Introduction
Colorectal cancer (CRC) is the fourth most lethal
cancer worldwide, following lung, liver and gastric cancer
mortality, with ~700,000 individuals succumbing to CRC annually
(1). Chemotherapy is the most
common treatment for recurrent and metastatic colon cancer, and
chemotherapeutic resistance is the largest limitation to cancer
treatment efficacy. 5-FU is one of the oldest chemotherapeutics and
it acts by suppressing tumor cell proliferation via the inhibition
of DNA replication and RNA synthesis (2). Amongst patients receiving 5-FU, ~50%
develop drug resistance, which is the main cause of the poor
prognosis of patients with colon cancer.
Long non-coding (lnc)RNAs are an RNA subclass of
>200 bp in length, lacking a protein-coding sequence. The lncRNA
plasmacytoma variant translocation 1 (PVT1) gene has been reported
to be highly expressed in esophageal, gastric, liver, colorectal,
pancreatic, and other gastrointestinal cancers (3). PVT1 was first identified as a MYC
agonist in mice (4). Subsequent
analyses revealed that PVT1 could potentially upregulate
epithelial-mesenchymal transition in esophageal cancer (5). PVT1 has also been reported to promote
cell proliferation, activate the cell cycle and support a tumor
stem cell-like population in hepatocellular carcinoma cells
(6). This gene has been also
reported to exert anti-apoptotic effects, and promote CRC tumor
cell proliferation and CRC metastasis (7). In addition, PVT1 has been suggested
to promote resistance to chemotherapeutics in a variety of
gastrointestinal cancers and to enhance multidrug resistance in
gastric cancers (8). Furthermore,
PVT1 has been reported to promote resistance to gemcitabine by
regulating the Wnt/β-catenin and autophagy signaling pathways in
pancreatic cancer (9), and also to
regulate 5-FU resistance in gastric cancer (10) which could be reversed by the
knockdown of PVT1 in 5-FU-resistant CRC cells (11). However, the mechanisms through
which PVT1 confers resistance to 5-FU remain unclear.
The aim of the present study was to reveal the
mechanisms underlying PVT1-mediated 5-FU resistance in colon
cancer, and to determine whether PVT1 may be a promising possible
therapeutic target in 5-FU-resistant cells, with the potential to
improve the survival rate of patients who are resistant to 5-FU
treatment.
Materials and methods
Cells, cell culture and
transfection
The colon cancer cell line, HCT116 (CCL-247), and
293T (CRL-3216) cells were obtained from the American Type Culture
Collection (ATCC). Cells were cultured in RPMI-1640 (Gibco; Thermo
Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS;
HyClone; Cytiva) at 37°C in a 5% CO2 incubator. RNA/DNA
was transfected into parental and 5-FU-resistant HCT116 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were harvested after 48 h of incubation at
37°C. For PVT1 knockdown, 20 nM siRNA targeting PVT1 or 20 nM
non-targeting control siRNA were transfected into cells. The siRNA
sequences are presented in Table
I. The siRNA sequences were synthesized at ribobio Technologies
Cells were transfected with 50 ng CDK4 overexpression plasmid
(Shanghai GenePharma Co., Ltd.) or 50 ng pcDNA3.1 empty vector
(Shanghai GenePharma Co., Ltd.), in order to upregulate CDK4
expression, The overexpression and inhibition of microRNA
(miRNA/miR)-486-5p was achieved by transfecting 20 nM miR-486-5p
mimic or an miR-486-5p inhibitor (Guangzhou RiboBio Co., Ltd.) into
parental and 5-FU-resistant HCT116 cells with
Lipofectamine® 2000. A corresponding scrambled
oligonucleotide sequence was used as a negative control. The miRNA
mimics/inhibitor sequences are presented in Table I.
 | Table I.Sequences of siRNAs targeting PVT1 and
miR-486-5p mimics/inhibitor. |
Table I.
Sequences of siRNAs targeting PVT1 and
miR-486-5p mimics/inhibitor.
| Name | Sequence (5′-3′) |
|---|
| siRNA-1 |
GGCACAUUUCAGGAUACUAAA |
| siRNA-2 |
GCUUAUUAUAGACUUAUAUGU |
| siRNA-3 |
GGGAUUUAGGCACUUUCAAUC |
| si-con |
AACCGCUGAGUUGUCUGAUAU |
| miR-486-5p
mimics |
UCCUGUACUGAGCUGCCCCGAG |
| Mimics nc |
GUCACUUGAUCCGAUGCGCCCG |
| miR-486-5p
inhibitor |
CUCGGGGCAGCUCAGUACAGGA |
| Inhibitor nc |
GGCCAAGUAGGUCCUAGCGCAG |
Construction of resistant strains
HCT116 cells (CCL-247, ATCC) were grown to 60–80%
confluency, followed by the addition of 0.2 mM 5-FU (Beijing
Solarbio Science & Technology Co., Ltd.) and incubation at 37°C
for 24 h. The culture medium was then discarded, and the cells were
washed twice with phosphate-buffered saline (PBS) (Gibco; Thermo
Fisher Scientific, Inc.). The conditioned medium was replaced with
5-FU-free medium. The cells were passaged after they resumed
proliferation, and the 5-FU shock procedure was repeated 6–8 times.
When a cell population had grown stably at this concentration, the
cells were exposed to a higher concentration of 5-FU and passaging
was continued, which resulted in the gradual increase of the drug
concentration toleration threshold. The induction of 5-FU
resistance lasted for 6 months until the cells could grow stably in
a significantly increased drug concentration.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from parental and
5-FU-resistant HCT116 cells using TRIzol reagent (MilliporeSigma).
RNA purity and integrity were assessed using RNA electrophoresis.
Using random primers, total RNA was reverse transcribed into cDNA
at 42°C for 1 h (RevertAid First Strand cDNA Synthesis kit; cat.
no. K1622; Thermo Fisher Scientific, Inc.). Target genes were
amplified in a 20 µl reaction volume using SYBR-Green qPCR Mix
(cat. no. AB1323A; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. The following conditions were
applied: 50°C for 2 min; 95°C for 2 min; followed by 40 cycles of
95°C for 15 sec and 60°C for 32 sec. The primer sequences used for
the RT-qPCR analyses are listed in Table II. The expression levels of
mRNA/lncRNA and miRNA were assessed using the 2−ΔΔCq
method (12), and experiments were
performed in triplicate.
 | Table II.Primer sequences used to analyze the
expression of various RNAs and miRNAs. |
Table II.
Primer sequences used to analyze the
expression of various RNAs and miRNAs.
| Gene symbol | Forward (5′-3′) | Reverse (5′-3′) |
|---|
| PVT1 |
GCCTTCCCTCCTTCTGGAAG |
GGTCCAGGTGGAGTCATG |
| CDK4 |
CCATCAGCACAGTTCGTGAGGT |
TCAGTTCGGGATGTGGCACAGA |
| GAPDH |
GTCTCCTCTGACTTCAACAGCG |
ACCACCCTGTTGCTGTAGCCAA |
| U6 |
CTCGCTTCGGCAGCACAT |
TTTGCGTGTCATCCTTGCG |
| miR-486-5p |
ACACTCCAGCTGGGTCCTGTAC |
CTCAACTGGTGTCGTGGA |
|
| TGAGCTGCCCCGAG |
|
Western blot analysis
HCT116 parental and 5-FU-resistant HCT116 cells were
lysed in lysis buffer (Beyotime Institute of Biotechnology)
containing a protease inhibitor cocktail (Bimake). Protein
concentration was determined using the bicinchoninic acid method.
Denatured proteins (20 µg) were then separated on a 1% gel via
SDS-PAGE, transferred to PVDF membrane (Millipore), and incubated
with a primary antibody at 4°C overnight, prior to washing and
incubation with an HRP-conjugated Affinipure Goat Anti-Rabbit
IgG(H+L) secondary antibody (1:5,000; SA00001-2; ProteinTech Group,
Inc.) at room temperature for 1 h. Finally, proteins were
visualized using an enhanced chemiluminescence reagent (ECL; Thermo
Fisher Scientific, Inc.). Gray value analysis of strips was
performed with ImageJ software (v.1.8.0; National Institutes of
Health). Anti-GAPDH antibody (1:2,000; cat. no. 10494-1-AP;
Proteintech Group, Inc.) was used as an internal reference. CDK4
was probed using anti-CDK4 antibody (1:1,000; cat. no. ab108357;
Abcam). All the experiments were performed in triplicate.
Cell Counting Kit-8 (CCK-8) assay
HCT116 parental and 5-FU-resistant HCT116 cells
(3×103 cells/well) were plated in 96-well plates.
Following a 72-h time period, 10 µl CCK-8 reagent (Beijing Solarbio
Science & Technology Co., Ltd.) was added to each well
containing cells, and the plates were incubated at 37°C for 60 min.
Proliferation was quantified at 450 nm using a Varioskan LUX
Multifunctional microplate reader (Thermo Fisher Scientific, Inc.).
All the experiments were performed in triplicate.
Clone formation assay
Cultured HCT116 parental and 5-FU-resistant HCT116
cells were digested with 0.25% trypsin (Gibco; Thermo Fisher
Scientific, Inc.) and pipetted to break up cell clusters. A total
number of 200 cells per dish were seeded in cell culture dishes
containing 10 ml of pre-warmed RPMI-1640 culture medium (Gibco;
Thermo Fisher Scientific, Inc.) and incubated at 4°C in 5%
CO2 for 2 weeks. The cells were then fixed for 15 min in
5 ml of 4% paraformaldehyde and an appropriate amount of 1X Giemsa
dye solution (G1015; Beijing Solarbio Science & Technology Co.,
Ltd.) was applied to the cells for 10–30 min at room temperature.
Dishes were air-dried, and the clones were counted directly with
the naked eye. Clone formation rate=(number of clones/number of
inoculated cells) ×100. Microscope photographs were acquired, and
cells were observed by using an optical microscope (MF52; Guangzhou
Micro-shot Technology Co., Ltd.). All the experiments were
performed in triplicate.
Cell cycle assays
Cells (1×106) were harvested in a flow
tube and washed with PBS. Pre-chilled 70–80% ethanol (5 ml) was
added in a dropwise manner to the cells and incubated in the dark
at 4°C overnight. The cells were washed twice to completely remove
the ethanol. Cells were incubated for 30 min in the dark at 37°C
with 0.5 ml PI/RNase staining solution (cat. no. C1052; Beyotime
Institute of Biotechnology). Samples were stored at 4°C in the
dark, detected using FACSAria (BD Biosciences) and analyzed with
FlowJo software (version 10.6.2; BD Biosciences). All experiments
were performed in triplicate.
Dual-luciferase assay
CDK4-WT (wild type), CDK4-MUT (mutant), PVT1-WT, and
PVT1-MUT sequences were synthesized and subcloned into the
psiCHECK2 vector (Synbio Technologies). Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to
co-transfect psiCHECK2 subcloning plasmid with miR-486-5p mimic or
mimic negative control into 293T cells. Following a 48-h incubation
at 37°C, Firefly and Renilla fluorescence values were
obtained using a dual-luciferase reporter assay (Promega
Corporation). Relative fluorescence values were Firefly
fluorescence value/Renilla fluorescence value ratios. The
miRNA mimic/mimic nc sequences used were as follows: miR-486-5p
mimics: 5′UCCUGUACUGAGCUGCCCCGAG3′, mimics nc:
5′GUCACUUGAUCCGAUGCGCCCG3′. Independent experiments were performed
in triplicate.
RNA immunoprecipitation (RIP)
assay
A total number of 1×107 5-FU-resistant
HCT116 cells were lysed with RIPA buffer (Beyotime Institute of
Biotechnology) and supplemented with a protease inhibitor cocktail
(Bimake); 10% of each 1 ml lysate was used as input. Protein A/G
magnetic beads (Millipore) were coupled with anti-AGO2 antibody
(1:50; cat. no. ab186733; Abcam) or normal rabbit IgG (negative
control) (1:50; cat. no. 2729; Cell Signaling Technology, Inc.) at
4°C for 4 h. Subsequently, these antibody-coupled magnetic beads
were incubated with the lysis mixtures for 6 h at 4°C, followed by
the elution of captured RNA (RNeasy Mini Kit; Qiagen) and RT-qPCR,
according to the instructions of the Magna RIP™ RNA-Binding Protein
Immunoprecipitation Kit (17–700; MilliporeSigma). All experiments
were performed in triplicate.
RNA pull-down assay
Sense strand probes and probes for mutated PVT1 were
synthesized in vitro and incubated with cell lysis complex
solution at 4°C for 4 h. Streptavidin magnetic beads were added and
incubated at 4°C for 1 h, RNA was extracted, and miR-486-5p was
detected by RT-qPCR. The RNA probe sequences are demonstrated in
Table III. All the experiments
were performed in triplicate.
 | Table III.RNA pull-down probe sequences. |
Table III.
RNA pull-down probe sequences.
| Name | Sequence
(5′-3′) |
|---|
| PVT1-WT |
CTCCGGGCAGAGCGCGTGTGGCGGCCGAGCACATGGGCCCGCGGGCCGGGCGGGCTCGGGG |
|
|
CGGCCGGGACGAGGAGGGGCGACGACGAGCTGCGAGCAAAGATGTGCCCCGGGACCCCCGG |
|
|
CACCTTCCAGTGGATTTCCTTGCGGAAAGGATGTTGGCGGTCCCTGTGACCTGTGGAGACACG |
|
|
GCCAGATCTGCCCTCCAGCCTGATCTTTTGGCCAGAAGGAGATTAAAAAGATGCCCCTCAAGA |
|
|
TGGCTGTGCCTGTCAGCTGCATGGAGCTTCGTTCAAGTATTTTCTGAGCCTGATGGATTTACAG |
|
|
TGATCTTCAGTGGTCTGGGGAATAACGCTGGTGGAACCATGCACTGGAATGACACACGCCCGG |
|
|
CACATTTCAGGATACTAAAAGTGGTTTTAAGGGAGGCTGTGGCTGAATGCCTCATGGATTCTTA |
|
|
CAGCTTGGATGTCCATGGGGGACGAAGGACTGCAGCTGGCTGAGAGGGTTGAGATCTCTGTT |
|
|
TACTTAGATCTCTGCCAACTTCCTTTGGGTCTCCCTATGGAATGTAAGACCCCGACTCTTCCTCT |
|
|
GGTGAAGCATCTGATGCACGTTCCATCCGGCGCTCAGCTGGGTGAGCTGACCATACTCCCTGG |
|
|
AGCCTTCTCCCGAGGTGCGCGGGTGACCTTGGCACATACAGCCATCATGATGGTACTTTAAGT |
|
|
GGAGGCTGAATCATCTCCCCTTTGAGCTGCTTGGCACGTGGCTCCCTTGGTGTTCCCCTTTTAC |
|
|
TGCCAGGACACTGAGATTTGGAGAGAGTCTCACTCTGTGGTCCAGGCTGAAGTACAGTGGCA |
|
|
TGATCCCAGGTCACTGCAACCCCCACCTCCCGGGTTCAAGTGATCCTCCTGCCTCAGCCTCCC |
|
|
GAGTAGCTGGTATTACAGGCGTGTGCCACAAAGCCTGGCTAAGTTTTGTATTTTTAGTAGAGAC |
|
|
GGGGTTTCACCATGTTGGCCAGGTTGGTCTCGAACTCCTGACCTCAAGTGATCCACTCACTTT |
|
|
GGCCTTTCAACGTGCTGGGATTACAGGCGAGAGTCACCGCACCCGGACGACTCTGACATTTTT |
|
|
GAAGAGTCCAGAATCCTGTTACACCTGGGATTTAGGCACTTTCAATCTGAAAAAATACATATCC |
|
|
TTTCAGCACTCTGGACGGACTTGAGAACTGTCCTTACGTGACCTAAAGCTGGAGTATTTTGAGA |
|
|
TTGGAGAATTAAGAGCCAGTCTTGGTGCTCTGTGTTCACCTGGTTCATCTGAGGAGCTGCATCT |
|
|
ACCCTGCCCATGCCATAGATCCTGCCCTGTTTGCTTCTCCTGTTGCTGCTAGTGGACATGAGAA |
|
|
GGACAGAATAACGGGCTCCCAGATTCACAAGCCCCACCAAGAGGATCACCCCAGGAACGCTT |
|
|
GGAGGCTGAGGAGTTCACTGAGGCTACTGCATCTTGAGACTCAGGATGAAGACCCAGCTTGGG |
|
|
GCTGTCAAAGAGGCCTGAAGAGGCAGAACACCCCAGAGGAGCCTGGGGCCACCACCCAGCA |
|
|
TCACTGTGGGAAAACGGCAGCAGGAAATGTCCTCTCGCCTGCGTGCTCCACCTCGGTCCACGC |
|
|
CTTCCCTCCTTCTGGAAGCCTTGCCTGACCACTGGCCTGCCCCTTCTATGGGAATCACTACTGA |
|
|
CCTTGCAGCTTATTATAGACTTATATGTTTTTTGCATGTCTGACACCCATGACTCCACCTGGACC |
|
|
TTATGGCTCCACCCAGAAGCAATTCAGCCCAACAGGAGGACAGCTTCAACCCATTACGATTTCA |
|
|
TCTCTGCCCCAACCACTCAGCAGCAAGCACCTGTTACCTGTCCACCCCCACCCCTTCCCCCAAA |
|
|
CTGCCTTTGAAAAATCCCTAACCTATGAGCTTTGAATAAGATGAGTACGAACTTCATCGCCCACG |
|
|
TGGCGTGGCCGGCCTCGTGTCTATTAAATTCTTTTTC
TACTAAAAAAAAAAAAAAAAAA |
| PVT1-WT |
CTCCGGGCAGAGCGCGTGTGGCGGCCGAGCACATGGGCCCGCGGGCCGGGCGGAGAGCCCCG |
|
|
TCGGTCCTGGTCCTGGGCGACGACGAGCTGCGAGCAAAGATGTGCCCCGGGACCCCCGGCAC |
|
|
CTTCCAGTGGATTTCCTTGCGGAAAGGATGTTGGCGGTCCCTGTGACCTGTGGAGACACGGCC |
|
|
AGATCTGCCCTCCAGCCTGATCTTTTGGCCAGAAGGAGATTAAAAAGATGCCCCTCAAGATGG |
|
|
CTGTGCCTGTCAGCTGCATGGAGCTTCGTTCAAGTATTTTCTGAGCCTGATGGATTTACAGTGA |
|
|
TCTTCAGTGGTCTGGGGAATAACGCTGGTGGAACCATGCACTGGAATGACACACGCCCGGCAC |
|
|
ATTTCAGGATACTAAAAGTGGTTTTAAGGGAGGCTGTGGCTGAATGCCTCATGGATTCTTACAGC |
|
|
TTGGATGTCCATGGGGGACGAAGGACTGCAGCTGGCTGAGAGGGTTGAGATCTCTGTTTACTT |
|
|
AGATCTCTGCCAACTTCCTTTGGGTCTCCCTATGGAATGTAAGACCCCGACTCTTCCTGGTGAA |
|
|
GCATCTGATGCACGTTCCATCCGGCGCTCAGCTGGGCTTGAGCTGACCATACTCCCTGGAGCCT |
|
| TCTCCCG
AGGTGCGCGGGTGACCTTGGCACATACAGCCATCATGATGGTACTTTAAGTGGAGGC |
|
|
TGAATCATCTCCCCTTTGAGCTGCTTGGCACGTGGCTCCCTTGGTGTTCCCCTTTTACTGCCAGG |
|
|
ACACTGAGATTTGGAGAGAGTCTCACTCTGTGGTCCAGGCTGAAGTACAGTGGCATGATCCCA |
|
|
GGTCACTGCAACCCCCACCTCCCGGGTTCAAGTGATCCTCCTGCCTCAGCCTCCCGAGTAGCT |
|
|
GGTATTACAGGCGTGTGCCACAAAGCCTGGCTAAGTTTTGTATTTTTAGTAGAGACGGGGTTTC |
|
|
ACCATGTTGGCCAGGTTGGTCTCGAACTCCTGACCTCAAGTGATCCACTCACTTTGGCCTTTCA |
|
|
ACGTGCTGGGATTACAGGCGAGAGTCACCGCACCCGGACGACTCTGACATTTTTGAAGAGTCC |
|
|
AGAATCCTGTTACACCTGGGATTTAGGCACTTTCAATCTGAAAAAATACATATCCTTTCAGCACT |
|
|
CTGGACGGACTTGAGAACTGTCCTTACGTGACCTAAAGCTGGAGTATTTTGAGATTGGAGAATT |
|
|
AAGAGCCAGTCTTGGTGCTCTGTGTTCACCTGGTTCATCTGAGGAGCTGCATCTACCCTGCCCA |
|
|
TGCCATAGATCCTGCCCTGTTTGCTTCTCCTGTTGCTGCTAGTGGACATGAGAAGGACAGAATAA |
|
|
CGGGCTCCCAGATTCACAAGCCCCACCAAGAGGATCACCCCAGGAACGCTTGGAGGCTGAGG |
|
|
AGTTCACTGAGGCTACTGCATCTTGAGACTCAGGATGAAGACCCAGCTTGGGGCTGTCAAAGA |
|
|
GGCCTGAAGAGGCAGAACACCCCAGAGGAGCCTGGGGCCACCACCCAGCATCACTGTGGGAA |
|
|
AACGGCAGCAGGAAATGTCCTCTCGCCTGCGTGCTCCACCTCGGTCCACGCCTTCCCTCCTTC |
|
|
TGGAAGCCTTGCCTGACCACTGGCCTGCCCCTTCTATGGGAATCACTACTGACCTTGCAGCTTA |
|
|
TTATAGACTTATATGTTTTTTGCATGTCTGACACCCATGACTCCACCTGGACCTTATGGCTCCAC |
|
|
CCAGAAGCAATTCAGCCCAACAGGAGGACAGCTTCAACCCATTACGATTTCATCTCTGCCCCAA |
|
|
CCACTCAGCAGCAAGCACCTGTTACCTGTCCACCCCCACCCCTTCCCCCAAACTGCCTTTGAA |
|
|
AAATCCCTAACCTATGAGCTTTGAATAAGATGAGTACGAACTTCATCGCCCACGTGGCGTGGCC |
|
|
GGCCTCGTGTCTATTAAATTCTTTTTCTACTAAAAAAAAAAAAAAAAAA |
Database analysis
IC50 values of all CRC cell lines against
5-FU were queried from the GDSC database (https://www.cancerrxgene.org/). For RNA screening, the
following was performed: The GSE100179 dataset contained data from
three CRC samples (GSM2674208, GSM2674209 and GSM2674210) and three
normal human tissue samples (GSM2674188, GSM2674189 and
GSM2674190). The online tool GEO2R (ncbi.nlm.nih.gov/geo/geo2r) was
used to recognize anomalously expressed lncRNA/mRNA that yielded a
log2 |fold change (FC)| >2 with a P-value <0.05.
Volcano plots was drawn to represent differentially expressed
lncRNA/mRNAs. miRNA data were derived from the GSE98406 dataset,
including three CRC samples (GSM2593633, GSM2593634 and GSM2593635)
and three normal human samples (GSM2593614, GSM2593615 and
GSM2593616). lncRNAdisease2 (http://www.rnanut.net/lncrnadisease/) was used for the
screening of lncRNAs related to colon cancer. BiBiServ2 (https://bibiserv.cebitec.uni-bielefeld.de/sa),
TargetScan (http://www.targetscan.org/) and miRDB (http://www.mirdb.org/) were used to analyze genes that
potentially interact with miR-486-5p, PVT1, miR-486-5p and
CDK4.
Statistical analyses
All data were analyzed using GraphPad Prism (version
8; GraphPad Software, Inc.). Data are expressed as the mean ±
standard deviation. Data comparing two groups were analyzed using
the unpaired Student's t-test, while data comparing multiple groups
were analyzed by using one-way ANOVA, followed by the Bonferroni
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
PVT1 is highly expressed in
drug-resistant colon cancer cells
5-FU is a chemotherapeutic drug commonly used for
the treatment of colon cancer; however, drug resistance often poses
a challenge in clinical settings. In the present study, to
elucidate the mechanisms through which cells acquire resistance to
5-FU, the GEO and lncRNADisease2 databases were first used to
screen for lncRNAs closely connected to the emergence and
metastasis of colon cancer, including urothelial
carcinoma-associated 1 (UCA1) and PVT1 (Fig. 1A and B). Among these, UCA1 has been
revealed to be associated with colon cancer 5-FU resistance in
previous studies (13,14). Thus, it was hypothesized that PVT1
may also be associated with 5-FU resistance in colon cancer.
Firstly, according to the GDSC database query results, HCT116 colon
cancer cells were highly sensitive to 5-FU (Fig. 1C). A 5-FU-resistant strain of
HCT116 cells was then established and a CCK-8 assay was used to
determine the IC50 values of parental and 5-FU-resistant
HCT116 cells. The IC50 value of the 5-FU-resistant
HCT116 cells was significantly higher than that of the parental
cells (Fig. 1D). PVT1 expression
was be elevated in the 5-FU-resistant HCT116 cells, as revealed
using RT-qPCR (Fig. 1E). Thus, it
was hypothesized that PVT1 may have contributed to the resistance
of HCT116 cells to 5-FU; subsequently, nuclear and cytoplasmic
separation was used in HCT116 and HCT116-5FU-resistant cells for
the detection of the distribution of PVT1 in the nucleus and
cytoplasm. The results demonstrated that PVT1 was evenly
distributed in the nucleus and cytoplasm of HCT116 parental cells.
However, it was more greatly distributed in the cytoplasm of
HCT116-5FU-resistant cells (Fig. 1F
and G). Thus, it was suspected that 5FU resistance may be
attributed mainly due to the cytoplasmic role of PVT1.
PVT1 knockdown inhibits 5-FU
resistance in colon cancer cells
To examine the effects of PVT1 expression on 5-FU
resistance in colon cancer, PVT1 was knocked down in 5-FU-resistant
HCT116 cells. Firstly, cells were transfected with three PVT1
siRNAs and the knockdown efficiency of each siRNA was detected
using RT-qPCR. Since siRNA-2 produced the most intense knockdown
effect (Fig. 2A), it was selected
for use in follow-up experiments. Subsequently, the
HCT116-5FU-resistant cells were treated with 1 mM 5-FU. CCK-8 assay
demonstrated that PVT1 knockdown markedly reduced the
IC50 value of 5-FU-resistant HCT116 cells (Fig. 2B) and reduced the proliferation of
HCT116-FU-resistant cells treated with or without 5-FU (Fig. 2C). Cell cycle analyses revealed
that PVT1 knockdown inhibited the cell cycle progression of
5-FU-resistant HCT116 cells treated with or without 5-FU (Fig. 2D). Clone formation assays
demonstrated that PVT1 knockdown reduced the number of clones
formed by 5-FU-resistant HCT116 cells treated with or without 5-FU
(Fig. 2E). It was thus concluded
that PVT1 knockdown notably reduced 5-FU resistance in colon cancer
cells.
PVT1 absorbs miR-486-5p to regulate
CDK4 expression
Subsequently, the mechanisms through which PVT1
confers drug resistance in cancer cells were investigated. An
established lncRNA mechanism involves the regulation of gene
expression by adsorbing miRNA (15). Therefore, miRNAs that were
downregulated in colon cancer using the GEO database were analyzed
first. The volcano plot shows the differential expression of the
whole transcriptome, where green indicates downregulated miRNAs and
red indicates upregulated miRNAs. Among these, as compared with
normal tissues, the downregulated expression of miR-486-5p has the
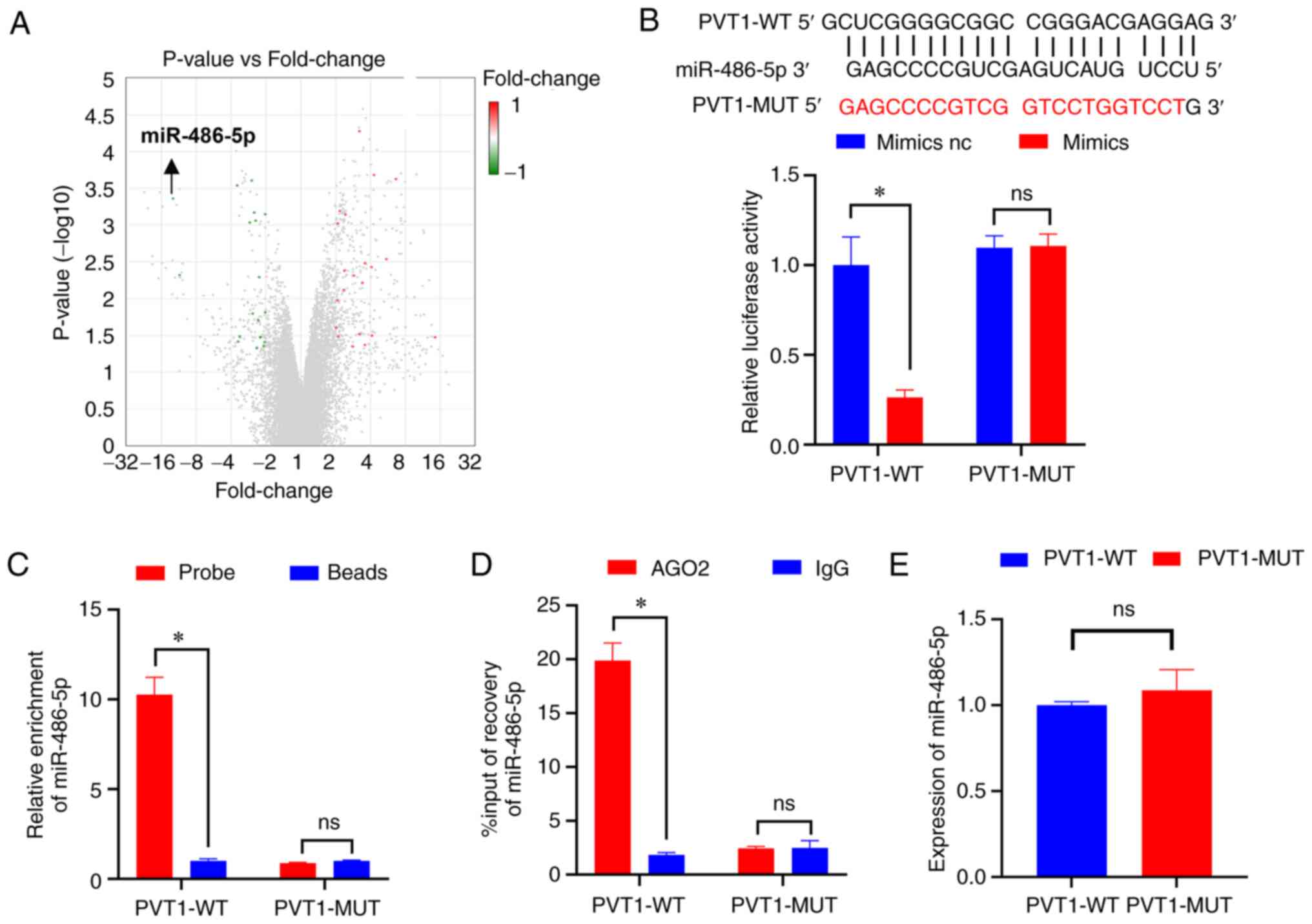
largest fold-change in colon cancer (Fig. 3A). Subsequently, BiBiServ2 software
analysis indicated that PVT1 strongly interacted with miR-486-5p.
Thus, dual-luciferase experiments were used to confirm miR-486-5p
binding to PVT1 (Fig. 3B).
Moreover, RNA pull-down and RIP assays were used to verify the
binding of miR-486-5p to PVT1 in HCT116-5FU-resistant cells
(Fig. 3C and D) and it was found
that PVT1 could adsorb miR-486-5p. Additionally, the results of
RT-qPCR demonstrated that PVT1 failed to affect the expression of
miR-486-5p (Fig. 3E). Since miRNAs
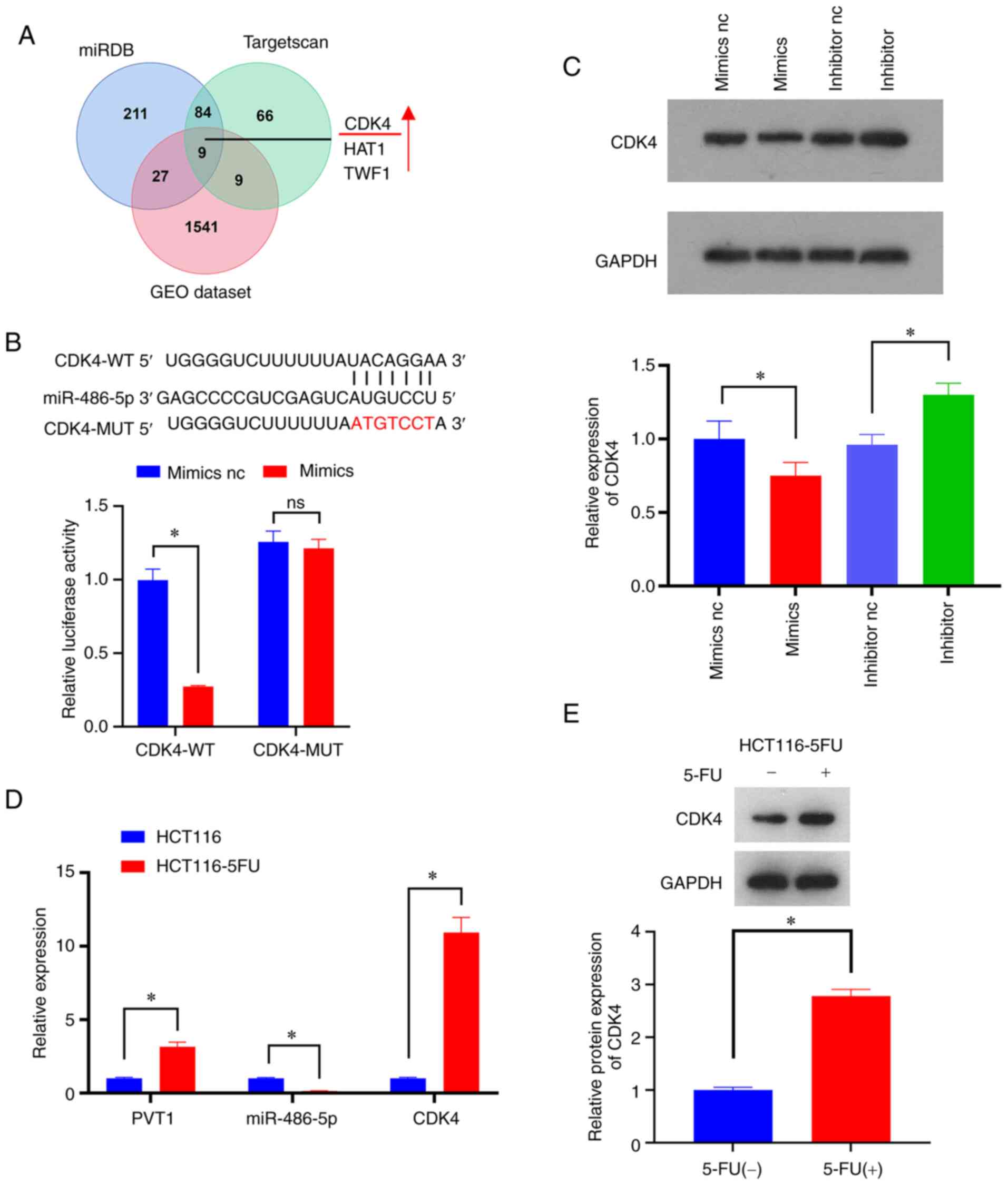
mainly act on the 3′UTR of their target functional gene mRNA, nine
possible miR-486-5p interacting mRNAs were obtained by using the
combined analysis of TargetScan, miRDB and RNA-chip differential
genes in the GEO database. Three of the upregulated miR-486-5p
targeted gene candidates were CDK4, histone acetyltransferase 1
(HAT1) and twinfilin actin binding protein 1 (TWF1) (Fig. 4A). CDK4 was selected as an
miR-486-5p target gene, which is closely related to the
proliferation cycle. Dual luciferase analysis verified that
miR-486-5p binds to the 3′UTR region of CDK4 (Fig. 4B). The results of western blot
analysis revealed that miR-486-5p suppressed CDK4 expression
(Fig. 4C). RT-qPCR was also used
for the detection of PVT1, miR-486-5p and CDK4 expression in
drug-resistant and non-drug-resistant cells (Fig. 4D). The results demonstrated that
the PVT1 and miR-486-5p expression levels were inversely
associated, which also applied for CDK4 expression. Furthermore,
western blot analysis revealed that 5-FU enhanced the expression of
CDK4 in HCT116-5FU-resistant cells (Fig. 4E). In summary, PVT1 may promote
colon cancer cell resistance to 5-FU via the miR-486-5p/CDK4
axis.
PVT1 regulates the resistance of colon
cancer cells to 5-FU via the miR-486-5p/CDK4 axis
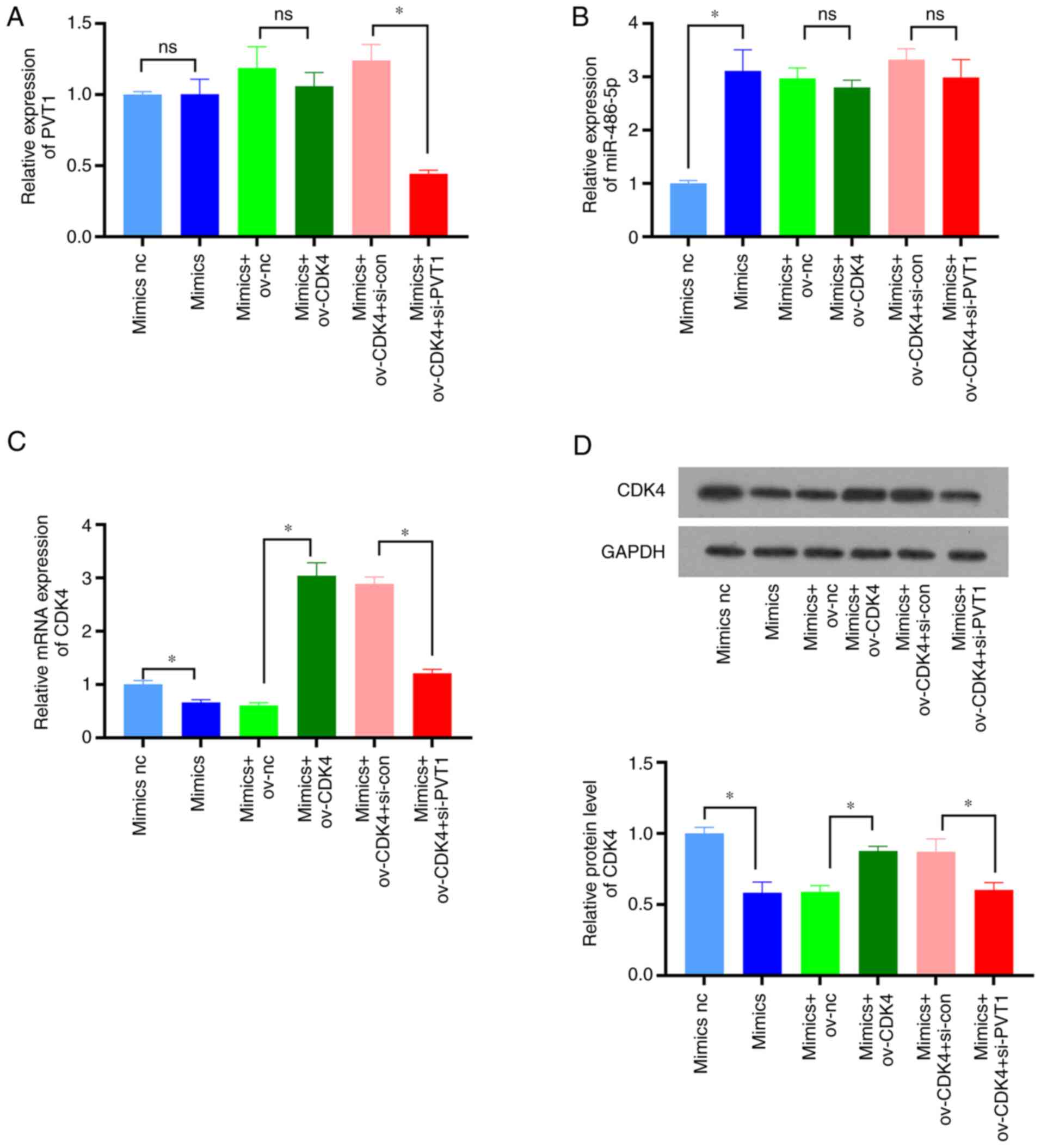
To further examine the effects of PVT1 on the
resistance of colon cancer cells to 5-FU through the modulation of
the miR-486-5p/CDK4 axis, and following the overexpression of
miR-486-5p, CDK4 was also overexpressed in order to observe the
regulatory effects of CDK4 and miR-286-5p on HCT116-5FU-resistant
cells. Subsequently, PVT1 was downregulated and the effect of the
PVT1, miR-286-5p and CDK4 interaction on the proliferation of
HCT116-5FU drug-resistant cells was observed. Firstly, by using
RT-qPCR and western blot analysis, PVT1, miR-486-5p and CDK4
expression was detected in each cell type. The results demonstrated
that miR-486-5p overexpression (Fig.
5B) hardly affected PVT1 expression (Fig. 5A), although it inhibited CDK4
expression (Fig. 5C and D).
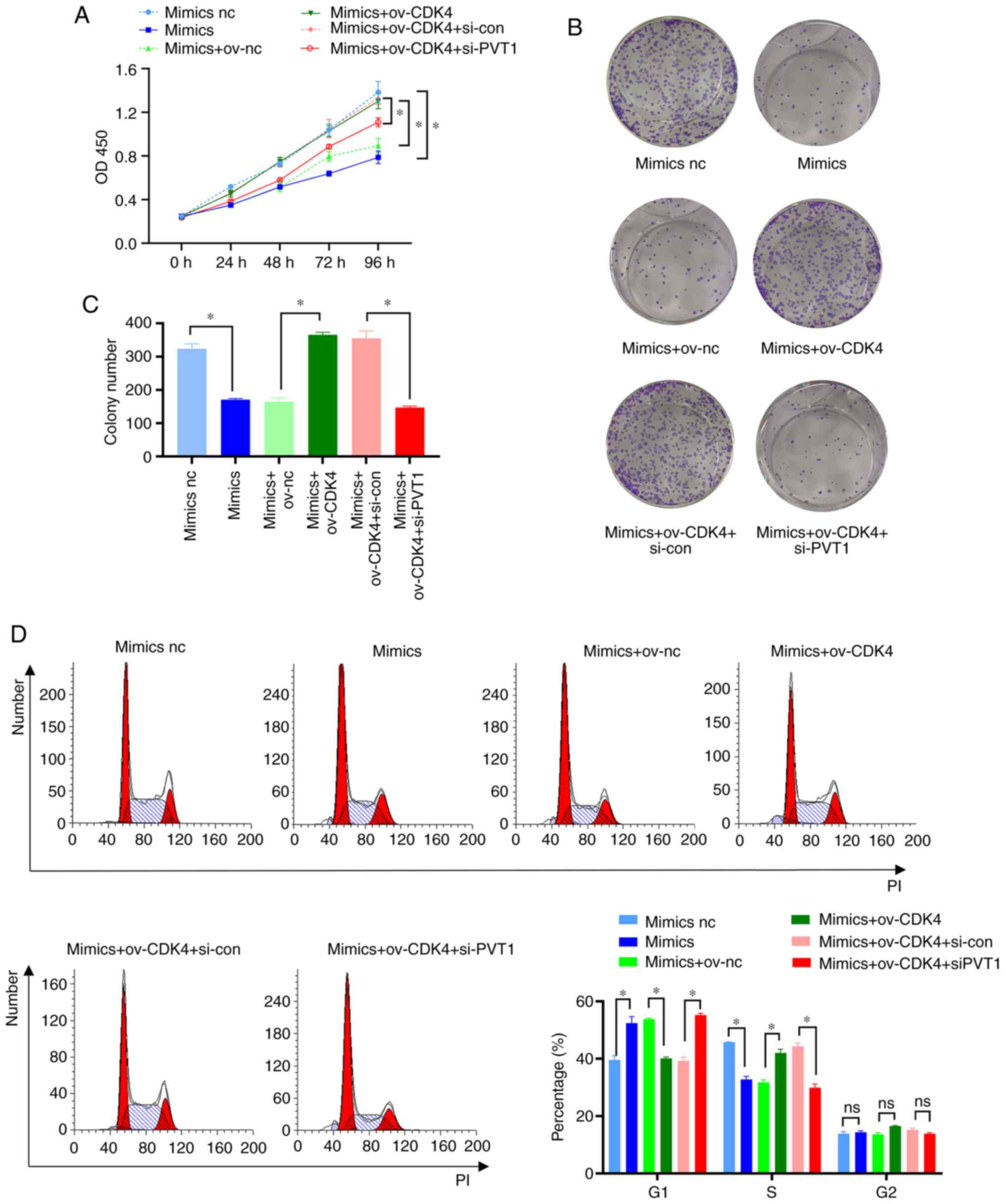
Additionally, PVT1 knockdown decreased CDK4 expression (Fig. 5C and D). CCK-8 and clone formation
assays demonstrated that CDK4 overexpression attenuated the
inhibitory effects of miR-486-5p on the proliferation of
HCT116-5FU-resistant cells, and PVT1 knockdown restored the
suppressive effects of miR-486-5p on the proliferation of
HCT116-5FU-resistant cells (Fig.
6A-C). Consistent with previous experimental results, flow
cytometry revealed that CDK4 overexpression attenuated the
inhibitory effects of miR-486-5p on the cell cycle progression of
HCT116-5FU-resistant cells, and PVT1 knockdown restored the
miR-486-5p-mediated sensitivity of HCT116-5FU cells (Fig. 6D). In summary, PVT1 may upregulate
the resistance of colon cancer cells to 5-FU through the
miR-486-5p/CDK4 axis.
Discussion
Due to an increase in the consumption of refined
foods and limited exercise, CRC has become the third most common
type of cancer worldwide, with ~1.8 million new cases diagnosed
each year (1). When a tumor
produces local or remote metastases, 5-FU is often used as a
chemotherapeutic agent (16).
However, patients often develop drug resistance, leading to tumor
recurrence (17). Therefore, the
further understanding of the mechanisms responsible for the
resistance of colon cancer to 5-FU is of utmost urgency.
In the present study, differentially expressed genes
in colon cancer were first identified using the GEO database and
the lncRNADisease2 database was then utilized to identify lncRNAs
associated with colon cancer (UCA1 and PVT1). The mechanism by
which UCA1 confers resistance op 5-FU in CRC has been previously
investigated. UCA1 has been reported to increase the resistance of
colon cancer cells to 5-FU by suppressing miR-204-5p (14). UCA1 has also been suggested to
promote the resistance of colon cancer to 5-FU via the
miR-23b-3p/ZNF281 axis (13). Fan
et al (11) demonstrated
that knocking down PVT1 also inhibited the resistance of CRC to
5-FU and at the same time, the expression of PVT1 was significantly
upregulated in 5-FU-resistant patients, compared to that in
5-FU-sensitive patients. However, the mechanisms by which PVT1
affects the resistance of colon cancer to 5-FU have not been
investigated, at least to the best of our knowledge. Therefore, the
present study examined the mechanisms through which PVT1 confers
resistance to 5-FU in CRC.
lncRNAs often act as endogenous competitive RNAs in
cells by sponging miRNAs to control target genes performing
biological functions (18). Thus,
differentially regulated miRNAs in colon cancer were analyzed using
the GEO database and miRNA-miR-486-5p was selected, which had the
largest fold-change among all downregulated miRNAs. Bibiserve2
software predicted and it was also experimentally verified that
PVT1 bound to miR-486-5p that can inhibit the proliferation and
cell cycle progression of HCT116-5FU-resistant cells. In line with
the present study, Zhang et al (19) revealed that miR-486-5p inhibited
colorectal cancer metastasis. Liu et al (20) demonstrated that lnc-NEAT1 may
adsorb miR-486-5p, activating the NR4A1/Wnt/β-catenin signaling
pathway, thereby promoting colorectal cancer proliferation.
Subsequently, the differential expression analysis of the GEO
database for the identification of miR-486-5p target genes was
performed and miRDB and TargetScan were screened. The results
demonstrated that PVT1 may promote the drug resistance and cell
cycle progression of colon cancer cells. Thus, a screening for cell
cycle-related differentially expressed genes was performed and
CDK4, a cyclin-dependent kinase that can combine with cyclin D to
form a heterodimer that phosphorylates and inactivates
retinoblastoma protein to drive the transition from G1 to S phase
was identified (21). It was
experimentally verified that miR-486-5p may regulate CDK4
expression and that CDK4-knockdown restored the inhibitory effects
of miR-486-5p on the proliferation and cell cycle progression of
HCT116-5FU cells. PVT1 inhibited CDK4 expression and inhibited
proliferation and cell cycle progression in HCT116-5FU cells. In
summary, it was observed that PVT1 promoted drug resistance in
colon cancer cells by upregulating the miR-486-5p target, CDK4.
A variety of CDK4/6 inhibitors have been used in
clinical treatment, among which palbociclib and ribociclib have
been approved for the clinical treatment of breast cancer. These
CDK4/6 inhibitors are small molecule chemical therapeutics that
mainly bind to CDK4/6 to inhibit cell cycle and proliferation.
These inhibitors have been reported to easily induce drug
resistance in cancer cells. However, PVT1 decreased the expression
of CDK4/6 via miR-486-5p, thus indicating that PVT1 may regulate
CDK4 mRNA expression. Therefore, PVT1 can still reduce the
expression of CDK4 when CDK4 resistance to inhibitors develops, and
PVT1 can be used as an adjuvant for CDK4 inhibitors.
The present study did not include in vivo
validation experiments, in order to further verify the association
between PVT1 and colon cancer drug resistance, and to clarify
further the mechanisms by which PVT1 affects colon cancer drug
resistance. Nevertheless, the present findings strongly support the
potential application of targeting PVT1 to counteract 5-FU
resistance in colon cancer in clinical practice.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZH and ZL conceived and designed the study. ZL, RC,
SH and XH performed the experiments. All authors have read and
approved the final manuscript. ZH and ZL confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
PVT1
|
plasmacytoma variant translocation
1
|
|
5-FU
|
5-fluorouracil
|
|
CDK4
|
cyclin dependent kinase 4
|
|
GEO
|
Gene Expression Omnibus
|
|
siRNA
|
small interfering RNA
|
|
ov
|
overexpression
|
|
NC
|
negative control
|
|
OD
|
optical density
|
References
|
1
|
Brody H: Colorectal cancer. Nature.
521:S12015. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vodenkova S, Buchler T, Cervena K,
Veskrnova V, Vodicka P and Vymetalkova V: 5-fluorouracil and other
fluoropyrimidines in colorectal cancer: Past, present and future.
Pharmacol Ther. 206:1074472020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou DD, Liu XF, Lu CW, Pant OP and Liu
XD: Long non-coding RNA PVT1: Emerging biomarker in digestive
system cancer. Cell Prolif. 50:e123982017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cory S, Graham M, Webb E, Corcoran L and
Adams JM: Variant (6;15) translocations in murine plasmacytomas
involve a chromosome 15 locus at least 72 kb from the c-myc
oncogene. EMBO J. 4:675–681. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng X, Hu H and Li S: High expression of
lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal
transition in esophageal cancer. Oncol Lett. 12:2357–2362. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W and Sun SH: Oncofetal long
noncoding RNA PVT1 promotes proliferation and stem cell-like
property of hepatocellular carcinoma cells by stabilizing NOP2.
Hepatology. 60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Br J Cancer.
110:164–171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang XW, Bu P, Liu L, Zhang XZ and Li J:
Overexpression of long non-coding RNA PVT1 in gastric cancer cells
promotes the development of multidrug resistance. Biochem Biophys
Res Commun. 462:227–232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X,
Zhang X, Huang Y, Zhang R, Wei J, et al: LncRNA PVT1 promotes
gemcitabine resistance of pancreatic cancer via activating
Wnt/β-catenin and autophagy pathway through modulating the
miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 19:1182020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Du P, Hu C, Qin Y, Zhao J, Patel R, Fu Y,
Zhu M, Zhang W and Huang G: LncRNA PVT1 mediates antiapoptosis and
5-fluorouracil resistance via increasing Bcl2 expression in gastric
cancer. J Oncol. 2019:93254072019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan H, Zhu JH and Yao XQ: Knockdown of
long noncoding RNA PVT1 reverses multidrug resistance in colorectal
cancer cells. Mol Med Rep. 17:8309–8315. 2018.PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xian Z, Hu B, Wang T, Zeng J, Cai J, Zou Q
and Zhu P: lncRNA UCA1 contributes to 5-fluorouracil resistance of
colorectal cancer cells through miR-23b-3p/ZNF281 axis. Onco
Targets Ther. 13:7571–7583. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng J, Lei W, Fu JC, Zhang L, Li JH and
Xiong JP: Targeting miR-21 enhances the sensitivity of human colon
cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys
Res Commun. 443:789–795. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nishida N, Yamashita S, Mimori K, Sudo T,
Tanaka F, Shibata K, Yamamoto H, Ishii H, Doki Y and Mori M:
MicroRNA-10b is a prognostic indicator in colorectal cancer and
confers resistance to the chemotherapeutic agent 5-fluorouracil in
colorectal cancer cells. Ann Surg Oncol. 19:3065–3071. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kartha RV and Subramanian S: Competing
endogenous RNAs (ceRNAs): New entrants to the intricacies of gene
regulation. Front Genet. 5:82014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Fu J, Zhang Z and Qin H:
miR-486-5p regulates the migration and invasion of colorectal
cancer cells through targeting PIK3R1. Oncol Lett. 15:7243–7248.
2018.PubMed/NCBI
|
|
20
|
Liu Z, Gu Y, Cheng X, Jiang H, Huang Y,
Zhang Y, Yu G, Cheng Y and Zhou L: Upregulation lnc-NEAT1
contributes to colorectal cancer progression through sponging
miR-486-5p and activating NR4A1/Wnt/beta-catenin pathway. Cancer
Biomark. 30:309–319. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao X, Leone GW and Wang H: Cyclin
D-CDK4/6 functions in cancer. Adv Cancer Res. 148:147–169. 2020.
View Article : Google Scholar : PubMed/NCBI
|