Introduction
Osteosarcoma (OS) is the most common type of primary
malignant bone tumor, which generally occurs in adolescents between
10 and 20 years old, and is associated with high malignancy and a
poor prognosis (1,2). Once OS occurs, due to its aggressive
growth, patients have to receive large doses of preoperative and
postoperative chemotherapy, which places a great psychological and
physical burden on the patients. Certain cases of OS are resistant
to chemotherapy and the tumors continue to grow rapidly. The limb
of the patient may need to be amputated, lung metastases may occur
and the mortality rate remains high (3,4).
The treatment of OS has not been significantly
updated in the past four decades (5,6).
Numerous targeted drugs are improving the treatment of other
tumors, but their development for the treatment of OS is
progressing slowly, highlighting the urgent need to study the
mechanisms driving the occurrence and progression of OS (5). If essential genes that play key roles
in the growth of OS can be identified, effective OS-targeted
therapy drugs could be screened or developed.
In mammalian cells, protein synthesis requires the
participation of amino acid synthetases, such as aminoacyl-tRNA
synthetases (ARSs), which can transfer amino acids to tRNAs
(7). ARSs have been reported to be
abnormally expressed in numerous tumors, such as colon cancer,
hepatoma and breast cancer (8).
Previous studies have revealed that ARSs play important roles in
triggering cancer cell proliferation, differentiation and RNA
splicing (9–11). Isoleucyl-tRNA synthetase 2 (IARS2),
also known as isoleucine-tRNA ligase, catalyzes the aminoacylation
of tRNA through its cognate amino acids and it is located in
mitochondria (9). IARS2 mutations
have been identified in patients with cataracts, growth hormone
deficiency with short stature and peripheral neuropathy (12). In addition, IARS2 has been reported
to be overexpressed in numerous types of cancer, such as
glioblastoma, non-small-cell lung cancer (NSCLC) and gastric
carcinoma (GC) (13). IARS2
knockdown inhibits cell proliferation and colony formation, and
promotes apoptosis in NSCLC and GC (14–16).
However, to the best of our knowledge, the expression and function
of IARS2 in OS have not been reported.
In the present study, the expression of IARS2 in OS
was analyzed using the Gene Expression Profiling Interactive
Analysis 2 (GEPIA2) database. In addition, the function of IARS2 in
cell proliferation and apoptosis was evaluated by knocking down its
expression in OS cell lines, which is helpful for exploring
potential therapeutic strategies for OS.
Materials and methods
Cell culture
MNNG/HOS Cl #5 [(R-1059-D) (MNNG/HOS)] and U-2 OS
(U2OS) cell lines were purchased from Shanghai Institutes for
Biological Sciences, Chinese Academy of Sciences. MNNG/HOS cells
were cultured in MEM (Gibco; Thermo Fisher Scientific, Inc.), and
U2OS cells were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) and incubated at 37°C in a humidified
chamber containing 5% CO2.
Lentivirus (LV) packaging and
infection
The short hairpin (sh)RNA IARS2 (shIARS2) LV and
empty plasmid control (shControl) were constructed by GeneChem,
Inc, which all bearing GFP gene. The shRNA was synthesized and
inserted into the GV115 vector (GeneChem, Inc.) at the AgeI
and EcoRI restriction sites. IARS2 gene shRNA
sequence was:
5′-CCGGGTACTTGCAGTCATCCATTAATTCAAGAGATTAATGGATGACTGCAAGTACTTTTTG-3′.
The insertion of the shRNA cassette was verified by restriction
mapping and direct Sanger DNA sequencing. The 2nd generation system
recombinant Lentivirus plasmids (4 µg), together with packaging (2
µg) and envelope (4 µg) plasmids were transfected into 293T cells
(Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences) at 40% density in a 10-cm dish using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. The ratio used for the lentivirus,
packaging and envelope plasmids is 2:1:2. Infectious LVs were
harvested at 48 h post-transfection and filtered through 0.45-mm
cellulose acetate filters. The infectious titer was determined by a
hole-by-dilution titer assay.
MNNG/HOS and U2OS cells were seeded in six-well
plates at a density of 5×104 cells/well for 24 h and
then infected with recombinant lentiviral vectors shIARS2 or
shCtrl. The infection MOI was 20. After 48 h, the MNNG/HOS and U2OS
cells were collected and the efficiency of lentiviral infection was
evaluated by observing the green signal under a fluorescence
microscope (both shIARS2 or shCtrl were all bearing the GFP gene),
reverse transcription-quantitative PCR (RT-qPCR) and western
blotting.
RNA extraction and RT-qPCR
analysis
Total RNA was extracted from MNNG/HOS and U2OS cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and the mature mRNA was reverse transcribed into
cDNA using MMLV reverse transcriptase (Promega Corporation)
according to the manufacturer's instructions. RT-qPCR was performed
in a Real-Time PCR Detection System (Agilent Technologies, Inc.)
using a SYBR Master Mixture Kit (Takara Bio, Inc.); the PCR mixture
contained 10 µl SYBR premix Ex Taq, 0.5 µl sense and antisense
primers (2.5 µM), 1 µl cDNA and 8 µl RNase-free H2O. The
reaction was conducted with initial pre-denaturation at 95°C for 15
sec, followed by 42 cycles of denaturation for 5 sec and extension
at 60°C for 30 sec. β-actin was used as the internal control. The
primers used for amplification were as follows: IARS2 forward,
5′-TGGACCTCCTTATGCAAACGG-3′ and reverse,
5′-GGCAACCCATGACAATCCCA-3′; and β-actin forward,
5′-GCGTGACATTAAGGAGAAGC-3′ and reverse,
5′-CCACGTCACACTTCATGATGG-3′. The relative expression levels were
determined by the 2−ΔΔCq method (17). All experiments were performed at
least in triplicate.
Western blot analysis
MNNG/HOS and U2OS cells were lysed in RIPA buffer
(Beyotime Institute of Biotechnology) on ice for 30 min. The lysate
was centrifuged at 15,000 × g, 4°C for 15 min, and the supernatant
was heated at 100°C for 5 min. The concentration of the protein
samples was determined using a BCA Protein Assay Kit (Beyotime
Institute of Biotechnology). Following separation by 10% SDS-PAGE
(20 µg protein/lane), the proteins were transferred onto
polyvinylidene fluoride membranes (MilliporeSigma). Subsequently,
the membranes were blocked at room temperature with 5% skim milk
for 1 h and incubated with primary antibodies against IARS2
(1:2,000; MilliporeSigma; SAB4502342), cleaved-caspase-3 (1:500;
Cell Signaling Technology, Inc; cat. no. # 9664S), BAX (1:500;
Abcam; ab32503) or β-actin (1:5,000; Abcam; ab8226) at 4°C
overnight. Subsequently, the membranes were washed with
Tris-buffered saline-0.5% Tween and incubated at room temperature
with goat anti-mouse IgG-HRP secondary antibody (1:2,000; Santa
Cruz Biotechnology, Inc; cat. no. #7076) for 2 h. Protein bands on
the membranes were then detected using an ECL-PLUS/Kit (Thermo
Fisher Scientific, Inc.).
Cell proliferation assay
After infection with either shCtrl or shIARS2 LV,
~3,000 infected MNNG/HOS and U2OS cells/well were seeded into
96-well plates. The plates were incubated at 37°C in 5%
CO2 for 5 days. During this period, the number of cells
was counted and images were captured by a Celigo Image Cytometer
(Nexcelom Bioscience) every day.
Additionally, cell proliferation was measured every
day using the MTT reagent. Cells infected with shIARS2 or shCtrl
were cultured at 37°C with 5% CO2 for 24 h, detached
with 0.25% trypsin solution and placed in 96-well plates at a final
concentration of 2,000 cells/ml. MTT assays were performed at 24,
48, 72, 96 and 120 h following infection. Briefly, the culture
medium was replaced and 10 µl MTT solution/well was added. The
cells with MTT were incubated for 4 h at 37°C in 5% CO2.
After incubation, the cells were washed and 200 µl dimethyl
sulfoxide/well was added. Finally, the cells were incubated at room
temperature for 10 min. The absorbance of the stained supernatants
was detected at 490 nm using a spectrophotometer (Thermo Fisher
Scientific, Inc.). Cell proliferation is directly proportional to
the absorbance rate. The experiment was performed in
triplicate.
Colony formation assay
shCtrl- and shIARS2-infected MNNG/HOS and U2OS cells
were plated (1,000 cells/well) in a six-well culture plate in
triplicate and maintained at 37°C. The culture medium was refreshed
every 3 days for 10 days or until colonies were formed. Following
colony formation, they were washed twice with PBS, fixed at room
temperature with 4% paraformaldehyde for 30 min and stained at room
temperature with 0.4% crystal violet for 15 min. The number of
colonies containing ≥50 cells was counted under a light
microscope.
Flow cytometric analysis
Early cell apoptosis was evaluated by flow cytometry
according to the manufacturer's protocol. After MNNG/HOS and U2OS
cells infected with shIARS2-LV or shCtrl-LV were cultured in
six-well plates for 5 days, they were collected via centrifugation
at 1,000 × g at 4°C for 3 min and washed with cold PBS.
Subsequently, a 100 µl cell suspension was prepared and stained at
4°C with 5 µl Annexin V-APC (BD Biosciences) for 15 min. The FITC
channel indicated for the GFP signal. Signal intensity
>1×104 was regarded as positive signal. The ratio of
apoptotic cells was analyzed (Flow Jo 7.6.1, Tree Star, Inc) using
a flow cytometer (Accuri C6 plus; BD Biosciences).
GEPIA2 database analysis
IARS2 expression and survival curves were analyzed
in the GEPIA2 database via the GEPIA database website (http://gepia2.cancer-pku.cn/) Expression DIY (Box
Plot) or Survival analysis function module, with SARC (sarcoma) as
the specific type of cancer.
Statistical analysis
Statistical analysis was performed using SPSS 16.0
(SPSS, Inc.). The statistical data for each group are presented as
the mean ± SD of 3 independent experimental repeats. Unpaired
Student's t-tests were used for comparisons between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Association of IARS2 with prognosis in
sarcoma
To explore the role of IARS2 in human sarcoma
development, the GEPIA2 database was first used to analyze the
expression of IARS2 and its relationship with survival. There was
no significant difference in IARS2 expression between tumor tissues
and normal tissues (Fig. 1A).
Notably, high IARS2 expression tended to be associated with poor
overall and disease-free survival but this was not statistically
significant (Fig. 1B). These
findings indicated that IARS2 tends to be high expressed in OS
tissue and has the tendency to be associated with survival but this
was not statistically significant.
Knockdown of IARS2 expression in OS
cell lines
To further investigate the function of IARS2 in the
human OS cell lines MNNG/HOS and U2OS, a shIARS2 LV was prepared
and MNNG/HOS and U2OS cells were infected. The MNNG/HOS and U2OS
cells were successfully infected with the shCtrl and shIRSA2 LVs
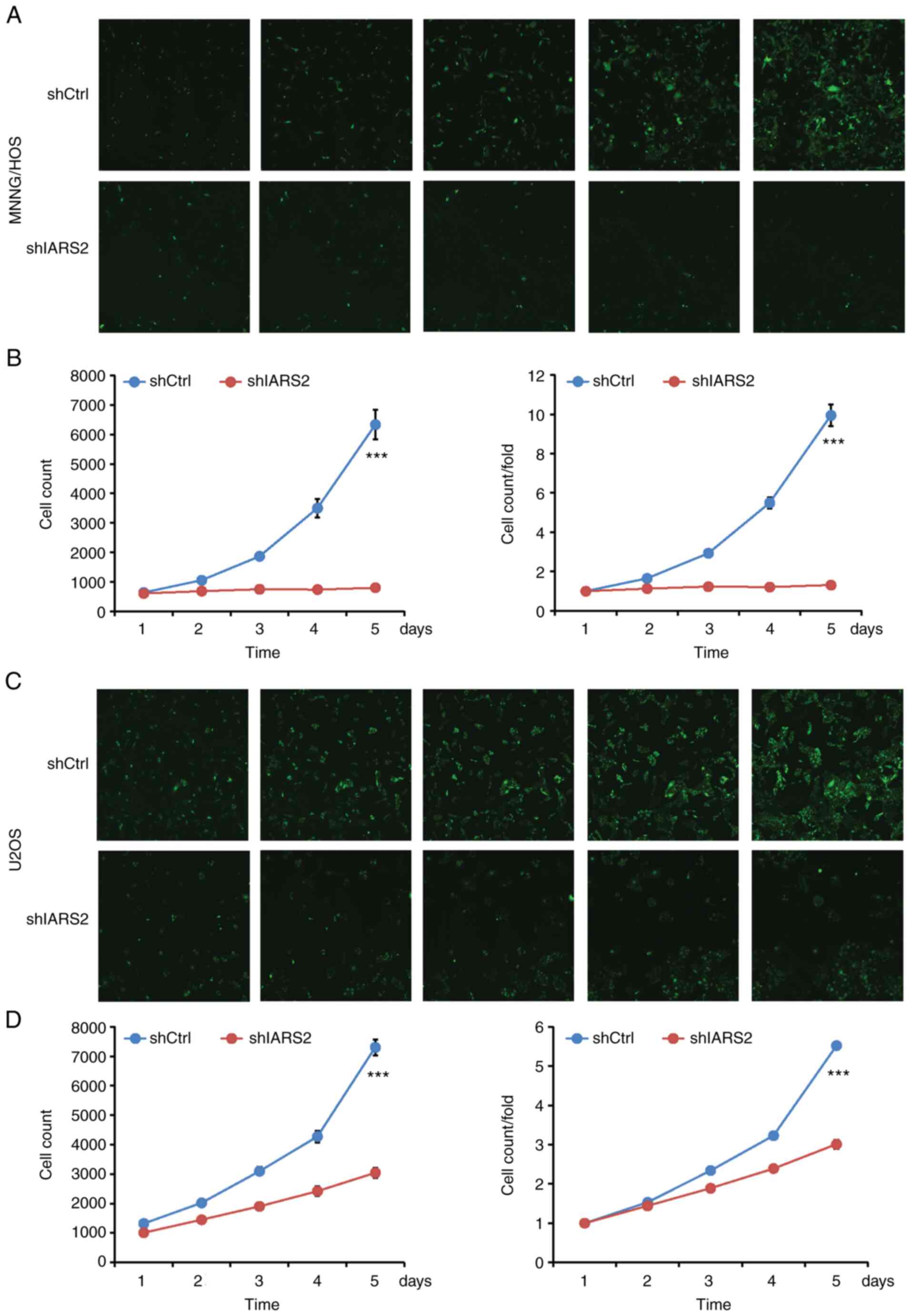
using GFP fluorescence as the indicator (Fig. 2A and B). Subsequently, the
expression levels of IARS2 were detected by RT-qPCR and western
blot analysis. The mRNA (Fig. 2C and
D) and protein (Fig. 2E and F)
expression levels of IARS2 were markedly decreased in MNNG/HOS and
U2OS cells infected with shIARS2 LV compared with in the shCtrl
cell groups. These data indicated that the expression of IARS2 was
effectively knocked down in OS cell lines.
IARS2 knockdown inhibits proliferation
of OS cell lines
To determine the effect of IARS2 knockdown on cell
proliferation, MNNG/HOS and U2OS cells were cultured, images were
captured and the number of cells with green fluorescence signals
was calculated by Celigo every day for 5 days. Proliferation of
shIARS2 MNNG/HOS (Fig. 3A and B)
and U2OS (Fig. 3C and D) cells was
significantly decreased compared with in the shCtrl cell
groups.
To confirm the inhibitory effect of IARS2 on human
OS cell proliferation, the proliferation of MNNG/HOS and U2OS cells
was next detected using the MTT assay. The MTT assays revealed that
MNNG/HOS (Fig. 4A) and U2OS
(Fig. 4B) cell proliferation was
significantly inhibited in shIARS2-infected cells compared with in
shCtrl-infected cells. These results demonstrated that IARS2
knockdown inhibited proliferation of the OS cell lines.
IARS2 knockdown suppresses colony
formation in OS cell lines
In cell proliferation, cell density is a vital
influencing factor. A colony formation assay is an easy approach to
detect whether cell proliferation is affected by cell density and
thus a colony formation assay was performed to investigate the
effect of IARS2 knockdown. As revealed in Fig. 5A, the colonies formed by MNNG/HOS
cells infected with shIARS2 were significantly smaller in size than
those infected with shCtrl. Consistently, the colonies formed by
U2OS cells infected with shIARS2 were also smaller than those
formed by U2OS cells infected with shCtrl (Fig. 5B). This result indicated that IARS2
knockdown suppressed colony formation by OS cell lines.
IARS2 knockdown promotes the apoptosis
of OS cell lines
Apoptosis plays an important role in tumor cell
proliferation and survival, and is often decreased in tumors
(18) IARS2 knockdown markedly
inhibited cell proliferation. Subsequently, the apoptosis of OS
cells with IARS2 knockdown was investigated. Apoptosis was measured
by single staining with Annexin-V-APC and cell infection efficiency
was detected by FITC. As shown in Fig.
6, IARS2 knockdown increased cell apoptosis in MNNG/HOS
(Fig. 6A) and U2OS (Fig. 6C) cells. In order to further assess
apoptosis, the expression levels of apoptosis-related proteins
cleaved caspase-3 (the activated form of caspase-3) and BAX
(pro-apoptotic protein) were detected by western blotting. IARS2
knockdown also increased the expression levels of apoptosis-related
proteins in MNNG/HOS (Fig. 6B) and
U2OS (Fig. 6D) cells. These data
indicated that IARS2 knockdown promoted the apoptosis of OS cell
lines.
Discussion
Mitochondrial IARS is encoded by the IARS2 gene in
zone 4 band 1 of chromosome 1 (19). IARS2 is synthesized in the
cytoplasmic ribosome and transported to the mitochondria to execute
its function. IARS2 catalyzes the binding of isoleucine to specific
tRNAs for mitochondrial DNA translation (20). A number of studies have revealed
that IARS2 is associated with various types of cancer. IARS2
knockdown has been shown to inhibit the proliferation and promote
the apoptosis of human A375 melanoma cells and AGS GC cells
(16,21). In acute myeloid leukemia (AML)
cells, knockdown of IARS2 can also inhibit proliferation, which is
achieved by regulating the p53/p21/PCNA/Eif4E pathway (19). IARS2 overexpression has been
reported to promote NSCLC tumorigenesis by activating the AKT/mTOR
pathway, and IARS2 silencing can inhibit cell proliferation and
promote apoptosis (14,15). It has been reported that the
mutation or variation of IARS2 also leads to novel disorders, such
as cataracts, sensorineural hearing deficit and skeletal dysplasia
(13,22). A previous study concluded that
mitochondrial IARS2 not only participates in tumorigenesis but is
also involved in mitochondria-related disorders, which indicates
that IARS2 has an important role in the process of disease
(13,14,19–22).
Nevertheless, the function of IARS2 in OS has not been reported so
far.
The present study was, to the best of our knowledge,
the first to demonstrate that IARS2 knockdown inhibited cell
proliferation and promoted the apoptosis of human OS cells, which
is consistent with previous studies on the role in IARS2 in other
tumor types (19–21). In the present study, the expression
of IARS2, and its relationship with overall and disease-free
survival were analyzed using the GEPIA2 database. It was observed
that IARS2 tends to be high expressed in OS tissue and has the
tendency to be related with survival but this was not statistically
significant. To further investigate the effect of IARS2 in
vitro, two OS cell lines (MNNG/HOS and U2OS) were infected with
shIARS2 or shCtrl LV. Firstly, IARS2 knockdown significantly
suppressed the proliferation of the OS cell lines compared with in
the shCtrl cell groups. In addition, IARS2 knockdown suppressed
colony formation compared with in the shCtrl cell groups.
Apoptosis, an important process supporting tumor cell
proliferation, was detected by flow cytometric analysis; The
results is consistent with the hypothesis thatIARS2 knockdown
increases the apoptosis of OS cell lines.
In the present study, the effect of IARS2 expression
on cell function was investigated. However, the molecular mechanism
underlying the regulatory effects of IARS2 on the proliferation and
apoptosis of OS cell lines has not been studied. IARS2 has been
reported to regulate the p53/p21/PCNA/Eif4E pathway to promote cell
proliferation in AML, and the AKT/mTOR pathway to promote
tumorigenesis in NSCLC (14,15,19).
Thus, whether these signaling pathways are also involved in OS cell
proliferation and apoptosis driven by IARS2 should be analyzed in
future research. Furthermore, due to the normal sample limitation
in the GEPIA2 dataset and previous study has not reported an
association between survival and IARS2 expression so far (19–21),
the expression and survival curve should be further confirmed with
more healthy and OS clinical samples. In addition, the expression
of IARS2 in tumor and tumor-adjacent tissue needs to be detected by
western blotting and immunohistochemistry. Furthermore, protein
microarrays and RNA sequencing are useful technologies that should
be used to explore important pathways or molecules associated with
IARS2. Given the existence of off-target effects of shRNA, another
shRNA should be designed to investigate the function of IARS2 in OS
cell lines.
Rapid cell proliferation is one of the hallmarks of
tumor cells. Hence, tumor cells need more energy and substrates to
synthesize biological macromolecules, such as nucleic acids and
proteins (23). It was
hypothesized that IARS2, as a mitochondrial isoleucine-tRNA
synthetase could enable tumor cells to synthesize more proteins in
the mitochondria. Mitochondrial function is extremely active in the
tumor environment (24,25) and overexpression of IARS2 promotes
the synthesis of proteins, which may support the function of
mitochondria.
In conclusion, it was identified that IARS2 tends to
be high expressed in OS tissue and has the tendency to be related
with survival but this was not statistically significant. In
vitro, it was demonstrated that IARS2 knockdown inhibited cell
proliferation and colony formation, and increased apoptosis.
Consequently, further investigation of the mechanism and function
of IARS2 in OS may provide a potential treatment strategy for OS.
Given that IARS2 is an enzyme, the development of inhibitors that
target it could provide a new approach to OS therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Shanghai
Xuhui District Medical Peak Subject Project (grant no. SHXH201710)
and the Xuhui District Medical Science and Technology Project
(grant no. SHXU201709).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FL and QL conceived and designed the study. FL and
QL acquired, analyzed and interpreted the data. QL drafted the
manuscript. QL and FL confirm the authenticity of all the raw data.
Both authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer Am Cancer Soc. 115:1531–1543. 2009.PubMed/NCBI
|
|
2
|
Sadykova LR, Ntekim AI, Muyangwa-Semenova
M, Rutland CS, Jeyapalan JN, Blatt N and Rizvanov AA: Epidemiology
and risk factors of osteosarcoma. Cancer Invest. 38:259–269. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lilienthal I and Herold N: Targeting
molecular mechanisms underlying treatment efficacy and resistance
in osteosarcoma: A review of current and future strategies. Int J
Mol Sci. 21:68852020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen C, Xie L, Ren T, Huang Y, Xu J and
Guo W: Immunotherapy for osteosarcoma: Fundamental mechanism,
rationale, and recent breakthroughs. Cancer Lett. 500:1–10. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harrison DJ, Geller DS, Gill JD, Lewis VO
and Gorlick R: Current and future therapeutic approaches for
osteosarcoma. Expert Rev Anticancer Ther. 18:39–50. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang C, Tian Y, Zhao F, Chen Z, Su P, Li Y
and Qian A: Bone microenvironment and osteosarcoma metastasis. Int
J Mol Sci. 21:69852020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu YC, Han JM and Kim S: Aminoacyl-tRNA
synthetases and amino acid signaling. Biochim Biophys Acta Mol Cell
Res. 1868:1188892021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou Z, Sun B, Nie A, Yu D and Bian M:
Roles of Aminoacyl-tRNA synthetases in cancer. Front Cell Dev Biol.
8:5997652020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rajendran V, Kalita P, Shukla H, Kumar A
and Tripathi T: Aminoacyl-tRNA synthetases: Structure, function,
and drug discovery. Int J Biol Macromol. 111:400–414. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim D, Kwon NH and Kim S: Association of
aminoacyl-tRNA synthetases with cancer. Top Curr Chem. 344:207–245.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim YW, Kwon C, Liu JL, Kim SH and Kim S:
Cancer association study of aminoacyl-tRNA synthetase signaling
network in glioblastoma. PLoS One. 7:e409602012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schwartzentruber J, Buhas D, Majewski J,
Sasarman F, Papillon-Cavanagh S, Thiffault I, Sheldon KM,
Massicotte C, Patry L, Simon M, et al: Mutation in the
nuclear-encoded mitochondrial isoleucyl-tRNA synthetase IARS2 in
patients with cataracts, growth hormone deficiency with short
stature, partial sensorineural deafness, and peripheral neuropathy
or with Leigh syndrome. Hum Mutat. 35:1285–1289. 2014.PubMed/NCBI
|
|
13
|
Mazaris P, Hong X, Altshuler D, Schultz L,
Poisson LM, Jain R, Mikkelsen T, Rosenblum M and Kalkanis S: Key
determinants of short-term and long-term glioblastoma survival: A
14-year retrospective study of patients from the Hermelin Brain
Tumor Center at Henry Ford Hospital. Clin Neurol Neurosurg.
120:103–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin J, Liu W, Li R, Liu J, Zhang Y, Tang W
and Wang K: IARS2 silencing induces non-small cell lung cancer
cells proliferation inhibition, cell cycle arrest and promotes cell
apoptosis. Neoplasma. 63:64–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Di X, Jin X, Ma H, Wang R, Cong S, Tian C,
Liu J, Zhao M, Li R and Wang K: The oncogene IARS2 promotes
non-small cell lung cancer tumorigenesis by activating the AKT/MTOR
Pathway. Front Oncol. 9:3932019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang Z, Wang X, Yan Q, Zhang S and Li Y:
Knockdown of IARS2 suppressed growth of gastric cancer cells by
regulating the phosphorylation of cell cycle-related proteins. Mol
Cell Biochem. 443:93–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pistritto G, Trisciuoglio D, Ceci C,
Garufi A and D'Orazi G: Apoptosis as anticancer mechanism: function
and dysfunction of its modulators and targeted therapeutic
strategies. Aging (Albany NY). 8:603–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H, Tian Y, Li X, Wang B, Zhai D, Bai Y,
Dong C and Chao X: Knockdown of IARS2 inhibited proliferation of
acute myeloid leukemia cells by regulating p53/p21/PCNA/eIF4E
Pathway. Oncol Res. 27:673–680. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhong L, Zhang Y, Yang JY, Xiong LF, Shen
T, Sa YL, O'Yang YM, Zhao SH and Chen JY: Expression of IARS2 gene
in colon cancer and effect of its knockdown on biological behavior
of RKO cells. Int J Clin Exp Pathol. 8:12151–12159. 2015.PubMed/NCBI
|
|
21
|
Ma D, Li S, Nie X, Chen L, Chen N, Hou D,
Liu X and Gao B: RNAi-mediated IARS2 knockdown inhibits
proliferation and promotes apoptosis in human melanoma A375 cells.
Oncol Lett. 20:1093–1100. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vona B, Maroofian R, Bellacchio E, Najafi
M, Thompson K, Alahmad A, He L, Ahangari N, Rad A, Shahrokhzadeh S,
et al: Expanding the clinical phenotype of IARS2-related
mitochondrial disease. BMC Med Genet. 19:1962018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burke PJ: Mitochondria, bioenergetics and
apoptosis in cancer. Trends Cancer. 3:857–870. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ippolito L, Giannoni E, Chiarugi P and
Parri M: Mitochondrial Redox Hubs as Promising Targets for
Anticancer Therapy. Front Oncol. 10:2562020. View Article : Google Scholar : PubMed/NCBI
|