Introduction
Blood vessels are essential elements for supplying
oxygen and nutrients to maintain normal tissues (1). Angiogenesis, a process in which new
blood vessels are developed, begins in embryogenesis. In adulthood,
the process maintains a static state, except in certain conditions,
such as inflammatory diseases. Vascular endothelial growth factor
(VEGF) is a crucial component involved in promoting angiogenesis
and can initiate angiogenesis by binding to VEGF receptor 2
(VEGFR2) in the endothelial cells (ECs) (2). The regulation of VEGFR2 by a positive
feedback mechanism increases reliance on VEGF-induced angiogenesis.
It can promote endothelial proliferation and sprouting (3).
The induction of angiogenesis in a static vascular
state is termed ‘angiogenic switch’, which is one of the hallmarks
of cancer (4,5). As the size of the tumor increases
with cancer progression, the demand for blood vessels is
indispensable. Hypoxia is a condition involving the absence of
blood vessels observed during the typical progression of cancer. To
overcome hypoxia and induce angiogenesis, cancer cells secrete
VEGF, a pro-angiogenic factor. Hence, VEGF is a critical factor for
cancer progression and growth (6).
The first anti-angiogenesis therapy involved
administering the monoclonal antibody ‘bevacizumab’, which binds to
the VEGFA isoform and interrupts the interaction between VEGFA and
VEGFR. Bevacizumab was first approved by the FDA in 2004 for the
treatment of metastatic colorectal cancer. it has since been used
to treat various cancers, including breast cancer, non-small cell
lung cancer, metastatic breast cancer, ovarian cancer, renal cell
carcinoma, and recurrent glioblastoma (GBM) (7). Although bevacizumab has shown
favorable outcomes in treating many cancers, no significant effect
has been observed on GBM. Bevacizumab improves the quality of life
but not the overall survival of patients with GBM (8). Induction of hypoxia via the
inhibition of angiogenesis and VEGF-independent angiogenesis
mechanisms has been reported. The precise reason remains primarily
unknown.
Endothelin 1 (EDN1) was initially identified as a
peptide hormone secreted from ECs and is known to act on smooth
muscle cells (SMCs). Exposure of SMCs to EDN1 promotes
vasoconstriction, and excessive EDN1 levels are a major factor
contributing to the development of hypertension (9). At present, EDN1 functions in various
types of cells, including cancer cells as well as in SMCs. In
particular, it influences malignancy by increasing proliferation,
suppressing apoptosis, and promoting the endothelial to mesenchymal
transition (10). However, the
effect of EDN1 on ECs and the relationship between ECs and cancer
malignancy remain unknown.
According to a previous study conducted by our
group, the inhibitor of differentiation 1 (ID1) may be associated
with VEGF-independent angiogenesis in GBM. The growth rate
increases in ID1-expressing GBM cells and promotes tumor
angiogenesis (11). Therefore, we
examined the association between ID1 and EDN1 and elucidated the
angiogenic capability of EDN1.
Materials and methods
Cell culture and culture
conditions
The human glioma cell line U87MG (TP53wt,
PTENmut, p14ARF/p16del) was purchased from the
American Type Culture Collection (ATCC, Cat. HTB-14; glioblastoma
of unknown origin). It was authenticated that the cell line was
U87MG cells through short tandem repeat profiling. U87MG cells were
cultured in Dulbecco's modified Eagle's medium-high glucose (DMEM;
Lonza, Cat. SH30243.01) supplemented with 10% fetal bovine serum
(FBS; Hyclone, Cat. SH30919.03), 1% penicillin/streptomycin (P/S;
Hyclone, Cat. SV30010), and 2 mM L-glutamine (Cat. SH30034.01) at
37°C in an atmosphere containing 5% CO2 and 95%
humidity.
Human umbilical vein endothelial cells (HUVECs) were
purchased from Lonza (Lonza, Cat. CC-2517). HUVECs were cultured on
0.2% gelatin-coated plates (Sigma-Aldrich, Cat. G1890) and grown in
an endothelial cell growth medium (EGM-2, Lonza, Cat. CC-3162). All
experiments were conducted until passage five.
Plasmid construction and virus
infection
U87MG cells were infected with lentivirus produced
using the HEK293FT cell line (Life Technologies, Cat. R70007) that
was transfected with a lentiviral vector (pLL-CMV-GFP,
pLL-CMV–ID1-GFP) and packaging vectors (third-generation:
pMDLg/pRRE, pRSV-Rev, and pMD2.G).
Western blot analysis
Western blotting was performed to analyze protein
expression. Briefly, cell extracts were prepared using the RIPA
lysis buffer (150 mM sodium chloride, 1% NP-40, 0.1% SDS, 50 mM
Tris, pH 7.4) containing 1 mM β-glycerophosphate, 2.5 mM sodium
pyrophosphate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and
a protease inhibitor (Roche, Cat. 11836170001). The protein
concentration was quantified using the Bradford assay reagent
(Bio-Rad) according to the manufacturer's instructions. Proteins
were resolved by SDS-PAGE and then transferred to a polyvinylidene
fluoride membrane (Pall Corporation, Cat. BSP0861). The membranes
were blocked with 5% non-fat milk and incubated with the following
antibodies at indicated dilutions: anti-ID1 (1:1,000; Biocheck,
Cat. BCH-1/195-14-50) and anti-β-actin (1:10,000; Santa Cruz
Biotechnology, Cat. sc-47778). Membranes were then incubated with
horseradish peroxidase-conjugated anti-IgG secondary antibody
(Pierce Biotechnology) and visualized using the SuperSignal West
Pico Chemiluminescent Substrate (Pierce Biotechnology, Cat.
34580).
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed to determine mRNA levels.
Briefly, total RNA was isolated from cells using the QIAzol lysis
reagent (QIAGEN, Cat. 79306) according to the manufacturer's
instructions. Then, 1 U of DNase I (RNase-free; Thermo Fisher
Scientific, Cat. EN0525) was added to 1 µg of template RNA,
followed by incubation for 30 min at 37°C. For inactivating DNase
I, 50 mM of EDTA was added, followed by heating at 65°C for 10 min.
DNase I-treated RNA was used as a template for synthesizing
complementary DNA (cDNA) using a RevertAid First Strand cDNA
Synthesis Kit (Thermo Fisher Scientific, Cat. K1622) according to
the manufacturer's instructions. RT-qPCR analysis was performed
using Takara Bio SYBR Premix Ex Taq (Takara, Cat. RR420A) and
CFX096 (Bio-Rad, Hercules, CA, USA) using the following
thermocycling conditions: Initial denaturation at 95°C for 30 sec,
followed by 45 cycles at 95°C for 5 sec and 60°C for 30 sec for
annealing and elongation. The expression of each target gene was
normalized to that of GAPDH and quantified using the
2−ΔΔCq method (12).
The following primer sequences were used: human GAPDH
forward: 5′-CTACACTGAGCACCAGGTGGTCTC-3′, reverse:
5′-GATGGATACATGACAAGGTGCGGC-3′; human EDN1 forward:
5′-CGAGCACATTGGTGACAGAC-3′, reverse:
5′-GAAGATGGTTGGGGGAACTC-3′.
Transepithelial electrical resistance
(TEER) value
To measure the TEER values of the monolayers, 5×104
HUVECs were seeded into Transwell inserts (6.5 mm diameter, 5.0 µm
pore size, Costar, Cat. 3421). Insert was pre-coated with
fibronectin from human plasma (Sigma-Aldrich, Cat. F0895) in PBS
for 2 h at 37°C in an incubator and air-dried for 45 min. One hour
after seeding the cells, the insert was transferred to an empty
well containing 850 µl EGM-2 medium. The TEER value was measured
one day after seeding using an ERS-2 epithelial volt-ohm meter
(Millicell, Cat. MERS00002). The STX3 electrodes were introduced
into the apical and basolateral compartments of the inserts. For
each experiment, the TEER value was measured in a cell-free
Transwell insert (blank). The final TEER values were multiplied by
the surface area of the inserts and expressed as
Ωcm2.
Mouse aortic ring assay
Aortas obtained from two-week-old C57/BL/6J mice
were cut into approximately 1.0 mm pieces, as previously described
(13). Briefly, following a chest
incision, the lungs and liver are removed, which reveals the aorta.
Using forceps and scalpel, separate the aorta from the spine. The
fibroadipose tissue around the aorta was carefully removed.
Transfer the isolated aorta to an ice-cold culture dish containing
EGM-2 medium. Using a scalpel, cut the aorta into 1.0 mm pieces.
The aortic rings were cultured in 48-well Matrigel-coated plates
(Corning, Cat. 354234). Next, 500 µl of EGM-2 medium containing
VEGF (100 µg/ml; R&D Systems, Cat. 293-VE-050) and EDN1 (100
µg/ml; TOCRIS, Cat. 1160/100 U) was added to each well. After five
days, the aortic rings were fixed with 4% PFA and images were
captured by microscopy (Olympus, Cat. CKK53, magnification ×10).
The image analysis was performed by Image J (software v.1.52a,
plugin; Angiogenesis Analyzer, http://imagej.nih.gov/ij/). The Korea University
Institutional Animal Care & Use Committee approved the animal
experiments, which were carried out in accordance with governmental
and institutional guidelines as well as Korean regulations
(approval no. KUIACUC-2019-0040). A total of 10 male C57/BL/6J mice
(1 week old; average weight, 3 g) were purchased by Orient Bio,
Inc. Mice were bred with under conditions that included an average
temperature of 20–24°C, humidity of 45–65%, and a 12-h light/dark
cycle. The mice were received a continuous supply of food and
water. When the mice were 2 weeks old, they were anesthetized by
inhalation of 30% CO2 (4.5 l/min) for 2 min in a
CO2 gas chamber.
HUVEC spheroid sprouting assay
HUVECs (1×103) were mixed with a 0.25%
methylcellulose solution (Sigma-Aldrich, Cat. M0512) and EBM-2
(Lonza, Cat. CC-3156). A total of 20 µl of the mixture containing
1,000 cells was dispensed using the hanging-drop method and
incubated overnight at 37°C in an atmosphere containing 5% Co2 for
spheroid formation. Next, 61 µl of collagen was added, and the
total volume of the collagen mixture was 100 µl (4.07 mg/ml stock,
Corning, Cat. 354236). The spheroids were gently washed off the
hanging-drop plate with phosphate-buffered saline (PBS),
centrifuged at 500 rpm using a swing bucket rotor for 5 min, and
re-suspended in 60 µl of 0.25% methylcellulose solution. A total of
60 µl of 0.25% methylcellulose solution containing HUVEC spheroids
was added to the aforementioned mixture. A total of 150 µl of the
final mixture was dispensed into 48-well plates and polymerized at
37°C in an incubator for 30 min. The appropriate wells were then
overlaid with 500 µl of complete EGM-2 containing VEGF (100 µg/ml;
R&D system, Cat. 293-VE-050) and EDN1 (100 µg/ml; TOCRIS, Cat.
1160/100U) recombinant protein. Endothelial sprouting was observed
and fixed with 4% PFA for immunofluorescence (IF) analysis after
five days. The image was analyzed by Image J (software v.1.52a,
plugin; Angiogenesis Analyzer, Analyze Particles, http://image.nih.gov/ij/).
Preparation of conditioned medium
(CM)
For the collection of CM, U87MG-GFP, and
U87MG-ID1-GFP cells (7.5×105) were seeded in a 100-mm culture plate
with DMEM containing 10% FBS. After 48 h, the medium was replaced
with an EGM-2 medium. After 24 h, the CM was harvested and filtered
using 0.2-µm filters (Sartorius, Cat. 16534).
Fluorescence images
Fluorescence image analysis was performed as
previously described (14). HUVEC
spheroids were fixed in 4% PFA for 15 min at room temperature (RT).
The spheroids were washed thrice with PBS, permeabilized with 0.3%
Triton X-100 in PBS for 10 min at RT, and then blocked with 3% BSA
(Merck, Cat. 82-100-6) in PBS for 1 h at RT. The spheroids were
incubated with CD31 antibody (1:200, Thermo Fisher, Cat.
IHC-00055), EDNRA antibody (1:200, Alomone Labs, Cat. AER-001), and
EDNRB antibody (1:200, Alomone Labs, Cat. AER-002) overnight at
4°C, followed by incubation with Alexa Fluor 488- or 568-conjugated
secondary antibodies (1:400, ThermoFisher, Cat. A10042, A21202) for
2 h at RT. Nuclei were then stained with DAPI (1 µg/ml,
Sigma-Aldrich, Cat. D9542) for 5 min. For phalloidin staining
(Invitrogen, Cat. A12379), HUVECs were fixed in 4% PFA for 15 min
at RT. They were washed thrice with PBS, permeabilized with 0.3%
Triton X-100 (Sigma-Aldrich, Cat. T9281) in PBS for 10 min at RT,
and then blocked with 3% BSA in PBS for 1 h at RT. The spheroids
were incubated with phalloidin (6.6 µM) for 2 h at RT. Nuclei were
then stained with DAPI for 5 min.
Fluorescence images were obtained using confocal
laser scanning microscopy [LSM800; Carl Zeiss, ZEN acquisition
software version 2018 (blue edition)] at RT (magnification
×10).
In silico analysis
To analyze region-specific transcriptome profiles of
patients with GBM, we obtained fragments per kilobase of
transcripts per million mapped reads data from the Ivy glioblastoma
atlas project (Ivy GAP) website (http://glioblastoma.alleninstitute.org/). Regional
information was divided into seven types: hyperplastic blood
vessels in the cellular tumor (n=22), microvascular proliferation
(n=28), leading-edge (n=19), infiltrating tumor (n=24), cellular
tumor (n=111), perinecrotic zone (n=26), and pseudopalisading cells
around necrosis (n=40). The relative genes were converted to
z-scores, and the mean ± SD value of the z-score was
calculated.
The ID1 RNA-seq data obtained by eBiogen were
grouped based on changes in the expression higher than two-fold of
the baseline value to establish an ID1 signature determined by
comparing the mean expression values (Student's t-test; P<0.05)
(GEO: GSE182670).
To analyze the clinical relevance, we obtained
transcriptome profiles of patients with GBM with or without
bevacizumab treatment (15).
Analyses were performed for all datasets with the exception of
maximum and minimum values in bevacizumab responders and
non-responders.
Statistical analysis
All statistical analysis was performed using an
unpaired Student's t-test and one-way ANOVA test followed by
Bonferroni post hoc test. When comparing two groups, the
significance was determined using a student's t-test. Comparison
between multiple groups was determined one-way ANOVA test followed
by Bonferroni post hot test. All statistical analysis was performed
using GraphPad Prism (Version 9.3.1). Values of P<0.05 or
P<0.01 were considered statistically significant for different
experiments, as indicated in figure legends. Data are presented as
mean ± SEM.
Results
mRNA expression of ID1 and EDN1 is
primarily observed in the vascular-related region in tissues of
patients with GBM
To investigate the genes regulated by ID1 that
affect ECs, we performed RNA sequencing using the
ID1-overexpressing U87MG cell line. Next, we merged upregulated
differentially expressed genes (DEGs) from RNA sequencing data
(>two-fold, <FDR value 0.05, n=433) using a gene set (Module
92: secreted signaling molecules, n=149) to identify factors that
can affect ECs. Three genes (NMU, WISP2, and EDN1)
were selected (Fig. 1A).
Subsequently, we used the Ivy GAP project (http://glioblastoma.alleninstitute.org/), which
consists of the transcriptome data of seven anatomic structures
isolated via laser microdissection from patients with GBM, to
validate the genes that influence the synthesis of ECs among the
three genes. Interestingly, only ID1 and EDN1 were
highly expressed in the vascular-related region (microvascular
proliferation and hyperplastic blood vessels). Particularly,
EDN1 was scarcely expressed, except in the vascular-related
region (Fig. 1B and C).
Collectively, we verified the possibility that EDN1 could
function in the ECs of patients with GBM.
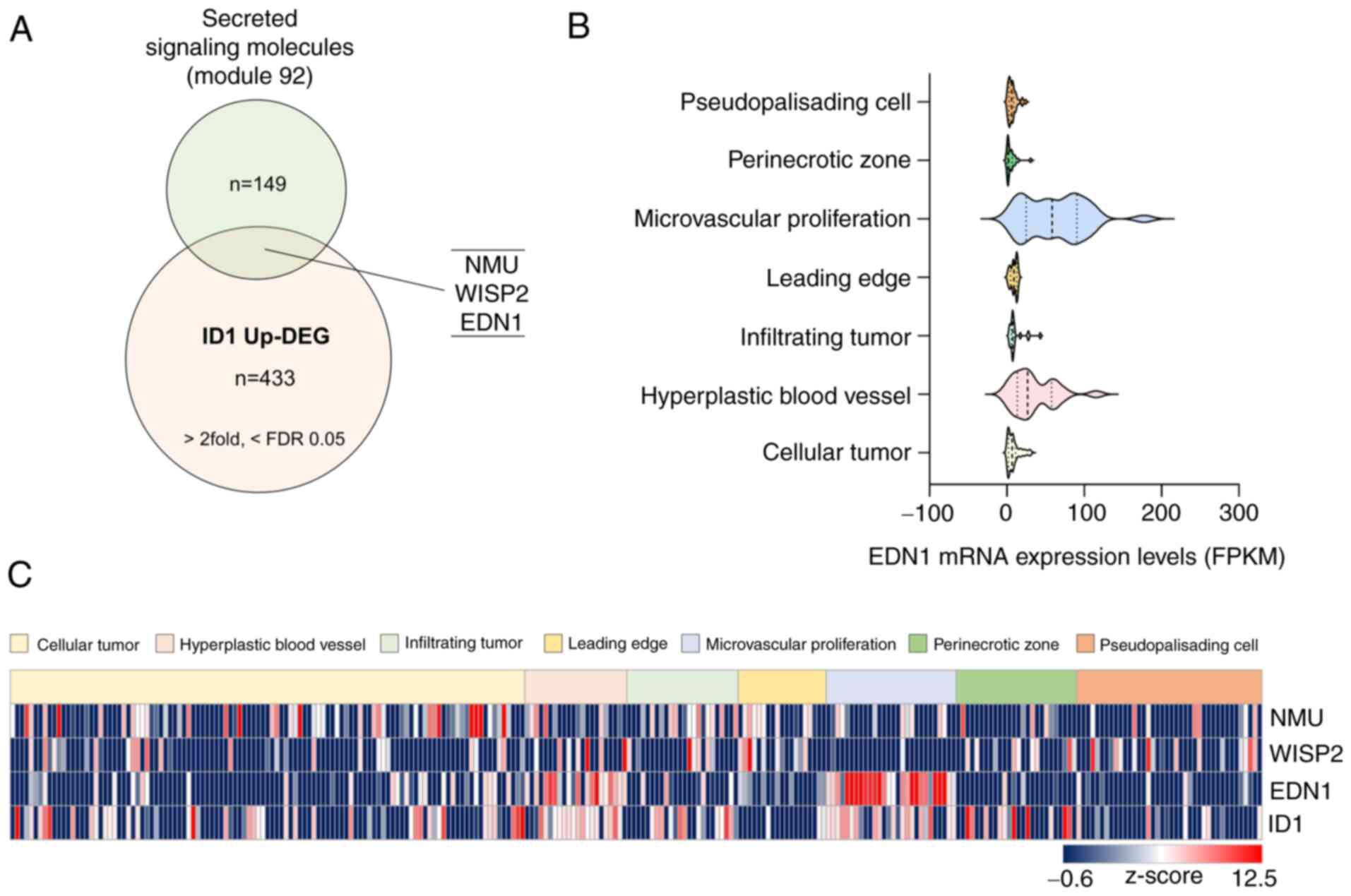 | Figure 1.EDN1 expression, which is upregulated
by ID1, is elevated in the vascular-related region in tissues
derived from patients with GBM. (A) Venn diagram obtained after
merging two datasets to identify vascular regulating factors
associated with ID1 (Module 92 refers to a list containing 149
secreted signaling molecules in humans, n=149, https://www.gsea-msigdb.org/gsea/msigdb/cards/module_92;
ID1 Up-DEG, n=433, >2 fold, <FDR value 0.05). (B) mRNA
expression levels of EDN1 in tissues of patients with GBM were
determined using the Ivy GAP database. (C) Identification of mRNA
expression of candidate genes (NMU, WISP, EDN1 and ID1) using the
Ivy GAP database (http://glioblastoma.alleninstitute.org/). The relative
mRNA levels were converted to z-score and are presented using a
heat-map. EDN1, endothelin1; FDR, false discovery rate; FPKM,
fragments per kilobase of exon per million mapped fragments; GBM,
glioblastoma; ID1, inhibitor of differentiation 1; ID1 Up-DEG, ID1
Upregulated-Differentially Expressed Genes; Ivy GAP, Ivy
glioblastoma atlas project; NMU, neuromedin u; WISP2, wnt1
inducible signaling pathway protein 2. |
EDN1 promotes endothelial sprouting
ability
We investigated whether the expression of
EDN1 was upregulated after ID1 overexpression in
U87MG cells. The mRNA expression of EDN1 was increased by six-fold
in U87MG cells with ID1 overexpression compared to that in control
cells (Fig. 2A). Next, we analyzed
the maintenance of barrier function and sprouting ability, which
are basic properties of ECs that could be influenced by EDN1.
First, we observed no changes in the barrier function when HUVECs
were treated with EDN1 (Fig. 2B).
Second, we performed ex-vivo culture using mouse arteries to
analyze the sprouting ability. Upon treatment with EDN1, the
sprouting ability was predominantly promoted compared to that in
the control and VEGF-treated groups (Fig. 2C). EDN1 is a more potent
pro-angiogenic factor than VEGF. The tendency of EDN1 to increase
the sprouting ability of ECs was also observed in the HUVEC
spheroid sprouting model (Fig.
2D). When EDN1 was added, the sprouting length and sprouting
cell number increased. Consequently, EDN1 did not affect the
barrier function but increased sprouting ability, which is critical
in the early stages of angiogenesis.
Putative positive feedback of EDNRB
modulates endothelial sprouting
Endothelial culture media contains various growth
factors. Therefore, the determination of the function of specific
factors is limited. To overcome this limitation, we compared the
endothelial sprouting ability induced by VEGF and EDN1 in basal
media without growth factors. Results showed that EDN1 induced
superior endothelial sprouting compared to VEGF, and no synergistic
effect was observed between the two factors (Fig. 3A). EDN1 signaling is regulated by
two receptors, EDNRA and EDNRB, and each receptor functions
depending on the location of the tissue. We used bosentan, which
can inhibit both EDN receptors, to determine whether EDNR regulated
the increase in the EDN1-mediated sprouting in ECs. EDN1 and
bosentan combinedly demonstrated a sprouting tendency similar to
that of the control group (Fig.
3B). This finding implies that the EDN1-EDNR axis promoted the
sprouting ability of HUVECs. VEGF, a representative angiogenic
factor, induces angiogenesis upon binding to VEGFR2. In this
process, VEGFR2 maintains and amplifies VEGF signaling via positive
feedback in tip cells rather than in stalk cells. To determine
whether this phenomenon could also be attributed to EDNR, we
compared EDNR expression after treatment with EDN1. Notably, the
EDNR expression was extremely low when the cells were not treated
with EDN1. However, EDNRB expression increased in the presence of
EDN1 (Fig. 3C). In addition, EDNRB
expression was primarily observed in the tip cells. Hence EDN1
induced endothelial sprouting by binding to EDNRB, and EDNRB may be
regulated by positive feedback.
 | Figure 3.EDN1-EDNRB axis promotes endothelial
sprouting, and its pro-angiogenesis function is more potent than
that of VEGF. (A) HUVEC spheroids were cultured in the presence or
absence of VEGF (50 ng/ml) and EDN1 (50 ng/ml) in EBM basal medium
(red, phalloidin; blue, DAPI). Quantification of the sprouting
length in HUVEC spheroids. ***P<0.001. Data are presented as the
mean ± SEM. Scale bar, 100 µm. (B) HUVEC spheroids were cultured in
the presence or absence of EDN1 (100 ng/ml) and bosentan (10 µM)
(green, CD31). Quantification of the sprouting length in HUVEC
spheroids. ***P<0.001. Data are presented as the mean ± SEM.
Scale bar, 100 µm. (C) HUVEC spheroids were cultured in the
presence or absence of EDN1 (100 ng/ml; green, EDNRA and EDNRB;
red, phalloidin; blue, DAPI). Quantification of the fluorescence
intensity (EDNRA and EDNRB) in the sprouting region. ***P<0.001.
Data are presented as the mean ± SEM. Scale bar, 100 µm. EDN1,
endothelin1; EDNRA, endothelin receptor type a; EDNRB, endothelin
receptor type b; VEGF, vascular endothelial growth factor. |
Promotion of endothelial sprouting by
the ID1-EDN1-EDNRB axis and its clinical relevance
To confirm the association between ID1 and EDN1, we
used a CM collected from glioblastoma cells that overexpressed ID1
(ID1_CM) and those that did not demonstrate overexpression
(GFP_CM). The CM was obtained using the EGM-2 medium, which was
used for the HUVEC spheroid sprouting assay (Fig. 4A). Upon confirming the expression
of EDN1 when obtaining CM, we observed that the number of cells
overexpressing ID1 was approximately 40 times higher than that of
the control cells (Fig. 4B).
However, sprouting was not observable on the spheroid when only CM
was treated because of lack of basic nutrients (data not shown). So
the CM was mixed with fresh EGM-2 medium in half and treated every
day. When treated with ID1_CM, the sprouting ability was higher
than that observed with GFP_CM (Fig.
4C). Next, we used bosentan to determine whether sprouting was
increased by EDN1 in ID1_CM. The increase in sprouting by ID1_CM
was inhibited by bosentan (Fig.
4D). This result suggests that the EDN1 in ID1_CM induced
sprouting. Finally, to assess the clinical relevance of EDN1, we
compared the transcriptomes of bevacizumab-responders and
non-responders in the cohort of patients with GBM (14). Patients responsive to bevacizumab
had low EDN1 expression, whereas non-responsive patients had higher
EDN1 expression (Fig. 4E). This
finding implies that EDN1, which is upregulated by ID1, can
potentially be an angiogenic factor that can be targeted instead of
VEGF during the treatment of patients with GBM (Fig. 4F).
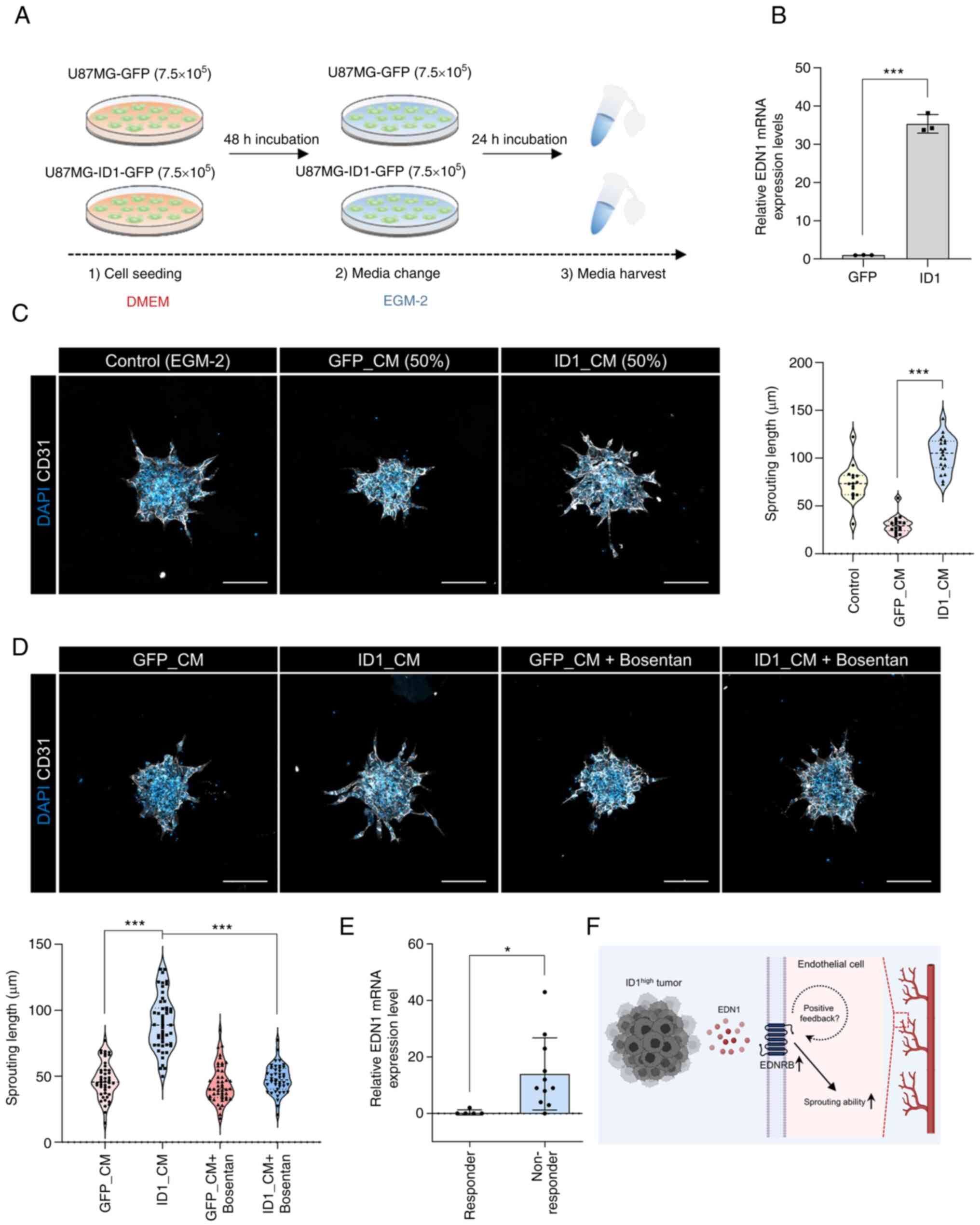 | Figure 4.Relationship between ID1 and EDN1 and
its clinical relevance. (A) A schematic diagram of the CM
experiment. (B) mRNA expression levels of EDN1 in ID1
overexpressing U87MG cells were determined by reverse
transcription-quantitative PCR using CM. ***P<0.001. Data are
presented as the mean ± SEM. (C) HUVEC spheroids were cultured in
the presence or absence of CM (white, CD31; blue, DAPI).
Quantification of the sprouting length in HUVEC spheroids.
***P<0.001. Data are presented as the mean ± SEM. Scale bar, 100
µm. (D) HUVEC spheroids were cultured in the presence or absence of
GFP_CM, ID1_CM and bosentan (10 µM) (white, CD31; blue, DAPI).
Quantification of the sprouting length in HUVEC spheroids
(bottom-left graph). ***P<0.001. Data are presented as the mean
± SEM. Scale bar, 100 µm. (E) mRNA expression levels of EDN1 in
bevacizumab-responders or non-responders in the cohort of patients
with GBM (responder, n=5; non-responder, n=11). *P<0.05. Data
are presented as the mean ± SEM. (F) Graphical summary of the study
(created with BioRender.com). CM, conditioned medium; EDN1,
endothelin1; EDNRB, endothelin receptor type b; EGM-2, endothelial
cell growth medium-2; GFP, green fluorescent protein; ID1,
inhibitor of differentiation 1. |
Discussion
The role of blood vessels in cancer maintenance and
progression has been recognized for a long time. Identifying
factors that promote angiogenesis, a hallmark of cancer, serves as
a foundation for anticancer strategies. VEGF, a master regulator of
angiogenesis, has been identified, and bevacizumab has been
developed to inhibit VEGF (1,4,5).
However, unresponsiveness has been observed in patients decades
after bevacizumab administration. The VEGF-independent angiogenesis
pathway is a representative cause of resistance to anti-VEGF
therapy (16). Therefore,
identifying new pro-angiogenic factors is critical.
In this study, we demonstrated a novel
pro-angiogenic, namely, EDN1. EDN1 increased sprouting ability at
an early stage of angiogenesis and was more potent than VEGF. In
addition, the EDNR demonstrated a positive feedback mechanism
similar to that of VEGFR2, a characteristic of tip cells, which are
known as sprouting cells in angiogenesis. To maintain a continuous
VEGF signal, the tip cell induces lateral inhibition, suppressing
VEGFR2 expression in the stalk cell, and VEGFR2 expression is
sustained through the positive feedback of VEGFR2. Although the
positive feedback mechanism of EDNRB mediated by EDN1 has not been
precisely elucidated, such regulation via positive feedback might
exist based on the fact that the expression of EDNRB is increased
by the action of EDN1 in tip cells.
EDN1 is a vasoconstrictor peptide produced by
endothelial cells. EDN1 induces vasoconstriction as a paracrine
effect by acting on mural cells, such as pericytes and smooth
muscle cells (9). Therefore, it
contributes to the onset and progression of many
hypertension-related diseases. Bosentan, which is capable of
inhibiting EDNR, has also been developed (17). Consequently, the paracrine effect
of EDN1 on mural cells has been well defined, but the paracrine
effect on endothelial cells mediated by cancer cells is unknown.
Most blood vessels in cancer are leaky. The primary cause of this
phenomenon is the detachment of mural cells from endothelial cells
(18). Kim et al (19) demonstrated that in a GBM mouse
model, the coverage of pericytes surrounding the endothelial cells
was reduced. Results showed that the expression of EDN1 was high in
the vascular-related region in the Ivy GAP database. Collectively,
the paracrine effect of EDN1 on endothelial cells from cancer cells
may prevail over that on mural cells in cancer blood vessels.
Although the paracrine effect of EDN1 on endothelial cells was not
verified in vivo, it is planned to examine the effect in the
future using a mouse model.
Results suggest that ID1 is an upstream factor for
EDN1. However, the detailed mechanism underlying the regulation of
EDN1 by ID1 has not yet been elucidated. ID1 has a helix-loop-helix
structure and lacks a basic DNA-binding domain; therefore, it
cannot independently bind to DNA. A well-known ID1 signaling
mechanism forms a heterodimer with E protein, a basic
helix-loop-helix structure, and suppresses cellular differentiation
(20). Therefore, finding the
binding partner of ID1 in the future is critical for understanding
the ID1-EDN1 regulatory mechanism.
In summary, we suggest a target that can be used to
develop treatment strategies for patients with GBM who are
resistant to anti-angiogenic therapy, which is currently used to
eradicate cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Research Foundation of Korea (NRF) (2020R1A2C2099668), College of
Life Sciences and Biotechnology, Korea University (K2207511) and
Brain Korea 21 PLUS.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The RNA-seq data generated and/or analyzed during the
current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182670.
Authors' contributions
SHC, MJP and HK designed the experiments and wrote
the manuscript. SHC conducted most experiments and analyzed the
data. MJP performed molecular experiments. This study was directed
by HK. All authors read and approved the final manuscript. SHC, MJP
and HK confirm the authenticity of the raw data.
Ethics approval and consent to
participate
Animal experiments were performed in the
specific-pathogen-free facility with the approval of the Korea
University Institutional Animal Care & Use Committee (approval
no. KUIACUC-2019-0040; Seoul, Republic of Korea) and according to
the governmental and institutional guidelines and Korean
regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Adams RH and Alitalo K: Molecular
regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell
Biol. 8:464–478. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shibuya M: Vascular endothelial growth
factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A
crucial target for anti- and pro-angiogenic therapies. Genes
Cancer. 2:1097–1105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Page DJ, Thuret R, Venkatraman L,
Takahashi T, Bentley K and Herbert SP: Positive feedback defines
the timing, magnitude, and robustness of angiogenesis. Cell Rep.
27:3139–3151.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Bock K, Mazzone M and Carmeliet P:
Antiangiogenic therapy, hypoxia, and metastasis: Risky liaisons, or
not? Nat Rev Clin Oncol. 8:393–404. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garcia J, Hurwitz HI, Sandler AB, Miles D,
Coleman RL, Deurloo R and Chinot OL: Bevacizumab
(Avastin®) in cancer treatment: A review of 15 years of
clinical experience and future outlook. Cancer Treat Rev.
86:1020172020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gramatzki D, Roth P, Rushing EJ, Weller J,
Andratschke N, Hofer S, Korol D, Regli L, Pangalu A, Pless M, et
al: Bevacizumab may improve quality of life, but not overall
survival in glioblastoma: An epidemiological study. Ann Oncol.
29:1431–1436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dhaun N and Webb DJ: Endothelins in
cardiovascular biology and therapeutics. Nat Rev Cardiol.
16:491–502. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosanò L, Spinella F and Bagnato A:
Endothelin 1 in cancer: Biological implications and therapeutic
opportunities. Nat Rev Cancer. 13:637–651. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin X, Jeon HM, Jin X, Kim EJ, Yin J, Jeon
HY, Sohn YW, Oh SY, Kim JK, Kim SH, et al: The ID1-CULLIN3 axis
regulates intracellular SHH and WNT signaling in glioblastoma stem
cells. Cell Rep. 16:1629–1641. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park MH, Kim AK, Manandhar S, Oh SY, Jang
GH, Kang L, Lee DW, Hyeon DY, Lee SH, Lee HE, et al: CCN1
interlinks integrin and hippo pathway to autoregulate tip cell
activity. Elife. 8:e460122019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee SY, Choi SH, Lee MS, Kurmashev A, Lee
HN, Ko YG, Lee K, Jeong S, Seong J, Kang JH and Kim H: Retraction
fibers produced by fibronectin-integrin α5β1 interaction promote
motility of brain tumor cells. FASEB J. 35:e219062021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Urup T, Staunstrup LM, Michaelsen SR,
Vitting-Seerup K, Bennedbæk M, Toft A, Olsen LR, Jønson L,
Issazadeh-Navikas S, Broholm H, et al: Transcriptional changes
induced by bevacizumab combination therapy in responding and
non-responding recurrent glioblastoma patients. BMC Cancer.
17:2782017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ferrara N: Pathways mediating
VEGF-independent tumor angiogenesis. Cytokine Growth Factor Rev.
21:21–26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gabbay E, Fraser J and McNeil K: Review of
bosentan in the management of pulmonary arterial hypertension. Vasc
Health Risk Manag. 3:887–900. 2007.PubMed/NCBI
|
|
18
|
Lugano R, Ramachandran M and Dimberg A:
Tumor angiogenesis: Causes, consequences, challenges and
opportunities. Cell Mol Life Sci. 77:1745–1770. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim IK, Kim K, Lee E, Oh DS, Park CS, Park
S, Yang JM, Kim JH, Kim HS, Shima DT, et al: Sox7 promotes
high-grade glioma by increasing VEGFR2-mediated vascular
abnormality. J Exp Med. 215:963–983. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lasorella A, Benezra R and Iavarone A: The
ID proteins: Master regulators of cancer stem cells and tumour
aggressiveness. Nat Rev Cancer. 14:77–91. 2014. View Article : Google Scholar : PubMed/NCBI
|


















