Introduction
According to an analysis based on cancer estimates
from GLOBOCAN 2020 and population estimates from the United
Nations, in 2022 there will be ~4,820,000 and 2,370,000 new cases
of cancer, and 3,210,000 and 640,000 cancer-associated deaths in
China and the USA, respectively (1). The most common cancer in China is
lung cancer, while in the USA, breast cancer is most common.
However, in both China and the USA, lung cancer is the leading
cause of cancer-associated mortality (1). The rapid progress of molecular
genetics has brought the treatment of tumors into a new era.
Non-small cell lung cancer (NSCLC) is characterized by the
accumulation of changes in multiple genotypes and includes
different histological subtypes. These changes in genotypes can be
used as effective therapeutic targets, and a series of promising
molecular inhibitors have been developed with potential in the
treatment of NSCLC.
The clinical methods for the treatment of tumors are
diverse, and require selection according to the clinical stage and
pathological type. It is generally considered that patients with
primary lung cancer can benefit from early surgery. However, there
is great debate about the clinical treatment of stage III lung
cancer. The American Society of Clinical Oncology guidelines
(2) recommend that for patients
with suspected or confirmed NSCLC, a multidisciplinary discussion
should be conducted before the initiation of treatment planning,
and that patients with epithelial growth factor receptor (EGFR) 19
exon deletions or exon 21 L858R mutations may be offered adjuvant
osimertinib after platinum-based chemotherapy. For the treatment of
advanced NSCLC with EGFR-sensitive mutations, EGFR-targeted drug
therapy is currently predominant. In the past, patients with NSCLC
with other target mutations had no relevant targeted drug options
and were required to undergo treatment with conventional
chemotherapy regimens. In recent years, with the development of
molecular medicine, targeted drugs have become available for
non-EGFR mutations, including anaplastic lymphoma kinase (3), c-ros oncogene 1 (4) and MET proto-oncogene (5) mutations. To date, three generations
of EGFR inhibitors have been approved by the US Food and Drug
Administration for the treatment of patients with NSCLC with
EGFR-activating mutations or secondary T790M mutations. Among them,
the third-generation EGFR inhibitor osimertinib has attracted
considerable attention. It has been reported that although patients
with NSCLC with EGFR T790M mutations exhibit a positive response
and prolonged survival time after treatment with osimertinib,
acquired resistance also occurs after ~10 months (6). Mechanisms of third-generation EGFR
inhibitor resistance have been widely explored; the C797S mutation
is considered representative, and targeted drugs are under
development (7). Considering the
evolutionary properties of organisms, the problem of targeted drug
resistance is inevitable. The continuous genetic exploration of
drug resistance mechanisms and preparation of the next generation
of targeted drugs is likely to be increasingly complex. Therefore,
the environment on which tumor survival depends, known as the tumor
microenvironment (TM), has become an alternative target of
interest.
The TM is the internal environment that the tumor
tissue creates and relies upon for survival and development, and
has become a popular topic of tumor research. Studies have
demonstrated that the TM can not only promote immune escape but
also induce resistance to tumor formation (8–10).
Macrophages are highly heterogeneous cells that exhibit unique
phenotypes and functions in complex microenvironments within the
body. Mantovani et al (11)
suggested that macrophages exist in a series of continuous
functional states, the extremes of which are type M1 and M2
macrophages. Type M1 macrophages participate in a positive immune
response via the production of inflammatory cytokines and
chemokines, and the presentation of antigens, while type M2
macrophages have weak antigen-presentation ability and serve an
important role in immune regulation through the secretion of the
inhibitory cytokines IL-10 or TGF-β. Macrophages in TM are derived
from immature monocytes in the blood and formed by the
microenvironment itself, where they exhibit the role and phenotype
of type M2 macrophages and are known as tumor-associated
macrophages (TAMs). TAMs are the major type of inflammatory cells
in the tumor matrix, representing 30–50% of all inflammatory cells
(12). In clinical studies, it was
shown that the degree of infiltration of M2 macrophages in NSCLC
tumors was positively associated with the progression of the tumor
and negatively associated with the therapeutic response to
EGFR-tyrosine kinase inhibitors (TKIs) (13,14).
Our previous study demonstrated that TAMs induced A549 lung
adenocarcinoma cells to exhibit stronger proliferation, invasion
and migration capabilities via upregulation of the AKT signaling
pathway (15). However, the study
failed to clarify the method of communication between TAMs and lung
adenocarcinoma cells. Wendler et al (16) hypothesized that matrix cells,
fibroblasts, endothelial cells and immune cells in the TM
communicate through interaction with extracellular vesicles (EVs)
to jointly promote the development of tumor resistance. Exosomes
are deemed to be a type of EV, with a membrane structure that
mostly originates from the endometrial system and a diameter of
40–100 nm. TAMs are considered to participate in the promotion of
angiogenesis of damaged tissue, cell proliferation and immune
regulation (17–19). Therefore, we hypothesize that TAMs
transmit proteins, factors or genetic substances through an exosome
bridge, thereby affecting the sensitivity of NSCLC to EGFR-TKI
drugs. However, this has not yet been confirmed by relevant
research. The aim of the present study was to explore the influence
of TAM-derived exosomes on the sensitivity of PC9 and HCC827 lung
adenocarcinoma cells to the primary targeted drug gefitinib. The
findings of the study may provide innovative ideas for delaying or
blocking the process by which resistance develops.
Materials and methods
Cell culture
The THP-1 human monocytic leukemia cell line (cat.
no. CL-0233) was purchased from Procell Life Science &
Technology Co., Ltd. The cells were cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 0.05 mM
β-mercaptoethanol (Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (P/S; Thermo Fisher Scientific, Inc.) (100
U/ml) then incubated in a 5% CO2 incubator at 37°C. The
PC9 (cat. no. SCSP-5085) and HCC827 (cat. no. SCSP-538) human lung
adenocarcinoma cell lines were purchased from The Cell Bank of the
Type Culture Collection of the Chinese Academy of Sciences. The
cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc.) or RPMI-1640 medium supplemented
with 10% FBS and 1% P/S (100 U/ml) and then incubated in a 5%
CO2 incubator at 37°C.
Induced differentiation of THP-1
cells
THP-1 cells were treated with 100 nM phorbol
12-myristate 13-acetate (PMA; cat. no. HY-D1056) for 24 h incubated
in a 5% CO2 incubator at 37°C, until they became
adherent, indicating that the M0 phenotype had been induced. Next,
using 20 ng/ml IL-4 (cat. no. HY-P70750) and 20 ng/ml IL-13 (cat.
no. HY-P7033) to treat the M0 macrophages, M2 macrophages were
obtained within 24 h after incubation in a 5% CO2
incubator at 37°C. The control group was treated with the same
amount of phosphate buffered saline (PBS) at the same time point.
The PMA, IL-4 and IL-13 were purchased from MedChemExpress.
Flow cytometric analysis of cellular
immunophenotype and annexin V/PI staining
Cellular immunophenotyping was performed to
determine the phenotypic characteristics of the cells after
treatment. The processed and untreated THP-1 cells were digested
with trypsin to prepare a single-cell suspension and counted to
1×106 cells/ml. Next, 5.0 µl CoraLite®488
anti-human CD163 (cat. no. CL488-65169; ProteinTech Group, Inc.)
was added to 100 µl cell suspension, incubated at 2–8°C for 30 min
and washed with PBS. Finally, the cells were examined by flow
cytometry (BD Accuri™ C6 Plus; BD Biosciences) and the data were
processed using BD FACSDiva™ 6.1 software (BD Biosciences).
Flow cytometric analysis was also performed to
detect the apoptosis characteristics of treated and untreated THP-1
cells. The cells in 6-well plates (1×106 cells/well)
were harvested after 24 h, resuspended in PBS and then washed with
PBS. Cells were stained with Annexin V (FITC)/PI (cat. no. APOAF;
MilliporeSigma) according to the manufacturer's protocol. The
apoptosis data were processed by flow cytometry using BD Accuri™ C6
Plus and analyzed with BD FACSDiva 6.1 software.
Extraction and identification of
exosomes
The cell supernatant of the M2 macrophages was
filtered using a microporous membrane (Meridian Bioscience, Inc.;
pore size, 0.22 µm) and then concentrated through an Amiconultra-15
ultrafiltration tube (MilliporeSigma). After ultracentrifugation
(120,000 × g, 2 h, 4°C) the supernatant was discarded and 10 ml
precooled PBS was added for resuspension and further
ultracentrifugation (120,000 × g, 2 h, 4°C). Next, the supernatant
was discarded, 200 µl PBS was added for resuspension and the
samples were sub-packed and frozen at −80°C until required. The
exosome protein content was measured by using a Pierce™
BCA protein assay kit (cat. no. A53226; Thermo Fisher Scientific,
Inc.). To identify the exosomes, the particle sizes were examined
by nanoparticle tracking analysis (NTA; ZetaView PMX 110; Particle
Metrix) and the samples were characterized by transmission electron
microscopy (HT-7700; Hitachi, Ltd.). The steps are as follows:
Freshly separated exosomes (10 µl) were absorbed and dropped onto a
copper grid with carbon-coated mesh for precipitation for 1 min.
The surface water was removed with filter paper. Next, 10 µl 2 wt%
aqueous uranyl acetate solution was used for positive staining at
room temperature for 1 min. The surface water was removed with
filter paper. Transmission electron microscopy was used after
natural air drying for 10 min at room temperature to obtain images,
operating at an acceleration voltage of 100 keV. In addition, the
expression levels of the exosomal marker proteins CD9, CD63 and
tumor susceptibility gene 101 protein (TSG101) were determined
using western blotting, as described in the western blot analysis
section.
Cell Counting Kit (CCK-8) assay
Different cells have different sensitivity to drugs.
HCC827 and PC9 are EGFR-sensitive mutant cell lines, which are
expected to be highly sensitive to gefitinib. A CCK-8 assay was
used to detect the IC50 of gefitinib in the two cell
lines. A counted cell suspension of 1×105 cells/ml was
evenly plated into a 96-well plate, and when the cell confluence
reached ~60%, 0.01, 0.03, 0.05, 0.07, 0.09, 0.11, 0.13, 0.15 and
0.17 µM gefitinib (cat. no. ZD1839; MedChemExpress) was added.
After 22 h of incubation at 37°C, 10 µl CCK-8 reagent (cat. no.
CA1210; Beijing Solarbio Science & Technology Co., Ltd.) was
added to each well for another 2 h. The absorbance at 450 nm was
determined using a microplate reader (Thermo Fisher Scientific,
Inc.). Finally, the IC50 of gefitinib against HCC827 and
PC9 cells was calculated.
Western blot analysis
Western blotting was used for the detection of the
exosomal specific marker proteins CD9, CD63 and TSG101. The protein
concentrations of the THP-1 and M2 cell supernatant exosomal
protein samples were determined by the bicinchoninic acid (BCA)
method. Approximately 50 µg protein/lane from each sample was
separated on 10% gels using SDS-PAGE electrophoresis with a 100
volt constant voltage and transferred to a PVDF membrane (GE
Healthcare). Next, 5% skimmed milk powder was added to block the
membrane at room temperature for 1 h. The primary antibodies,
namely mouse anti-CD63 (cat. no. sc-5275; 1:500; Santa Cruz
Biotechnology, Inc.), mouse anti-CD9 (cat. no. sc-13118; 1:500;
Santa Cruz Biotechnology, Inc.), mouse anti-TSG101 (cat. no.
sc-7964; 1:500; Santa Cruz Biotechnology, Inc.) and mouse
anti-β-actin (cat. no. ab8226; 1:10,000; Abcam), were added to the
membrane and incubated overnight at 4°C. The membrane was washed
with TBS with 0.1% Tween 20 (TTBS) 3 times (5 min each time), and
then goat anti-mouse IgG-horseradish peroxidase (HRP) secondary
antibody (cat. no. 32430; 1:5,000; Thermo Fisher Scientific, Inc.)
was added for incubation at room temperature for 1 h. The film was
washed 4 times with TTBS (5 min each time), SuperSignal™ West Dura
Extended Duration Substrate (cat. no. 34076; Thermo Fisher
Scientific, Inc.) was added dropwise, and the X-ray film was
exposed in the dark for development.
The possible mechanism by which M2-derived exosomes
influence the sensitivity of HCC827 and PC9 cells to gefitinib was
explored. Key proteins in the AKT/ERK1/2/STAT3 signaling pathway
were investigated. Gefitinib (IC50 concentration) and
M2-derived exosomes (100 µl, 100 µg/ml) were utilized to treat
HCC827 and PC9 cells separately and in combination for 24 h
incubated in a 5% CO2 incubator at 37°C. Total protein
was extracted from each group of cells and BCA was used for
quantification. Protein samples (30 µg/lane) were separated on 10%
gels using SDS-PAGE and transferred to PVDF membranes at 4°C for 90
min. After blocking with 5% non-fat milk at room temperature for 60
min, membranes were incubated with the following primary antibodies
overnight at 4°C: Mouse anti-phosphorylated (p)-AKT (cat. no.
23430), mouse anti-AKT (cat. no. 2920), mouse anti-p-ERK (cat. no.
9106), mouse anti-ERK (cat. no. 4696), mouse anti-p-STAT3 (cat. no.
4113), mouse anti-STAT3 (cat. no. 9139) (all 1:1,000; Cell
Signaling Technology, Inc.) and rabbit anti-GAPDH (cat. no.
ab181602; 1:10,000; Abcam). The membranes were then probed with IgG
HRP-conjugated goat anti-rabbit (cat. no. 32733) and goat
anti-mouse (both 1:5,000; Thermo Fisher Scientific, Inc.) secondary
antibodies for 1 h at room temperature. Finally, the proteins were
detected using SuperSignal West Dura Extended Duration Substrate.
Densitometric analysis was performed using Image J 1.44p software
(National Institutes of Health).
Statistical analysis
GraphPad Prism 6 software (GraphPad Software, Inc.)
was employed for statistical analysis. All data are obtained from
three replicate experiments and expressed as the mean ± standard
deviation (SD) of three independent experiments. Statistical
differences between multiple groups were analyzed using one-way
analysis of variance followed by Tukey's post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Formation and validation of TAMs
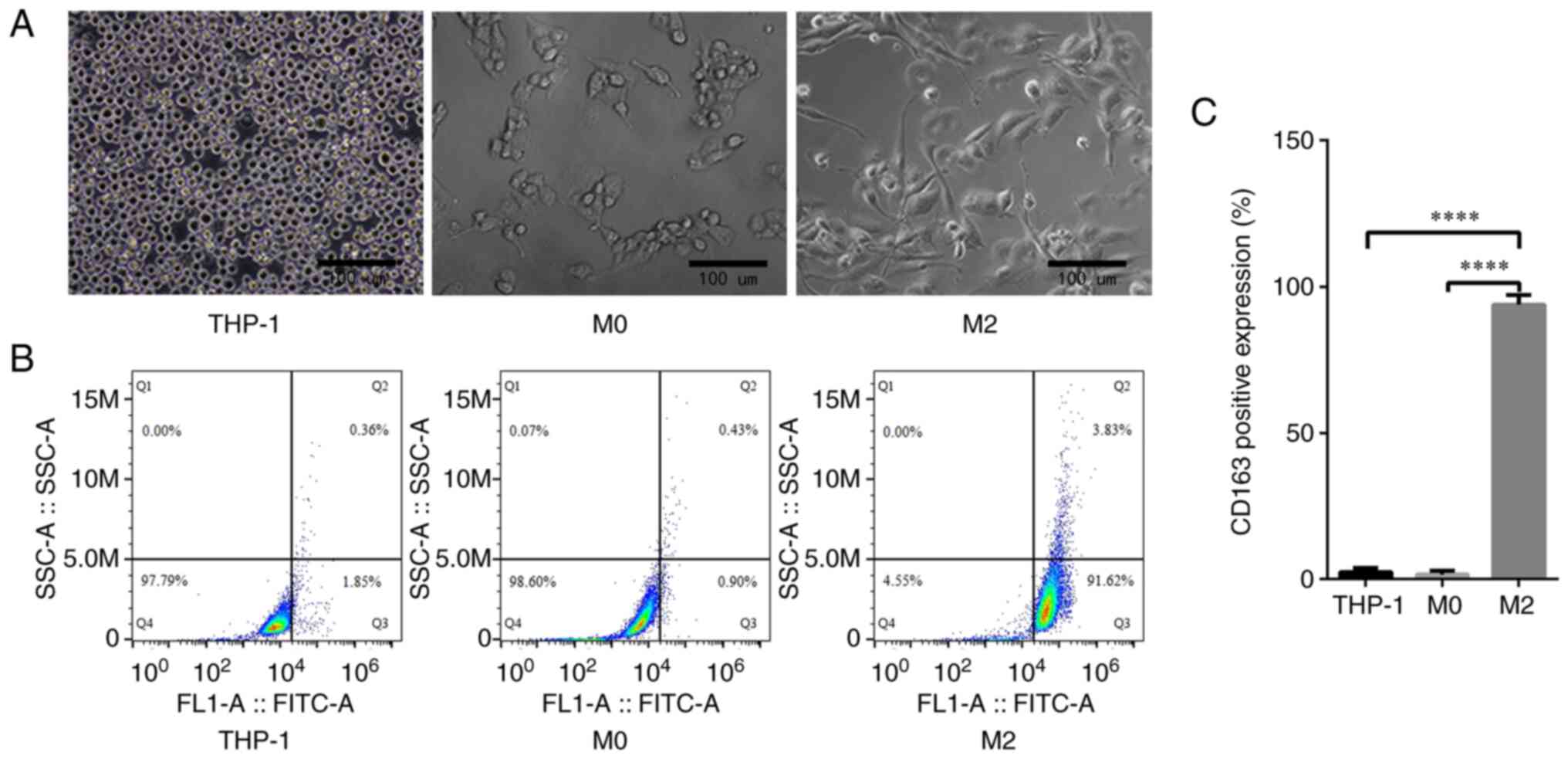
An inverted microscope was utilized to capture
images of the THP-1 human monocytic acute leukemia cell line and
the M0 and M2 macrophages derived from them. The THP-1 cells were
observed to be round and grow in suspension, while the M0
macrophages were fusiform with protruding pseudopods and short
antennae. However, the M2 macrophages exhibited a long fusiform or
polygonal morphology with longer antennae (Fig. 1A). By incubating the THP-1 cells,
M0 and M2 macrophages with CD163 antibodies and then using FACS, it
was shown that the fluorescence intensity of CD163 in the M2
macrophages was significantly higher than that in the THP-1 cells
and M0 macrophages (93.90±1.92 vs. 2.46±0.74 and 1.57±0.73%,
respectively, P<0.0001; Fig. 1B and
C).
Identification of TAM-derived
exosomes
To confirm whether TAM-derived exosomes affect
EGFR-TKI efficacy, exosomes were isolated from the conditioned
medium of M2 macrophages by filtration and high-speed
centrifugation, and were identified by electron microscopy, NTA and
the detection of specific proteins. The typical double-membrane
vesicle structure of the exosomes was clearly observed using
electron microscopy (Fig. 2A). The
NTA showed that the diameter of vesicle particles ranged between 50
and 150 nm, with an enrichment diameter of 90 nm (Fig. 2B). In addition, western blotting
was used to analyze the exosome-positive markers CD9, CD63 and
TSG101. The western blotting results showed that the three exosome
markers were highly abundant in the extracted material (Fig. 2C). These results indicate that
TAM-derived exosomes were successfully extracted.
TAM-derived exosomes affect the
killing effect of gefitinib on HCC827 and PC9 cells
Different concentrations of gefitinib were applied
to HCC827 and PC9 cells for 24 h. The results of the CCK-8 assay
demonstrated that gefitinib had a marked dose-dependent killing
effect on these two cell lines. The IC50 values of
gefitinib against HCC827 and PC9 cells were calculated to be 0.1086
and 0.1028 µM, respectively (Fig. 3A
and B). A gefitinib concentration of 0.10 µM was selected as
the concentration for use in subsequent experiments.
The effects of gefitinib and M2 macrophage-derived
exosomes on the early apoptosis of HCC827 and PC9 cell lines were
assessed by flow cytometry. The results showed that the total
apoptosis percentage in the gefitinib group was significantly
increased compared with that in the blank control group (PC9:
59.21±1.52 vs. 23.12±1.82%, respectively, P<0.0001; HCC827:
50.18±1.41 vs. 20.06±1.04%, respectively, P<0.0001), and the
total apoptosis percentage in the M2 macrophage-derived exosome
group was reduced compared with that in the blank control group
(PC9: 13.18±0.35 vs. 23.12±1.82, respectively, P<0.0001; HCC827:
7.40±0.14 vs. 20.06±1.04%, respectively; P<0.0001). The
percentage of total apoptosis in the M2 macrophage-derived exosome
and combination groups was significantly reduced compared with that
in the gefitinib group (PC9: 13.18±0.35 and 37.66±0.80 vs.
59.21±1.52%, respectively, P<0.0001; HCC827: 7.40±0.14 and
37.14±1.57 vs. 50.18±1.41%, respectively, P<0.0001; Fig. 4A and B). These data suggest that
gefitinib had a significant killing effect on the HCC827 and PC9
cell lines and that M2 macrophage-derived exosomes significantly
reduced this killing effect. This prompted an exploration of the
potential underlying molecular mechanism.
M2 macrophage-derived exosomes inhibit
cell apoptosis by regulating the AKT/ERK1/2/STAT3 signaling
pathway
Our previous research (15) explored the probable mechanism by
which M2 macrophages affect the biological activity of NSCLC cells;
in the present study, more possibilities were explored. Western
blot analysis was used to evaluate the protein levels of AKT,
p-AKT, ERK1/2, p-ERK1/2, STAT3 and p-STAT3 to verify the original
hypothesis that TAMs transmit proteins, factors or genetic
substances through an exosome bridge, thereby affecting the
sensitivity of NSCLC to EGFR-TKI drugs. The results indicated that
the levels of p-AKT, p-ERK1/2 and p-STAT3 relative to their
respective total proteins were markedly different between groups.
Compared with the control group, the levels of p-AKT, p-ERK1/2 and
p-STAT3 proteins in the gefitinib group were significantly
decreased. Furthermore, the protein levels of p-AKT, p-ERK1/2 and
p-STAT3 in the combination group were higher than those in the
gefitinib group, although not significantly in all cases (Fig. 5A and B). These results indicate
that M2 macrophage-derived exosomes attenuate the killing effect of
gefitinib on HCC827 and PC9 lung adenocarcinoma cell lines by
activating the AKT, ERK1/2 and STAT3 signaling pathways.
 | Figure 5.M2 macrophage-derived exosomes inhibit
cell apoptosis by regulating the AKT/ERK1/2/STAT3 signaling
pathway. Western blot analysis was used to evaluate the protein
levels of AKT, p-AKT, ERK1/2, p-ERK1/2, STAT3 and p-STAT3. (A)
Representative protein expression plots and (B) quantified
phosphorylated target protein data are presented. In comparison
with those in the control group, the levels of p-AKT, p-ERK1/2 and
p-STAT3 proteins in the gefitinib group were significantly
decreased. The levels of p-AKT, p-ERK1/2 and p-STAT3 in the
combination group were higher than those in the gefitinib group.
*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. ns, no
significant difference; p-, phosphorylated; Exo, exosome. Data are
displayed as the mean ± SD. |
Discussion
As aforementioned, a variety of components in the TM
are associated with the development of tumor drug resistance. Our
previous study also demonstrated that TAMs promote the
proliferation and migration of lung adenocarcinoma cells (15). Similarly, a model of TAMs was
successfully established using PMA, IL-4 and IL-13 to treat the
THP-1 cell line, which is a generally accepted model (20,21).
There are different subtypes of macrophages. At present, M2
macrophages and TAMs are considered to serve a similar role
(22), so this in vitro
research model is academically recognized; however, M2 macrophages
are not completely equivalent to TAMs, which is one of the
shortcomings of the present study.
Cells can communicate with each other by
cell-to-cell contact and the transmission of certain molecules. The
study of EV-mediated intercellular communication is an important
topic in cancer biology. Notably, tumor-derived EVs have been
indicated to be involved in various processes during tumor
progression, including immunosuppression and resistance to
anticancer therapy (23–25). Exosomes are a type of EV with
disc-shaped vesicles 40–100 nm in diameter. In the present study
the extracted particles had a diameter of 50–150 nm, an enrichment
diameter of 90 nm and a vesicle-like structure when observed by
electron microscopy; these findings confirm that the particles were
exosomes. In addition, the exosome-specific marker proteins CD9,
CD63 and TSG101 were clearly detected, further confirming that M2
macrophage-derived exosomes were successfully obtained.
The results of multiple studies suggest that M2
macrophage-derived exosomes promote the proliferation of lung
cancer cells and lead to antitumor drug resistance. For example,
Wei et al (26)
demonstrated that the infiltration of M2 macrophages was positively
associated with the metastasis of lung adenocarcinoma, and that M2
macrophage-derived exosomes were taken up by lung adenocarcinoma
cells and thereby promoted cell migration, invasion and
angiogenesis. In addition, Li et al (27) showed that exosomes derived from M2
TAMs were able to promote cell viability, migration and invasion
and the epithelial-mesenchymal transition in NSCLC, and that
microRNA (miR)-155 and miR-196a-5p in exosomes served an important
role in this process. Furthermore, Wang et al (28) demonstrated that M2
macrophage-derived exosomes are the main factor promoting cisplatin
resistance in lung cancer. The mechanism was revealed to comprise
the stabilization of c-Myc via the inhibition of E3
ubiquitin-protein ligase NEDD4-like, which increased the aerobic
glycolysis and chemoresistance of lung cancer. By contrast, the
present study explored the effect of TAM-derived exosomes on the
efficacy of EGFR-targeted drugs in lung adenocarcinoma. The results
suggest that TAM-derived exosomes inhibited the apoptosis of HCC827
and PC9 cells, and negatively influenced the killing effect of
gefitinib; in other words, these exosomes reduce the efficacy of
gefitinib. The precise mechanism may be via activation of the AKT,
ERK1/2 and STAT3 signaling pathways. The present study did not
include an M0-derived exosome group as a control when analyzing the
mechanism, so may have introduced some bias to the results, which
is the second limitation of the paper. Also, it did not identify
specific messengers or cytokines in TAM-derived exosomes, nor did
it explore potential mechanisms other than the AKT signaling
pathway. This the third limitation of the study and our direction
of future exploration. The present study is not alone in exploring
the field in which the TM leads to EGFR-TKI resistance. Zhou et
al (29) indicated that
crosstalk between cancer cells and macrophages plays a crucial role
in the development of cancer. The study results suggested that
NSCLC cells promote macrophage M2 polarization and suppress M1
polarization through targeting miR-627-3p/Smads signaling pathway
by transferring exosomes to THP-1 cells, These changes enhanced the
EGFR-TKI resistance in the NSCLC H1975 cell line. To explore
alternative treatment strategies following osimertinib resistance
in NSCLC, a similar study was performed by Wan et al
(30). The results suggested that
M2-type TAM-derived exosomes promoted osimertinib resistance in
NSCLC by regulating the MSTRG.292666.16/miR-6386-5p/MAPK 8
interacting protein 3 axis.
In conclusion, the communication between tumor cells
and other cells in the microenvironment is complex. With regard to
the contribution of macrophages, tumors can recruit monocytes from
the peripheral blood into the TM and then induce a transition into
TAMs that promote tumor growth and mediate the development of tumor
resistance. Therefore, any measure that interrupts this negative
feedback loop could theoretically have an antitumor effect. The
results of the present study suggest that exosomes derived from
type M2 TAMs promote resistance to gefitinib in NSCLC and that the
mechanism may be associated with reactivation of the AKT, ERK1/2
and STAT3 signaling pathways. The study may serve as a reference in
the exploration of alternative treatment strategies for NSCLC
following the development of resistance to EGFR-targeted drugs.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science Foundation
of Fujian Province (grant no. 2020J011312) and the National Natural
Science Foundation of China (grant nos. 81160294 and 81960425).
Availability of data and materials
The datasets used and/or analyzed during this study
are available from the corresponding author upon reasonable
request.
Authors' contributions
SY and JX designed the study. SY, WC and HX
performed the experiments. JY, YZ, WY and LD analyzed the data. SY
and HX wrote the paper. SY and JX confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Daly ME, Singh N, Ismaila N, Antonoff MB,
Arenberg DA, Bradley J, David E, Detterbeck F, Fruh M, Gubens MA,
et al: Management of stage III Non-small-cell lung cancer: ASCO
guideline. J Clin Oncol. 40:1356–1384. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin JJ, Zhu VW, Schoenfeld AJ, Yeap BY,
Saxena A, Ferris LA, Dagogo-Jack I, Farago AF, Taber A, Traynor A,
et al: Brigatinib in patients with alectinib-refractory
ALK-positive NSCLC. J Thorac Oncol. 13:1530–1538. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peled N, Gillis R, Kilickap S, Froesch P,
Orlov S, Filippova E, Demirci U, Christopoulos P, Cicin I, Basal
FB, et al: GLASS: Global Lorlatinib for ALK(+) and ROS1(+)
retrospective Study: Real world data of 123 NSCLC patients. Lung
Cancer. 148:48–54. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seto T, Ohashi K, Sugawara S, Nishio M,
Takeda M, Aoe K, Moizumi S, Nomura S, Tajima T and Hida T:
Capmatinib in Japanese patients with MET exon 14 skipping-mutated
or MET-amplified advanced NSCLC: GEOMETRY mono-1 study. Cancer Sci.
112:1556–1566. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schmid S, Li J and Leighl NB: Mechanisms
of osimertinib resistance and emerging treatment options. Lung
Cancer. 147:123–129. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shaikh M, Shinde Y, Pawara R, Noolvi M,
Surana S, Ahmad I and Patel H: Emerging approaches to overcome
acquired drug resistance obstacles to osimertinib in non-small-cell
lung cancer. J Med Chem. 65:1008–1046. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ostrand-Rosenberg S: Tolerance and immune
suppression in the tumor microenvironment. Cell Immunol. 299:23–29.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ruffell B and Coussens LM: Macrophages and
therapeutic resistance in cancer. Cancer Cell. 27:462–472. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kerkar SP and Restifo NP: Cellular
constituents of immune escape within the tumor microenvironment.
Cancer Res. 72:3125–3130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Solinas G, Germano G, Mantovani A and
Allavena P: Tumor-associated macrophages (TAM) as major players of
the cancer-related inflammation. J Leukoc Biol. 86:1065–1073. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chung FT, Lee KY, Wang CW, Heh CC, Chan
YF, Chen HW, Kuo CH, Feng PH, Lin TY, Wang CH, et al:
Tumor-associated macrophages correlate with response to epidermal
growth factor receptor-tyrosine kinase inhibitors in advanced
non-small cell lung cancer. Int J Cancer. 131:E227–E235. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang B, Zhang Y, Zhao J, Wang Z, Wu T, Ou
W, Wang J, Yang B, Zhao Y, Rao Z and Gao J: M2-polarized
macrophages contribute to the decreased sensitivity of EGFR-TKIs
treatment in patients with advanced lung adenocarcinoma. Med Oncol.
31:1272014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan S, Dong Y, Peng L, Yang M, Niu L, Liu
Z and Xie J: Tumor-associated macrophages affect the biological
behavior of lung adenocarcinoma A549 cells through the PI3K/AKT
signaling pathway. Oncol Lett. 18:1840–1846. 2019.PubMed/NCBI
|
|
16
|
Wendler F, Favicchio R, Simon T,
Alifrangis C, Stebbing J and Giamas G: Extracellular vesicles swarm
the cancer microenvironment: From tumor-stroma communication to
drug intervention. Oncogene. 36:877–884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Robbins PD and Morelli AE: Regulation of
immune responses by extracellular vesicles. Nat Rev Immunol.
14:195–208. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee JK, Park SR, Jung BK, Jeon YK, Lee YS,
Kim MK, Kim YG, Jang JY and Kim CW: Exosomes derived from
mesenchymal stem cells suppress angiogenesis by down-regulating
VEGF expression in breast cancer cells. PLoS One. 8:e842562013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Looze C, Yui D, Leung L, Ingham M, Kaler
M, Yao X, Wu WW, Shen RF, Daniels MP and Levine SJ: Proteomic
profiling of human plasma exosomes identifies PPARgamma as an
exosome-associated protein. Biochem Biophys Res Commun.
378:433–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu Z, Zhou J, Chen F, Yu J, Li H, Li Q and
Li W: 13-Methyl-palmatrubine shows an anti-tumor role in non-small
cell lung cancer via shifting M2 to M1 polarization of tumor
macrophages. Int Immunopharmacol. 104:1084682022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun D, Luo T, Dong P, Zhang N, Chen J and
Zhang S, Dong L, Janssen H and Zhang S: M2-polarized
tumor-associated macrophages promote epithelial-mesenchymal
transition via activation of the AKT3/PRAS40 signaling pathway in
intrahepatic cholangiocarcinoma. J Cell Biochem. 121:2828–2838.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Li D, Cang H and Guo B: Crosstalk
between cancer and immune cells: Role of tumor-associated
macrophages in the tumor microenvironment. Cancer Med. 8:4709–4721.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Goede KE, Driessen A and Van den
Bossche J: Metabolic cancer-macrophage crosstalk in the tumor
microenvironment. Biology (Basel). 9:3802020.PubMed/NCBI
|
|
24
|
Ruivo CF, Adem B, Silva M and Melo SA: The
biology of cancer exosomes: Insights and new perspectives. Cancer
Res. 77:6480–6488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pashazadeh M: The role of tumor-isolated
exosomes on suppression of immune reactions and cancer progression:
A systematic review. Med J Islam Repub Iran. 34:912020.PubMed/NCBI
|
|
26
|
Wei K, Ma Z, Yang F, Zhao X, Jiang W, Pan
C, Li Z, Pan X, He Z, Xu J, et al: M2 macrophage-derived exosomes
promote lung adenocarcinoma progression by delivering miR-942.
Cancer Lett. 526:205–216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Chen Z, Ni Y, Bian C, Huang J, Chen
L, Xie X and Wang J: Tumor-associated macrophages secret exosomal
miR-155 and miR-196a-5p to promote metastasis of non-small-cell
lung cancer. Transl Lung Cancer Res. 10:1338–1354. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Wang L, Pan H, Wang Y, Shi M, Yu
H, Wang C, Pan X and Chen Z: Exosomes derived from macrophages
enhance aerobic glycolysis and Chemoresistance in lung cancer by
stabilizing c-Myc via the inhibition of NEDD4L. Front Cell Dev
Biol. 8:6206032020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou D, Xia Z, Xie M, Gao Y, Yu Q and He
B: Exosomal long non-coding RNA SOX2 overlapping transcript
enhances the resistance to EGFR-TKIs in non-small cell lung cancer
cell line H1975. Hum Cell. 34:1478–1489. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Li D, Cang H and Guo B: Crosstalk
between cancer and immune cells: Role of tumor-associated
macrophages in the tumor microenvironment. Cancer Med. 8:4709–4721.
2019. View Article : Google Scholar : PubMed/NCBI
|