel W, Wortman M and Zheng Y:
Rational design of small molecule inhibitors targeting RhoA
subfamily Rho GTPases. Chem Biol. 19:699–710. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shang X, Marchioni F, Evelyn CR, Sipes N,
Zhou X, Seibel W, Wortman M and Zheng Y: Small-molecule inhibitors
targeting G-protein-coupled Rho guanine nucleotide exchange
factors. Proc Natl Acad Sci USA. 110:3155–3160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simpson KJ, Dugan AS and Mercurio AM:
Functional analysis of the contribution of RhoA and RhoC GTPases to
invasive breast carcinoma. Cancer Res. 64:8694–8701. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jackson B, Peyrollier K, Pedersen E, Basse
A, Karlsson R, Wang Z, Lefever T, Ochsenbein AM, Schmidt G,
Aktories K, et al: RhoA is dispensable for skin development, but
crucial for contraction and directed migration of keratinocytes.
Mol Biol Cell. 22:593–605. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fukuhara S, Chikumi H and Gutkind JS:
RGS-containing RhoGEFs: The missing link between transforming G
proteins and Rho? Oncogene. 20:1661–1668. 2001. View Article : Google Scholar : PubMed/NCBI
|