Introduction
Melanoma is one of the most malignant skin tumors
worldwide and the incidence of melanoma continues to increase, with
an age standardized incidence rate of 3.1 per 100,000/year,
particularly in Caucasian populations (1). The average age of onset of melanoma
is ~50 years (2). Although
melanoma is a rare disease, it has a high mortality rate of 1.7 per
100,000/year in Europe (1). As
melanocytes are located in the basal layer of the epidermis,
melanoma is most common on the skin, accounting for 75% of all skin
cancer-associated deaths (3). In
2019, 96,480 new cases of melanoma were estimated to have been
diagnosed, with a total of 7,230 deaths as a result of melanoma in
the United States alone (4). The
clinically distinguishable subtypes of melanoma are as follows:
Cutaneous, mucosal, uveal and unknown primary melanoma (5). At present, melanoma is mainly treated
via surgery, supplemented with radiotherapy and chemotherapy
(6). However, due to the toxicity
and side effects of radiotherapy and chemotherapy, novel
therapeutic approaches need to be identified. Therefore, the
present study aimed to identify novel therapeutic targets and novel
therapeutic strategies for the treatment of melanoma. However, the
molecular mechanisms that serve a role in melanoma remain to be
elucidated.
Kruppel-like factor 10 (KLF10), formerly known as
TGF-β induction early gene 1, is a DNA-binding transcriptional
regulator that contains a triple C2H2 zinc finger domain (7). In numerous types of cancer, KLF10
upregulation reduces cancer cell proliferation. For example, a
previous study demonstrated that KLF10 expression is negatively
associated with progression-free and overall survival of patients
with pancreatic cancer and could be used as a predictor of
pancreatic cancer stage (8). Under
the regulation of microRNA-106b-5p, KLF10 inhibits the
proliferation of several types of myeloma cells via the regulation
of pituitary tumor-transforming gene 1 (9). Jin et al (10) reported that KLF10 inhibits the
proliferation and invasion of breast cancer cells by inhibiting the
transcription of EGFR. Therefore, it can be hypothesized that KLF10
serves an important role in inhibiting cancer cell proliferation
and promoting apoptosis, which strongly suggests that it may be a
tumor suppressor. However, limited information is available
regarding its mechanism of action in melanoma cells.
Similar to KLF10, acyl-CoA medium-chain synthetase 3
(ACSM3) serves a role in several types of cancer. Previous studies
have used the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds), The Cancer Genome
Atlas (http://www.cbioportal.org/) and Human
Protein Atlas (https://www.proteinatlas.org/) databases to determine
the differential expression of ACSM3. It has previously been
demonstrated that ACSM3 expression is downregulated in malignant
melanoma and that low ACSM3 expression is associated with a poor
prognosis of melanoma (11).
Furthermore, upregulation of ACSM3 inhibits integrin β1/Akt
signaling, which inhibits the progression of ovarian cancer
(12). Additionally, upregulation
of ACSM3 reduces the migration and invasion of liver cancer cells
in vivo and in vitro, and downregulates the
phosphorylation of lysine-deficient protein kinase 1 and Akt
(13).
It was hypothesized that KLF10 might inhibit
melanoma progression by targeting ACSM3. Previous bioinformatics
analyses have demonstrated that the expression levels of KLF10 and
ACSM3 are decreased in melanoma cell lines (11,14).
However, to the best of our knowledge, the effects of KLF10 and
ACSM3 in melanoma on the proliferation, invasion, migration and
apoptosis of tumor cells, remain unclear. Therefore, the present
study investigated the effects of KLF10 and ACSM3 on the
proliferation, invasion, migration and apoptosis of tumor cells,
and their mechanisms, in order to find novel therapeutic targets
for melanoma.
Materials and methods
Cell culture and treatment
The human normal epidermal melanocyte (HEM; cat. no.
PCS-200-012) cell line and melanoma cell lines (SK-MEL-1, cat. no.
HTB-67; A2058, cat. no. CRL-11147; RPMI-7951, cat. no. HTB-66; and
A375, cat. no. CRL-1619) were obtained from the American Type
Culture Collection. All cell lines were cultured in DMEM (HyClone;
Cytiva) supplemented with 10% FBS (HyClone; Cytiva) and 1%
antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
Bioinformatics analysis
The expression levels of KLF10 and ACSM3 and their
correlation in the tissues of patients with cutaneous melanoma, as
well as the relationship between low ACSM3 expression and overall
and disease-free survival in cutaneous melanoma patients were
predicted using Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/). The
JASPAR database (http://jaspar.genereg.net/) was used to predict the
binding site of transcription factor KLF10 to the ACSM3
promoter.
Cell transfection
A375 cells (3×105 cells/well) were
inoculated onto 6-well plates and cultured for 24 h at 37°C with 5%
CO2. Following incubation, cells were transfected with
the pCDNA3.1 vector targeting KLF10 (Ov-KLF10), the negative
control (NC) empty vector (Ov-NC), the pGPH1 vector carrying short
hairpin RNA-ACSM3 (sh-ACSM3; 5′-GGTTTAGGATTATCTGTAA-3′) and its
negative control (sh-NC; 5′-TTCGGGTCATCCGATGGGCC-3′) at a
concentration of 25 nM. All plasmids were synthesized by Shanghai
GenePharma Co., Ltd. and transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Blank
control group cells were untransfected. Following transfection for
48 h at 37°C, the transfection efficiency was detected using
reverse transcription-quantitative PCR (RT-qPCR) and western
blotting.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed to assess cell
proliferation. Briefly, A375 cells were seeded into a 96-well plate
(5×103 cells/well), and incubated for 24, 48 and 72 h at
37°C with 5% CO2. Following incubation, 10 µl CCK-8
solution (cat. no. P0037; Beyotime Institute of Biotechnology) was
added into each well and the cells were cultured for another 2 h at
37°C with 5% CO2. The optical density at 450 nm was
detected using a microplate reader (BioTek Instruments, Inc.).
Colony formation assay
Transfected A375 cells (3×102 cells/well)
were digested with 0.25% trypsin to form a cell suspension, and
then cultured in DMEM and inoculated into 6-well plates, with
incubation at 37°C in 5% CO2 for 14 days. The cells were
fixed with 70% ethanol at room temperature (20–25°C) for 15 min and
stained with 0.05% crystal violet at 37°C for 20 min. The number of
colonies formed was counted (≥50 cells/colony) manually using an
Olympus BX40 light microscope (Olympus Corporation).
TUNEL assay
Apoptosis was detected using One Step TUNEL
Apoptosis Assay Kit (Beyotime Institute of Biotechnology) according
to the manufacturer's protocol at 37°C for 60 min. Briefly, the
A375 cells were washed with PBS three times and then fixed at room
temperature (20–25°C) with 4% paraformaldehyde (Beyotime Institute
of Biotechnology) for 15 min. Subsequently, 0.15% Triton-X-100 was
added to the cells at room temperature for a further 5 min.
Terminal deoxynucleotidyl transferase solution and
FITC-deoxyuridine triphosphate solution (Roche Diagnostics GmbH)
were added to the cells and the cells were incubated at 37°C for 60
min in the dark. DAPI (0.5 µg/ml) was adopted to stain the nuclei
for 5 min at room temperature. The detection solution was discarded
and cells were washed three times with PBS. Antifade Mounting
Medium was used to seal the cells. Cells were randomly selected
from 5 fields of view and an inverted fluorescence microscope
(Olympus Corporation) was used to observe excitation and emission
wavelengths within the range of 450–500 and 515–565 nm (green
fluorescence), respectively. Apoptosis index=number of apoptotic
cells/(number of apoptotic cells + normal cells).
Wound healing assay
A375 cells were inoculated in 6-well plates
(1×105 cells/well). When the cells reached 70–80%
confluence, the medium was replaced with serum-free DMEM and the
cells were incubated overnight at 37°C with 5% CO2.
Subsequently, a 200-µl sterile pipette tip was used to scratch the
cell monolayer. After washing with PBS three times, the plates were
maintained at 37°C with 5% CO2. Images were captured at
0 and 24 h using a BX51 inverted light microscope (magnification,
×100; Olympus Corporation). The area of wound closure in each
picture was determined by using ImageJ software (version. 1.52;
National Institutes of Health). The percentage of migration was
calculated as follows: % migration=scratch current width/scratch
original width ×100.
Cell invasion assay
Cell invasion was assessed using Transwell chambers
coated with Matrigel (BD Biosciences). The 24-well Transwell plates
(Corning, Inc.) with 8-µm pore inserts were coated with Matrigel
(BD Biosciences) at 37°C for 30 min. Subsequently, 4×104
A375 cells were placed in serum-free medium. The upper chamber was
precoated with matrix (Sigma-Aldrich; Merck KGaA) and the cells
were added to the upper chamber (0.1 ml cell suspension/well),
whereas the lower chamber was filled with cell culture medium
supplemented with 20% FBS. Following incubation at 37°C with 5%
CO2 for 24 h, the invading cells were fixed with 4%
formaldehyde at 25°C for 15 min and stained with 0.3% crystal
violet solution (Sigma-Aldrich; Merck KGaA) at room temperature for
30 min. The invading cells were observed under an inverted light
microscope (magnification, ×100; Olympus Corporation) in five
randomly selected fields of view. Percentage invasion was
calculated as follows: % invasion=invaded cells in lower
chamber/total cells added to top chamber ×100.
RT-qPCR
Total RNA was extracted from cells using
RNAzol® RT (Sigma-Aldrich; Merck KGaA) according to the
manufacturer's protocol. Subsequently, RNA was reverse transcribed
into complementary DNA (cDNA) using a QuantiTect Reverse
Transcription Kit (Qiagen GmbH) at 42°C for 1 h. The obtained cDNA
was amplified via qPCR using SYBR Select MasterMix (Takara Bio,
Inc.) on the ABI7500 Sequence Detection System (Applied Biosystems)
according to the detection system manufacturer's protocol. The
thermocycling conditions were 50°C for 2 min, followed by 95°C for
15 sec, 60°C for 15 sec and 72°C for 1 min for a total of 40
cycles. The 2−ΔΔCq method (15) was used to quantify relative mRNA
expression levels using GAPDH as the internal reference gene. The
sequences of the qPCR primers were as follows: KLF10 forward,
5′-ACCCAGGGTGTGGCAAGAC-3′ and reverse, 5′-AGCGAGCAAACCTCCTTTCA-3′;
ACSM3 forward, 5′-AGGAAGATGCTACGTCATGCC-3′ and reverse,
5′-ATCCCCAGTTTGAAGTCCTGT-3′; and GAPDH forward,
5′-ACCACAGTCCATGCCATCAC-3′ and reverse,
5′-TCCACCACCCTGTTGCTGTA-3′.
Western blotting
A375 cells were washed three times with pre-cooled
PBS, and total protein was extracted from the cells using RIPA
lysis buffer (Beyotime Institute of Biotechnology). The total
protein concentration was determined using a Pierce BCA Protein
Assay Kit (Thermo Fisher Scientific, Inc.). Following denaturation
in a 95°C metal bath, SDS-PAGE was performed with a 12% gel for
30-µg protein samples. Separated protein was transferred to PVDF
membranes (Beyotime Institute of Biotechnology), which were blocked
with 5% skimmed milk for 1 h at room temperature. Subsequently, the
membranes were incubated overnight at 4°C with the following
primary antibodies (all purchased from Abcam): KLF10 (dilution,
1:1,000; cat. no. ab184182), Bcl-2 (dilution, 1:1,000; cat. no.
ab194583), Bax (dilution, 1:1,000; cat. no. ab32503), caspase 3
(dilution, 1:5,000; cat. no. ab32351), cleaved-caspase 3 (dilution,
1:1,000; cat. no. ab32042), MMP2 (dilution, 1:1,000; cat. no.
ab92536), MMP9 (dilution, 1:1,000; cat. no. ab76003), ACSM3
(dilution, 1:1,000; cat. no. ab238682), phosphorylated (p)-PI3K
(dilution, 1:500; cat. no. ab154598), p-Akt (dilution, 1:1,000;
cat. no. ab38449), PI3K (dilution, 1:1,000; cat. no. ab191606), Akt
(dilution, 1:1,000; cat. no. ab8805) and GAPDH (dilution, 1:1,000;
cat. no. ab181602). After washing twice with PBS, membranes were
treated with goat anti-rabbit HRP-binding IgG secondary antibody
(dilution, 1:5,000; cat. no. ab6721; Abcam) at room temperature for
1 h. Protein bands were detected using ECL reagent
(MilliporeSigma). Bands were semi-quantified and normalized using
ImageJ v1.46 software (National Institutes of Health).
Chromatin immunoprecipitation
(ChIP)-PCR assay
According to the manufacturer's protocol, total
genomic DNA was isolated using the EZChip™ Kit (MilliporeSigma). A
total of 2×106 A375 cells were transfected with pCDNA3.1
or pGPH1 vectors and lysed with 250 µl SDS Lysis Buffer (Beyotime
Institute of Biotechnology). Next, the A375 cells were sonicated on
ice and fragments of DNA were then resolved using a 2% agarose gel.
For ChIP, samples were diluted in a 10X ChIP dilution buffer and
pre-cleared with 60 µl protein G-agarose beads mixed at 4°C for 1
h. During chromatin separation, the chromatin was centrifuged at
15,000 × g at 4°C for 10 min to remove the insoluble matter and
then pre-cleared chromatin was incubated with 1 µl antibodies
against KLF10 (dilution, 1:100; cat. no. sc-130408; Santa Cruz
Biotechnology, Inc.) at 4°C overnight. The precipitates were washed
with low-salt wash buffer, high-salt wash buffer and LiCI wash
buffer, and rinsed with TE buffer twice. Immunochromatin was
centrifuged at 15,000 × g for 10 min at 4°C and boiled, and then
was amplified via PCR as aforementioned.
Dual-luciferase reporter assay
A375 cells were inoculated in a 24-well plate
(1×105 cells/well). When cells reached 80% confluency,
luciferase activity was detected using the Promega Double
Fluorescence Detection Kit (Promega Corporation) according to the
manufacturer's protocol. ACSM3-wild-type (WT) and ACSM3 mutant
reporter plasmids were constructed in advance through the
PGL3-CMV-LUC-MCS provided by Genomeditech, A375 cells were
transiently co-transfected with Ov-KLF10 together with 0.1 µg
ACSM3-WT and ACSM3 mutant reporter plasmids using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h.
Subsequently, 120 µl cell lysate was added to each well and was
shaken on a horizontal oscillator for 45 min at 50 × g. Then, 10 µl
lysed cell mixture and 50 µl firefly luciferase reagent were added
to a 1.5-ml tube and mixed. Prior to assessment, 50 µl Stop/Glo
Sealing Luciferase Reagent was added. Firefly
luciferase/Renilla luciferase values were recorded and the
ACSM3 promoter transfer activity was analyzed. Each group was set
up with three wells and each experiment was repeated three
times.
Statistical analysis
Data are presented as the mean ± SD of three or more
independent experiments. GraphPad Prism 8.0.2 software (GraphPad
Software, Inc.) was used for data analysis. Statistical differences
were determined using one-way ANOVA followed by Tukey's post hoc
test for group comparisons. The correlation between ACSM3 and KLF10
was evaluated using Pearson's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
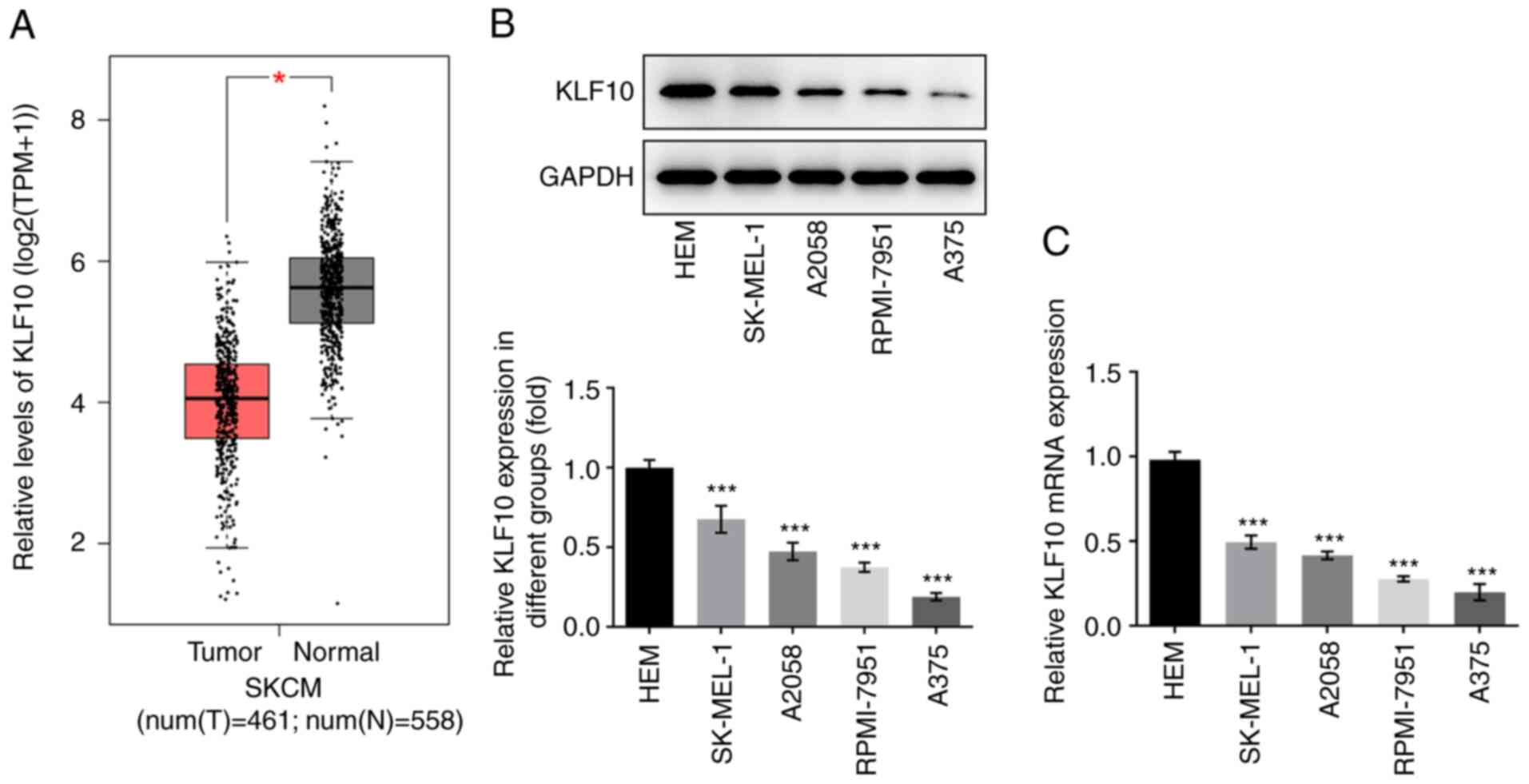
KLF10 expression is relatively low in
melanoma tissues and cell lines
KLF10 expression levels in melanoma were examined
using GEPIA. Compared with that in healthy tissues, KLF10
expression was significantly decreased in melanoma tissues
(Fig. 1A). Western blotting
(Fig. 1B) and RT-qPCR (Fig. 1C) were also used to detect KLF10
expression in melanoma cell lines. Compared with that of other
melanoma cell lines, including HEM, SK-MEL-1, A2058 and RPMI-7951,
KLF10 expression was lowest in A375 cells. Therefore, A375 cells
were selected for the subsequent experiments.
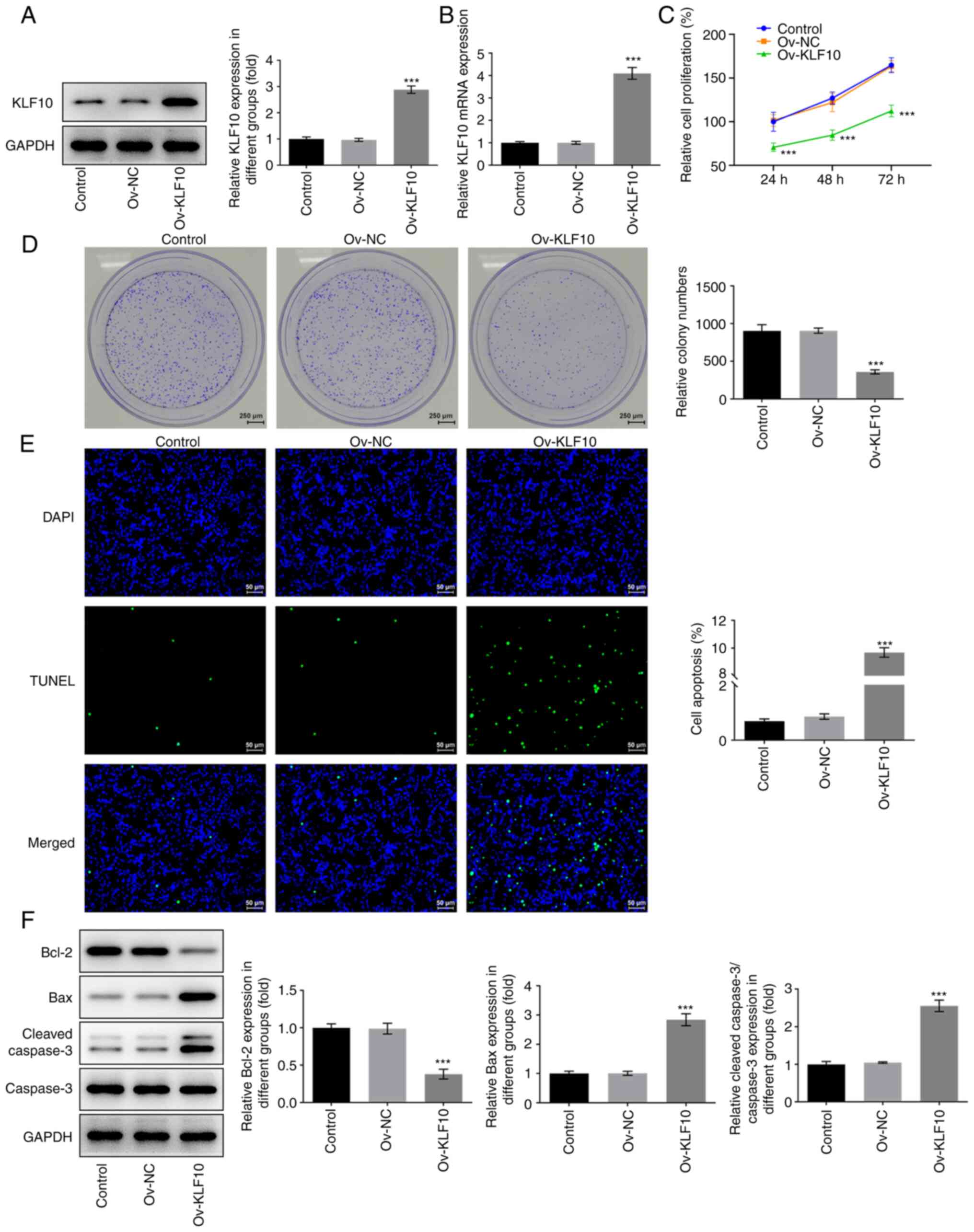
KLF10 overexpression inhibits melanoma
cell proliferation and induces apoptosis
To investigate the specific role of KLF10 in A375
cells, cells overexpressing KLF10 were analyzed using western
blotting (Fig. 2A) and RT-qPCR
(Fig. 2B). Compared with that in
the Ov-NC group, KLF10 expression was significantly increased in
the Ov-KLF10 group. Following KLF10 overexpression, CCK-8 (Fig. 2C) and colony formation (Fig. 2D) assays were used to detect cell
proliferation. The results demonstrated that KLF10 overexpression
significantly inhibited A375 cell proliferation compared with the
Ov-NC group. Furthermore, apoptosis was detected using a TUNEL
assay. The results demonstrated that the green fluorescence in the
Ov-KLF10 group was significantly enhanced compared with the Ov-NC
group (Fig. 2E), which indicated
that significant apoptosis had occurred in the cells with
overexpression. Subsequently, the expression levels of
apoptosis-related proteins were detected via western blotting
(Fig. 2F). Compared with those in
the Ov-NC group, the expression levels of the antiapoptotic protein
Bcl-2 were significantly decreased, whereas the expression level of
the proapoptotic proteins Bax and cleaved caspase-3/caspase 3 ratio
were significantly increased, in the Ov-KLF10 group. The results
suggested that overexpression of KLF10 may promote melanoma cell
apoptosis.
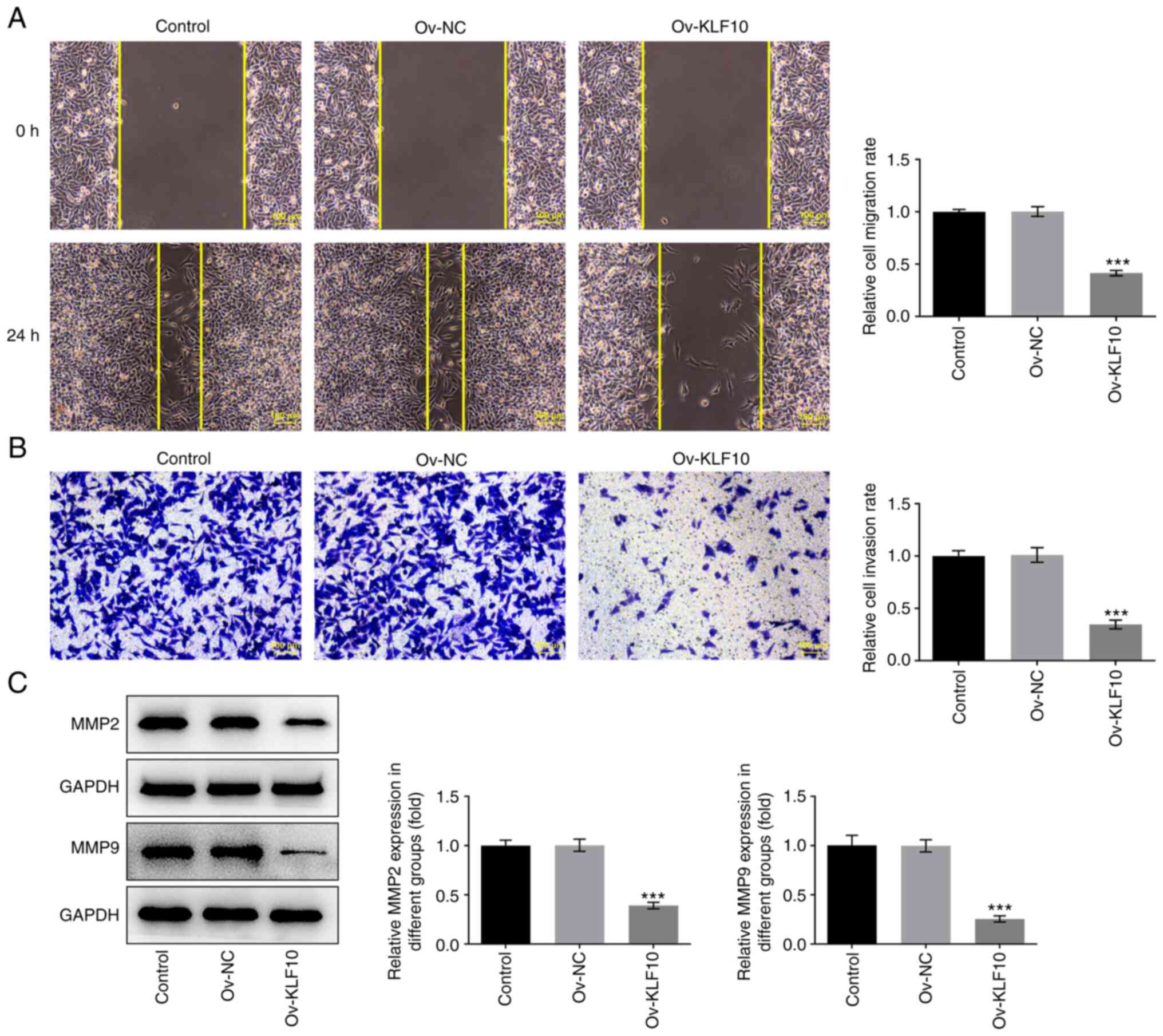
KLF10 overexpression inhibits melanoma
cell invasion and migration
Transwell and wound-healing assays were used to
detect cell invasion and migration. Compared with that in the Ov-NC
group, cell migration (Fig. 3A)
and invasion (Fig. 3B) were
significantly decreased in the Ov-KLF10 group. Additionally, KLF10
overexpression inhibited the expression of metastasis-related
proteins MMP2 and MMP9 in A375 cells (Fig. 3C). These results indicated that
KLF10 may inhibit A375 cell invasion and migration.
KLF10 promotes the transcription of
ACSM3
Results obtained using GEPIA indicated that ACSM3
expression was low in melanoma tissues (Fig. 4A). Furthermore, the overall
survival rate and disease-free survival rate of patients with
melanoma were positively associated with high ACSM3 expression
(Fig. 4B and C). Bioinformatics
analysis also demonstrated that the expression levels of KLF10 and
ACSM3 were positively associated in patients with melanoma
(Fig. 4D). Subsequently, western
blotting (Fig. 4E) and RT-qPCR
(Fig. 4F) were performed to detect
ACSM3 expression in HEM and A375 cell lines. Compared with that in
HEM cells, ACSM3 mRNA and protein expression was downregulated in
A375 cells. Furthermore, the binding sites between KLF10 and ACSM3
were predicted using the JASPAR database (Fig. 4G). In the presence of the wild-type
ACSM3 promoter, KLF10 overexpression induced a significant increase
in luciferase activity, whereas there was no significant change in
the presence of the mutant promoter (Fig. 4H). These results suggested that
KLF10 directly upregulated ACSM3 by binding to the ACSM3 promoter.
Considering the positive association between KLF10 and ACSM3
expression, a ChIP assay was performed to further verify the
binding of KLF10 and ACSM3. It was found that co-transfected ACSM3
wild-type and Ov-KLF10 A375 cells showed more relative luciferase
activity (Fig. 4H). ACSM3 was
present in the KLF10 fraction, which revealed that ACSM3 may bind
to KLF10 (Fig. 4I). Furthermore,
it was demonstrated that ACSM3 mRNA and protein expression was
upregulated in A375 cells overexpressing KLF10 compared with in the
Ov-NC group (Fig. 4J and K).
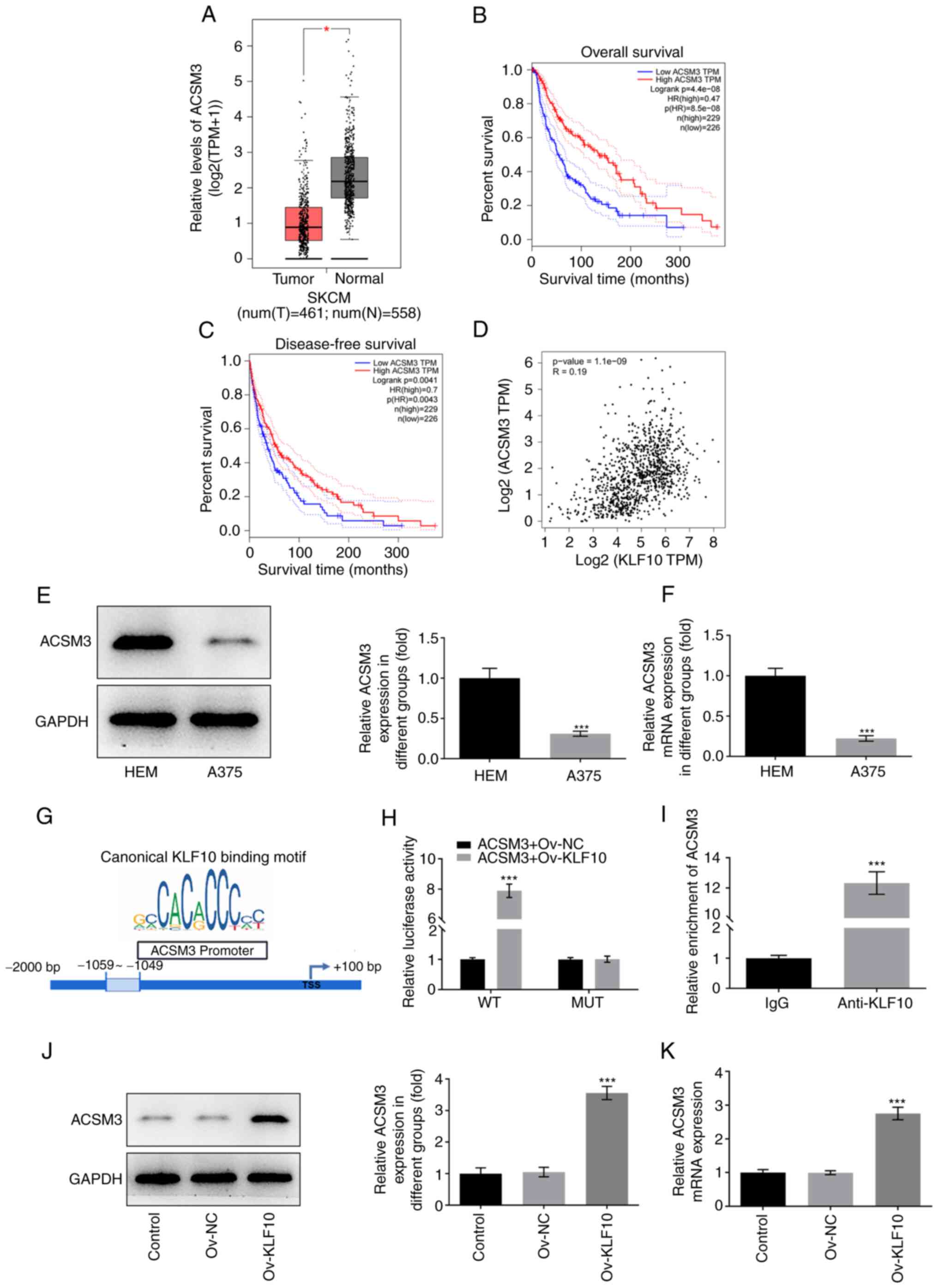 | Figure 4.KLF10 binds to ACSM3 and promotes the
transcription of ACSM3. (A) The Gene Expression Profiling
Interactive Analysis website revealed the expression levels of
ACSM3. (B) The association between ACSM3 expression and overall
survival rate in melanoma patients, (C) the association of ACSM3
and disease-free survival rate in patients with melanoma, and (D)
the association between KLF10 expression and ACSM3 expression in
patients with melanoma were examined. (E) Western blotting and (F)
RT-qPCR were used to detect the expression levels of ACSM3 in
melanoma cells. (G) JASPAR predicted the binding sites of KLF10 and
the ACSM3 promoter. (H) Luciferase reporter gene assay was used to
detect the ACSM3 promoter activity. (I) Immunoprecipitation further
indicated that KLF10 could bind with ACSM3. (J) Western blotting
and (K) RT-qPCR were used to detect the expression levels of ACSM3
after KLF10 overexpression. *P<0.05, ***P<0.001 vs. HEM,
ACSM3 + Ov-NC, IgG or Ov-NC. ACSM3, acyl-CoA medium-chain
synthetase 3; KLF10, Kruppel-like factor 10; MUT, mutant; N,
normal; NC, negative control; Ov, overexpression; RT-qPCR, reverse
transcription-quantitative PCR; SKCM, skin cutaneous melanoma; T,
tumor; TPM, transcripts per million; WT, wild-type. |
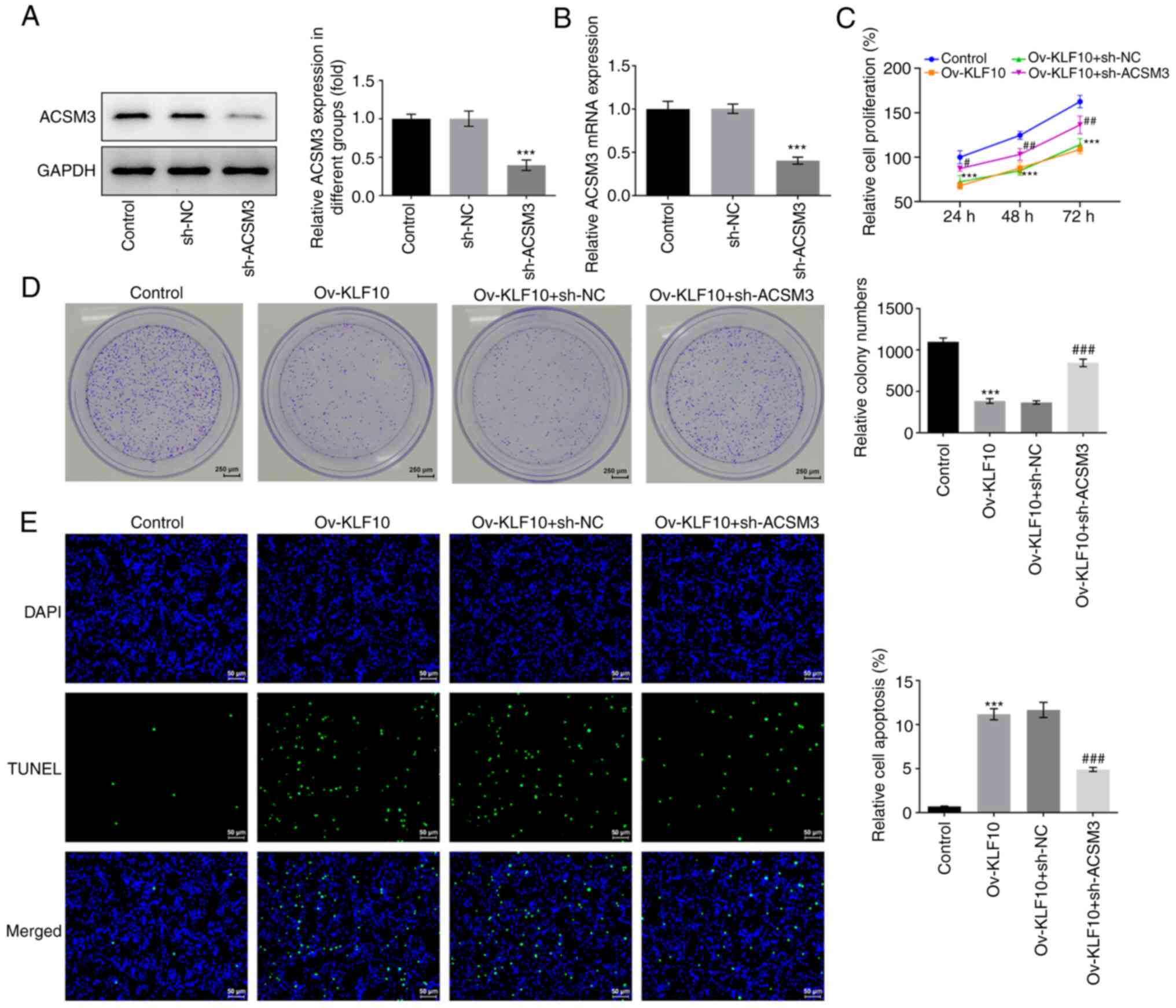
KLF10 upregulates ACSM3 and inhibits
the malignant progression of melanoma via the PI3K/Akt signaling
pathway
To elucidate the mechanism of the inhibition of
malignant melanoma progression via the binding of KLF10 to ACSM3,
sh-ACSM3 was constructed and transfected into A375 cells. The
transfection efficiency was analyzed using western blotting and
RT-qPCR. The results demonstrated that ACSM3 mRNA and protein
expression in the sh-ACSM3 group was significantly decreased
compared with that in the sh-NC group (Fig. 5A and B). CCK-8 and colony formation
assays were performed to assess cell proliferation. The results
demonstrated that cell proliferation was inhibited by KLF10
overexpression compared with that in the control group, which was
reversed by sh-ACSM3 as compared with that in the Ov-KLF10 + sh-NC
group (Fig. 5C and D).
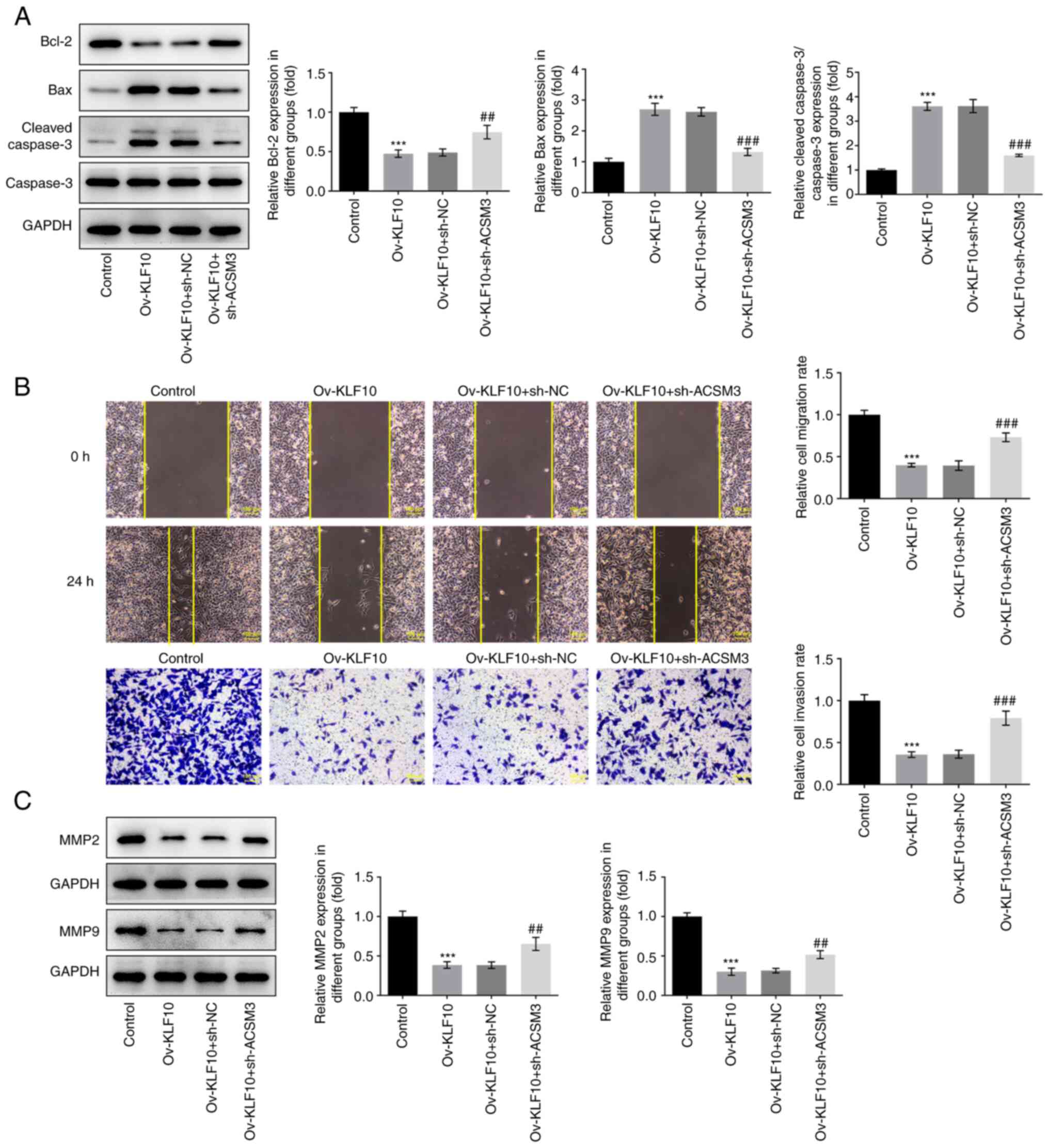
Furthermore, apoptosis and apoptosis-related
proteins were analyzed using the TUNEL assay and western blotting,
respectively. Overexpression of KLF10 significantly induced the
apoptosis of A375 cells compared with that in the control group,
whereas sh-ACSM3 decreased apoptosis in A375 cells transfected with
Ov-KLF10 in comparison with that in the Ov-KLF10 + sh-NC group
(Fig. 5E). The results of western
blotting demonstrated that Bcl-2 protein expression was upregulated
and the expression levels of proapoptotic proteins Bax and the
cleaved caspase 3/caspase 3 ratio were downregulated in the
Ov-KLF10+sh-ACSM3 group compared with those in the Ov-KLF10 + sh-NC
group (Fig. 6A). Transwell and
wound healing assays were used to detect cell invasion and
migration. The results demonstrated that KLF10 overexpression
inhibited the invasion and migration of A375 cells, whereas
silencing ACSM3 expression enhanced cell invasion and migration
(Fig. 6B). Furthermore, KLF10
overexpression inhibited the expression of metastasis-related
proteins MMP2 and MMP9 in A375 cells, whereas sh-ACSM3 increased
MMP2 and MMP9 expression (Fig.
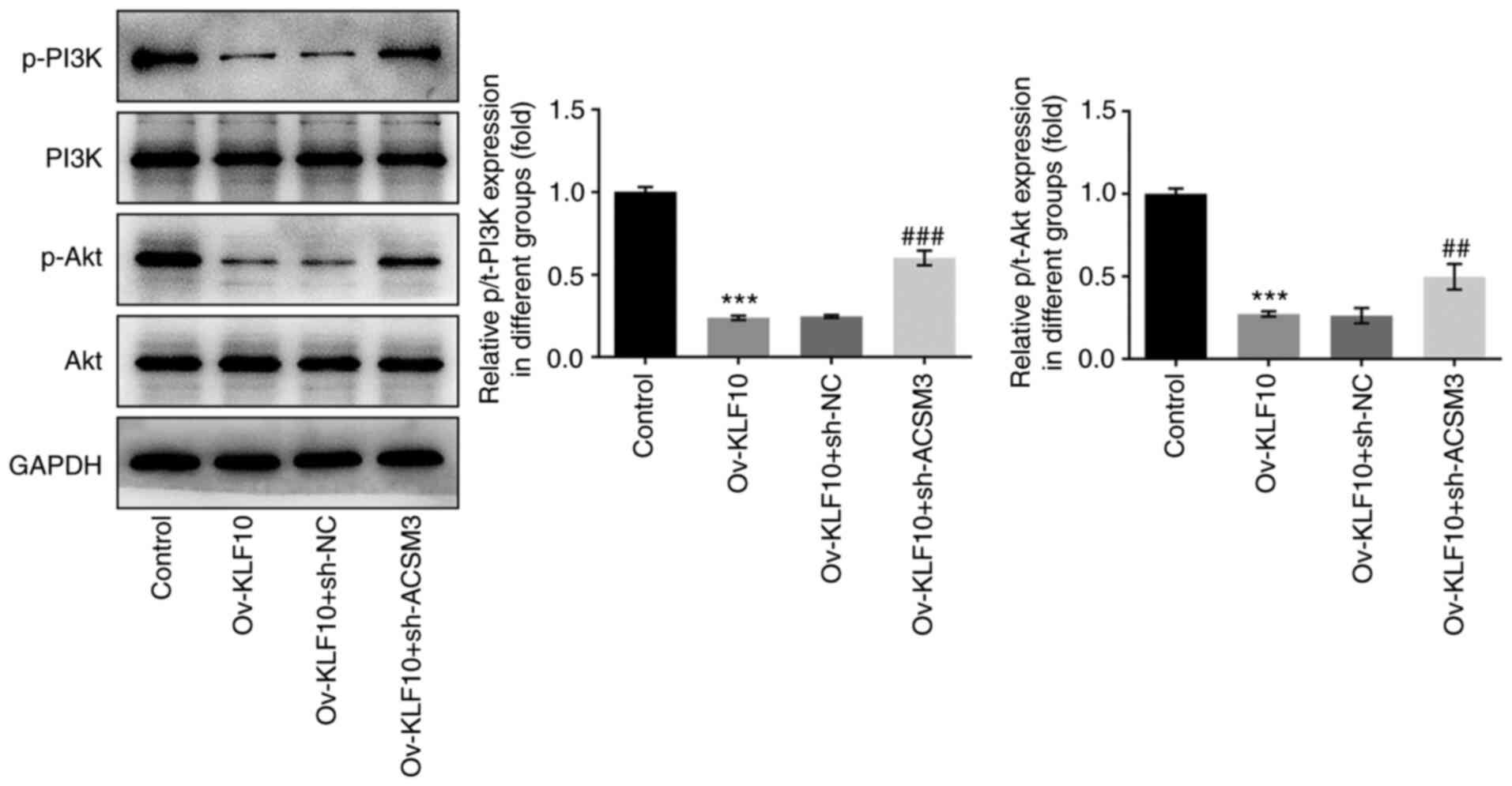
6C). It has previously been reported that ACSM3 can negatively
regulate the Akt signaling pathway (10). Therefore, western blotting was
performed to detect the protein expression levels of PI3K/Akt
signaling pathway proteins. Compared with the control group, the
overexpression of KLF10 significantly reduced the levels of p-PI3K
and p-Akt, which was reversed by sh-ACSM3, in contrast to the
Ov-KLF10 + sh-NC group (Fig. 7).
Overall, these results indicated that KLF10 may upregulate ACSM3
and inhibit the malignant progression of A375 cells via the
PI3K/Akt signaling pathway.
Discussion
Melanoma is considered to be the deadliest skin
cancer (16–18). In its early stages, melanoma can be
successfully treated by surgery alone, and it has a high 5-year
survival rate of 92%; however, the 1-year survival rate markedly
decreases to 55% following metastasis (19). Understanding the mechanisms that
lead to the occurrence of melanoma will provide novel strategies
for the diagnosis and treatment of this disease (20). In the present study, KLF10 and
ACSM3 expression levels were significantly downregulated in
melanoma cell lines, and ACSM3 was closely associated with low
overall and disease-free survival in patients with cutaneous
melanoma. ACSM3 expression levels were associated with KLF10.
Furthermore, KLF10 overexpression significantly inhibited the
proliferation, migration and invasion of A375 cells, and induced
apoptosis, which were reversed by knockdown of ACSM3.
The KLF10 transcription factor was originally cloned
from human osteoblasts and acts as a major response gene in TGF-β
therapy (21). Increasing evidence
suggests that KLF10 serves an important role in mimicking TGF-β
function in a number of types of cancer. For example, Baroy et
al (22) reported that LSAMP
reduces osteosarcoma cell proliferation by indirectly upregulating
one or more genes (HES1, CTAG2 or KLF10). Jin et al
(21) reported that KLF10 is
upregulated in apoptosis induced by tetrotylenine or vancomycin,
and that overexpression of KLF10 induces apoptosis of K562 cells.
Furthermore, Hsu et al (23) demonstrated that KLF10 regulation of
Bax inhibitor-1 expression and Ca2+ release may be a
pathway of estrogen-induced apoptosis. This evidence supports the
role of KLF10 as a suppressor in several types of cancer; however,
to the best of our knowledge, the role of KLF10 in melanoma has not
previously been reported. In the present study, the inhibitory
effect of KLF10 on the proliferation and survival of A375 cells and
its mechanism were investigated. The results determined that KLF10
may also act as a tumor suppressor in melanoma.
ACSM3 has previously been reported to be involved in
numerous biological processes, including tumor migration, invasion,
fat accumulation and the butyrate oxidation pathway (13,24,25),
and particularly in cancer. Gopal et al (26) demonstrated that ACSM3 gene
expression is decreased in hepatocellular carcinoma (HCC) tissues,
whereby the loss of ACSM3 expression is associated with advanced
HCC stage and a low survival rate. Zhu et al (11) demonstrated that ACSM3 is
downregulated in melanoma cells and is associated with a poor
prognosis and immune rejection of malignant melanoma. Furthermore,
in vitro and in vivo, the study identified that ACSM3
overexpression could reduce the proliferation, invasion and colony
formation of malignant melanoma (11). Consistent with the aforementioned
report, the present study demonstrated that ACSM3 expression was
downregulated in melanoma cells compared with healthy cells.
Furthermore, survival analysis using the GEPIA database
demonstrated that low levels of ACSM3 were significantly associated
with poor overall survival. Therefore, it can be hypothesized that
ACSM3 is associated with the clinical malignancy of melanoma.
ACSM3 was therefore considered to be an effective
target for the treatment of melanoma and was selected for further
study. According to the results of the GEPIA and ChIP assay, KLF10
was positively associated with ACSM3 expression in cutaneous
melanoma. KLF10 was demonstrated to bind to and promote the
transcription of ACSM3. As KLF10 was demonstrated to be involved in
the progression of melanoma, it was hypothesized that KLF10 may
target ACSM3 to inhibit the proliferation, migration and invasion
of melanoma cells and promote cell apoptosis. Therefore, silencing
of ACSM3 was performed. The results demonstrated that silencing
ACSM3 reversed the inhibitory effects of KLF10 overexpression on
the viability, proliferation, migration and invasion of A375 cells,
which indicated that there may be an important interaction between
the two molecules.
Furthermore, the mechanism of KLF10 as a tumor
suppressor gene of melanoma was preliminarily explored. Akt is
considered to be an important component of the cell cycle and in
cell survival and apoptosis, and is positively associated with PI3K
(27,28). Yang et al (29) reported that KLF10 inhibits the
PTEN/PI3K/Akt signaling pathway and inhibits the malignant
progression of myeloma. Furthermore, Li et al (30) demonstrated that LINC00641 inhibits
the activation of the PTEN/PI3K/Akt signaling pathway via KLF10 and
that activation of this signaling pathway promotes bladder cancer.
In the present study, it was demonstrated that KLF10 overexpression
in A375 cells decreased the levels of p-PI3K and p-Akt, which
indicated that the PI3K/Akt signaling pathway may be negatively
regulated by KLF10 expression. These results suggested that the
inhibition of A375 cell proliferation, migration and invasion may
be dependent on the inactivation of the PI3K/Akt signaling pathway
induced by KLF10. Furthermore, PI3K/Akt signaling pathway
inactivation induced by KLF10 overexpression was reversed by
sh-ACSM3. These results indicated that KLF10 upregulation of ACSM3
may inhibit the malignant progression of A375 cells via the
PI3K/Akt signaling pathway. Finally, one limitation of the present
study was that only the effect of KLF10 overexpression of KLF10 on
melanoma cells in vivo was investigated. Further
verification experiments involving knockdown of KLF10 would enrich
the current research results.
In conclusion, to the best of our knowledge, the
present study was the first to elucidate the role of KLF10 in
melanoma progression. The results demonstrated that the
KLF10/ACSM3/PI3K/Akt axis was associated with melanoma cell
proliferation. It was also indicated that KLF10 may inhibit A375
cell proliferation and migration by binding to ACSM3, resulting in
the inactivation of the PI3K/Akt signaling pathway. Overall, the
present study highlighted a novel mechanism underlying the
pathogenesis of melanoma.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ, YZ and LJ conceptualized and designed the
current study. ZZ, YZ, LJ and HZ acquired, analyzed and interpreted
the data. LJ and HZ drafted the manuscript and revised it
critically for important intellectual content. All authors agreed
to be held accountable for the current study in ensuring questions
related to the integrity of any part of the work are appropriately
investigated and resolved. All authors read and approved the final
manuscript. ZZ and HZ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Raimondi S, Suppa M and Gandini S:
Melanoma epidemiology and sun exposure. Acta Derm Venereol.
100:adv001362020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Situm M, Buljan M, Kolić M and Vučić M:
Melanoma-clinical, dermatoscopical, and histopathological
morphological characteristics. Acta Dermatovenerol Croat. 22:1–12.
2014.PubMed/NCBI
|
|
3
|
Davis LE, Shalin SC and Tackett AJ:
Current state of melanoma diagnosis and treatment. Cancer Biol
Ther. 20:1366–1379. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soenksen LR, Kassis T, Conover ST,
Marti-Fuster B, Birkenfeld JS, Tucker-Schwartz J, Naseem A, Stavert
RR, Kim CC, Senna MM, et al: Using deep learning for
dermatologist-level detection of suspicious pigmented skin lesions
from wide-field images. Sci Transl Med. 13:eabb36522021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Teramoto Y, Keim U, Gesierich A, Schuler
G, Fiedler E, Tüting T, Ulrich C, Wollina U, Hassel JC, Gutzmer R,
et al: Acral lentiginous melanoma: A skin cancer with unfavourable
prognostic features. A study of the German central malignant
melanoma registry (CMMR) in 2050 patients. Br J Dermatol.
178:443–451. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Longvert C and Saiag P: Melanoma update.
Rev Med Interne. 40:178–183. 2019.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Turner J and Crossley M: Mammalian
Kruppel-like transcription factors: More than just a pretty finger.
Trends Biochem Sci. 24:236–240. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang VH, Chu PY, Peng SL, Mao TL, Shan
YS, Hsu CF, Lin CY, Tsai KK, Yu WC and Ch'ang HJ: Kruppel-like
factor 10 expression as a prognostic indicator for pancreatic
adenocarcinoma. Am J Pathol. 181:423–430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou M, Chen J, Zhang H, Liu H, Yao H,
Wang X, Zhang W, Zhao Y and Yang N: KLF10 inhibits cell growth by
regulating PTTG1 in multiple myeloma under the regulation of
microRNA-106b-5p. Int J Biol Sci. 16:2063–2071. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin W, Chen BB, Li JY, Zhu H, Huang M, Gu
SM, Wang QQ, Chen JY, Yu S, Wu J and Shao ZM: TIEG1 inhibits breast
cancer invasion and metastasis by inhibition of epidermal growth
factor receptor (EGFR) transcription and the EGFR signaling
pathway. Mol Cell Biol. 32:50–63. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu Z, Wang D and Shen Y: Loss of ACSM3
confers worsened prognosis and immune exclusion to cutaneous
melanoma. J Cancer. 11:6582–6590. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan L, He Z, Li W, Liu N and Gao S: The
overexpression of Acyl-CoA Medium-Chain Synthetase-3 (ACSM3)
suppresses the ovarian cancer progression via the inhibition of
integrin β1/AKT signaling pathway. Front Oncol. 11:6448402021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ruan HY, Yang C, Tao XM, He J, Wang T,
Wang H, Wang C, Jin GZ, Jin HJ and Qin WX: Downregulation of ACSM3
promotes metastasis and predicts poor prognosis in hepatocellular
carcinoma. Am J Cancer Res. 7:543–553. 2017.PubMed/NCBI
|
|
14
|
Kim J, Shin S, Subramaniam M, Bruinsma E,
Kim TD, Hawse JR, Spelsberg TC and Janknecht R: Histone demethylase
JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem Biophys Res
Commun. 401:412–416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pavri SN, Clune J, Ariyan S and Narayan D:
Malignant melanoma: Beyond the basics. Plast Reconstr Surg.
138:330e–340e. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rastrelli M, Tropea S, Rossi CR and
Alaibac M: Melanoma: Epidemiology, risk factors, pathogenesis,
diagnosis and classification. In Vivo. 28:1005–1011.
2014.PubMed/NCBI
|
|
18
|
Elder DE, Bastian BC, Cree IA, Massi D and
Scolyer RA: The 2018 World health organization classification of
cutaneous, mucosal, and uveal melanoma: Detailed analysis of 9
distinct subtypes defined by their evolutionary pathway. Arch
Pathol Lab Med. 144:500–522. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Namikawa K and Yamazaki N: Targeted
therapy and immunotherapy for melanoma in Japan. Curr Treat Options
Oncol. 20:72019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jin W, Di G, Li J, Chen Y, Li W, Wu J,
Cheng T, Yao M and Shao Z: TIEG1 induces apoptosis through
mitochondrial apoptotic pathway and promotes apoptosis induced by
homoharringtonine and velcade. FEBS Lett. 581:3826–3832. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baroy T, Kresse SH, Skårn M, Stabell M,
Castro R, Lauvrak S, Llombart-Bosch A, Myklebost O and Meza-Zepeda
LA: Reexpression of LSAMP inhibits tumor growth in a preclinical
osteosarcoma model. Mol Cancer. 13:932014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsu CF, Sui CL, Wu WC, Wang JJ, Yang DH,
Chen YC, Yu WC and Chang HS: Klf10 induces cell apoptosis through
modulation of BI-1 expression and Ca2+ homeostasis in
estrogen-responding adenocarcinoma cells. Int J Biochem Cell Biol.
43:666–673. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Junkova K, Mirchi LF, Chylíková B, Janků
M, Šilhavý J, Hüttl M, Marková I, Miklánková D, Včelák J, Malínská
H, et al: Hepatic transcriptome profiling reveals lack of Acsm3
expression in polydactylous rats with high-fat diet-induced
hypertriglyceridemia and visceral fat accumulation. Nutrients.
13:14622021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
De Preter V, Arijs I, Windey K, Vanhove W,
Vermeire S, Schuit F, Rutgeerts P and Verbeke K: Impaired butyrate
oxidation in ulcerative colitis is due to decreased butyrate uptake
and a defect in the oxidation pathway. Inflamm Bowel Dis.
18:1127–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gopal R, Selvarasu K, Pandian PP and
Ganesan K: Integrative transcriptome analysis of liver cancer
profiles identifies upstream regulators and clinical significance
of ACSM3 gene expression. Cell Oncol (Dordr). 40:219–233. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu F, Na L, Li Y and Chen L: Roles of the
PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and
tumours. Cell Biosci. 10:542020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen H, Zhou L, Wu X, Li R, Wen J, Sha J
and Wen X: The PI3K/AKT pathway in the pathogenesis of prostate
cancer. Front Biosci (Landmark Ed). 21:1084–1091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang N, Chen J, Zhang H, Wang X, Yao H,
Peng Y and Zhang W: LncRNA OIP5-AS1 loss-induced microRNA-410
accumulation regulates cell proliferation and apoptosis by
targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple
myeloma. Cell Death Dis. 8:e29752017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Hong S and Liu Z: LncRNA LINC00641
predicts prognosis and inhibits bladder cancer progression through
miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun.
503:1825–1829. 2018. View Article : Google Scholar : PubMed/NCBI
|