Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
seventh most common cancer in the United States, with ~65,000 new
cases in 2019 (1). Despite
advancements in diagnostics and therapeutic strategies, the 5-year
survival rate remains poor at ~50 % (2). All solid tumors typically consist of
cancer cells and the surrounding, non-malignant tumor
microenvironment (TME) (3). This
TME consists of a mixture of the extracellular matrix, endothelial
cells, fibroblasts, immune cells and human mesenchymal stem cells
(hMSCs) (3). There is a complex
interaction between tumor cells and the TME, which has the overall
effect of regulating cancer cell proliferation and tumor growth
(3). This interaction has been
proposed to also regulate the potential for metastasis and
resistance to cancer therapy (3).
These hallmarks are predominantly mediated by cytokines,
chemokines, growth factors and cell-cell contacts (4). Therefore, research focus is
increasingly being placed on the TME to deepen the understanding in
the complexity of interactions in addition to developing novel
targeted therapies (5).
One of the main components in this interaction
between tumor cells and TME are hMSCs, which are pluripotent cells
with broad self-renewal capacities (6) and are able to differentiate into
osteogenic (osteo-hMSCs), chondrogenic or adipogenic (adipo-hMSCs)
lineages (7,8). The possibility of in vitro
expansion rendered this cell type to be major targets of scientific
investigation, especially in the field of regenerative medicine and
treatment of various diseases. For example autologous
transplantation of adipose-derived mesenchymal stem cells were
evaluated for the treatment of knee osteoarthritis (9). To date, >800 clinical trials
exploring the therapeutic potential of hMSCs are already underway
(https://clinicaltrials.gov/ct2/results?cond=&term=mesenchymal+stem+cell&cntry=&state=&city=&dist=).
The differentiation capacity of hMSC can be modified by
manipulating the profile of external factors, such as growth
factors, activation and inhibition of signaling pathways or
metabolic processes (10,11). Corre et al (12) previously characterized hMSCs
isolated from healthy donors and from patients with multiple
myeloma (MM). In total, 145 genes were found to be differentially
expressed between MM and healthy hMSCs (12). Among these, 46% were involved in
tumor-microenvironment cross-talk (12). Therefore, it was hypothesized that
hMSCs can create a highly favorable niche for supporting the
survival and proliferation of the MM cells (12). In another study, Fairfield et
al (13) investigated the
effects of MM cells on the differentiation capacity and gene
expression profile of hMSCs. It was shown that MM cells altered the
gene expression profiles of hMSCs (13). In addition, a marked increase in
the expression of MM-supporting genes, including IL-6 and C-X-C
motif chemokine ligand 12, was detected (13). This previous study also indicated
that MM cells inhibited adipogenic differentiation whilst inducing
the expression of senescence-associated secretory phenotype and
pro-myeloma proteins including IL-6 and Cxcl12 (13). Battula et al (14) observed that acute myeloid leukemia
(AML) can attract hMSCs through chemotaxis and subsequently induce
osteogenic differentiation. Furthermore, it was shown that
osteo-hMSCs could enhance cancer progression (14). Another previous study revealed that
MSCs from the bone marrow of patients with primary myelofibrosis
exhibited increased osteogenic potential ex vivo (15), which appeared to serve a
particularly important role in the pathophysiology of this disease
(15). However, to the best of our
knowledge, the impact of osteogenic and adipogenic differentiation
of hMSCs on solid tumors has not been previously investigated.
Therefore, the aim of the present study was to
evaluate the effects of the interaction between HNSCC and hMSCs in
terms of the induction of differentiation and the effects of hMSC
differentiation on cancer cell proliferation in vitro.
Materials and methods
HNSCC cell lines HLaC78 and Cal27
To ensure that the most important types of HNSCC
tumors are adequately represented, the laryngeal tumor-based cell
line HLaC78 and the tongue tumor-based cell line Cal27 were chosen.
The HNSCC cell line HLaC78 was isolated from a larynx carcinoma of
a male patient by Professor Hans-Peter Zenner in the Department of
Oto-Rhino-Laryngology, Plastic, Aesthetic and Reconstructive Head
and Neck Surgery of the University Hospital (Würzburg, Germany)
(RRID: CVCL_6647) (16). The Cal27
cell line was first isolated from the tongue tumor of a 56-year-old
male patient (17), which was
purchased at American Type Culture Collection. The cells were
cultured in RPMI-1640 (Biochrom, Ltd.) containing 10% fetal calf
serum (FCS; Linaris Biologische Produkte GmbH), 1% penicillin and
streptomycin (Sigma-Aldrich, Merck KGaA) at 37°C with 5%
CO2. The medium was changed every 2 days. After reaching
70–80% confluence, cells were detached by trypsinization with 0.25%
trypsin (Gibco; Thermo Fisher Scientific, Inc.), washed with PBS,
counted before 1×106 cells were seeded into new 250-ml
culture flasks. Cells in the exponential growth phase were used for
subsequent experiments.
hMSC isolation, identification, and
culture
Bone marrow was donated by 10 voluntary patients (5
male and 5 female; mean age 63.2 years), who had undergone surgery
in the Department of Orthopedics, Koenig-Ludwig-Haus (University
Hospital Würzburg, Germany). All patients agreed by providing
written informed consent. The present study was approved by the
Ethics Committee of the Medical Faculty of the University of
Würzburg (approval no. 91/19). Bone marrow was harvested from
acetabular reaming material as waste material from patients
undergoing total hip arthroplasty surgery at the Department of
Orthopedic Surgery, under aseptic conditions, and patients with
clinical signs of osteoporosis, cancer or infectious disease were
excluded. hMSCs were isolated in accordance with the protocol of
Lee et al (18), which was
also described in detail previously (19). Briefly, hMSCs were isolated by
Ficoll (density=1.077 g/ml; Biochrom, Ltd) density gradient
centrifugation (30 min; at room temperature; 800 × g; brake and
acceleration levels set to the lowest value). After centrifugation,
a clear phase separation was observed with a clearly visible
optical dense interphase containing mononuclear cells. Cells from
this interphase were pipetted in a new 50-ml reaction tube and
subsequently washed with PBS twice. Cell culture was performed in
the expansion medium (DMEM-EM), which consisted of DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing 4.5 g/l D-Glucose, 10%
FCS (Linaris Biologische Produkte GmbH), 1% penicillin and
streptomycin (Sigma-Aldrich; Merck KGaA), whereas incubation was at
37°C and 5% CO2. hMSC morphology was evaluated by
capturing phase contrast images using an inverted light microscope
at 100× magnification (DMI 4000b Inverted Microscope, Leica
Microsystems GmbH).
Flow cytometry
According to the guidelines provided by the
International Society of Cellular Therapy (ISCT), hMSC should be
adherent to plastic surfaces and positive for the expression of
surface markers CD105, CD90 and CD73 but negative for the
expression of hematopoietic surface markers, including CD45 or CD34
(20,21). Furthermore, hMSC should demonstrate
multipotency in vitro (20,21).
Plastic adherence was assessed using inverted light microscopy at
×10-40 magnification (Leica DMI 4000b Inverted Microscope; Leica
Microsystems GmbH). hMSC surface marker expression was evaluated by
flow cytometry. After detachment, cells were washed with PBS and
cultured with 5% FCS on ice for 1 h. Afterwards, hMSCs
(1×106) were incubated with anti-CD90 (dilution 1:500;
conjugate APC; cat. no. 559869; BD Biosciences), anti-CD73
(dilution 1:50; conjugate PE; cat. no. 550257; BD Biosciences),
anti-CD45 (dilution 1:50; conjugate FITC; cat. no. 555482; BD
Biosciences), anti-CD44 (dilution 1:50; conjugate FITC; cat. no.
555478; BD Biosciences) and anti-CD34 (dilution 1:50; conjugate PE;
cat. no. 550761; BD Biosciences) antibodies for 1 h at 4°C and flow
cytometric analysis was performed (BD FACSCanto™; BD Biosciences)
and further analyzed by FACS Diva Software v5.0.3 (BD
Biosciences).
Osteogenic and adipogenic
differentiation of hMSCs
The pluripotency of hMSCs was evaluated by staining.
First, hMSC were cultured in osteogenic and adipogenic media. hMSC
control was cultured in DMEM-EM medium. The osteogenic
differentiation medium was comprised of DMEM-EM, supplemented with
10−7 M dexamethasone, 10−3 M
β-glycerophosphate and 10−4 M ascorbate-2-phosphate (all
Sigma-Aldrich; Merck KGaA). The adipogenic differentiation medium
was comprised of DMEM-EM, combined with 10−7 M
dexamethasone and 10−9 g/ml recombinant human insulin
(Sigma-Aldrich; Merck KGaA). hMSCs incubated with the osteogenic
medium were termed osteo-hMSCs whereas hMSCs incubated with the
adipogenic medium were termed adipo-hMSC at 37°C with 5%
CO2 for 3 weeks.
Staining
For the evaluation of the osteogenic
differentiation, von Kossa and Alizarin-Red staining were performed
to detect calcium mineral components. For von Kossa staining the
cells were first washed with distilled water, incubated with 1%
silver nitrate solution at room temperature (diluted in distilled
water; cat. no. #7908.1; Carl Roth) under UV-light for 20 min,
washed three times with distilled water, incubated with 5% sodium
thiosulfate pentahydrate (diluted in distilled water; cat. no.
#6516.0500; Merck KGaA), washed three times with distilled water,
incubated with Nuclear Fast Red solution for 5 min at room
temperature [5 g Aluminum sulfate hydrate (cat. no. #227617;
Sigma-Aldrich; Merck KGaA) in 100 ml distilled water, 0.1 g Nuclear
Fast Red (cat. no. #5188; Sigma-Aldrich; Merck KGaA)], washed three
times with distilled water, incubated with ascending alcohol series
and dried for microscopy. The Alizarin-Red stock solution was
prepared by dissolving 2 g of Alizarin-Red S (cat. no. #K00332679;
Merck KGaA) in 100 ml distilled water. The pH-value was adjusted at
4.1-4.3 by adding of glacial acetic acid (cat. no. #1000661000;
Merck KGaA). For staining the cells were incubated with the stock
solution for 5 min at room temperature. Before and after incubation
the cells were washed with distilled water. Adipogenic
differentiation was assessed using Oil Red O staining to reveal
intracellular lipid droplets. For preparing of the Oil Red O
staining stock solution 0.5 g Oil Red O (cat. no. #O0625;
Sigma-Aldrich; Merck KGaA) was dissolved in 100 ml Propylenglycol
(cat. no. #P4347; Sigma-Aldrich; Merck KGaA) at 60°C. For the
staining procedure the cells was washed with distilled water,
incubated with Propylenglycol for 5 min at room temperature, then
with 60°C warm Oil Red O stock solution for 10 min, washed with
Propylenglycol, washed three times with distilled water and stained
with Mayers Hematoxylin-solution (cat. no. #1.09249; Merck KGaA)
for 30 sec. Until microscopy the cells were covered with PBS. All
images were acquired with a light microscope (DMI 4000b Inverted
Microscope; Leica Microsystems GmbH).
Reverse transcription-quantitative
(RT-q) PCR
RT-qPCR was used to verify hMSC differentiation. The
following primers were chosen for osteogenic differentiation: i)
Alkaline phosphatase (ALPL; cat. no. 4331182; assay ID,
Hs01029144_m1); ii) osteocalcin (BGLP; cat. no. 4331182; assay ID,
Hs01587814_g1); iii) collagen 1 (Col 1; cat. no. 4331182; assay ID,
Hs0016004_m1); and iv) runt-related transcription factor 2 (RUNX-2;
cat. no. 4331182; assay ID, Hs00231692_m1). The following primers
were chosen for adipogenic differentiation: i) fatty acid binding
protein 4 (FABP4; cat. no. 4331182; assay ID, Hs01086177_m1); ii)
leptin (LEP; cat. no. 4331182; assay ID, Hs00174877_m1); and iii)
lipoprotein lipase (LPL; cat. no. 4331182; assay ID,
Hs00173425_m1). GAPDH (cat. no. 4331182; assay ID, Hs02758991_g1)
was used as the housekeeping gene. All primers were purchased from
Thermo Fisher Scientific, Inc., the primer sequences of which are
not publicly available. RT-qPCR was performed as follows: For total
RNA extraction from hMSCs an RNeasy Kit (Qiagen GmbH) was used
according to the manufacturer's protocol. For reverse
transcription, the isolated RNA was converted into cDNA using
SuperScript™ VILO™ Master Mix (cat. no. #11755-500; Invitrogen;
Thermo Fisher Scientific, Inc.). The following temperature protocol
was used for reverse transcription: 25°C for 10 min; 42°C for 59
min; 85°C for 5 min; 4°C for 2 min. Subsequent qPCR was performed
using SYBR Green Real-Time PCR Master Mix (Thermo Fisher
Scientific, Inc.) in a StepOnePlus™ thermocycler system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The first denaturation
step was 10 min at 95°C. Afterwards the following thermocycling
protocol was utilized for 40 cycles: 50°C for 2 min, 60°C for 1 min
and 95°C for 15 sec. The 2−ΔΔCq method was applied to
quantify the relative gene expression levels (22). The gene expression values are then
normalized to those on of the hMSC control.
HNSCC cells and hMSC co-culture
The co-culture experiments were performed in
Transwell systems with a polyester membrane and pore size of 0.4 µm
(Costar® Transwell®; Corning, Inc.). After
seeding 60,000 hMSCs into the lower chambers of 12-well plates in
DMEM-EM medium and microscopic confirmation of adherence, 60,000
HLaC78 and 60,000 Cal27 cells were seeded onto the Transwell
inserts in DMEM-EM medium. hMSC without co-cultivation served as
the control. Cells were kept in this co-culture system for 3 weeks
at 37°C and 5% CO2. The DMEM-EM medium was changed every
2 days. After a period of 21 days, hMSC differentiation into
osteogenic and adipogenic lineages were determined using staining
and RT-qPCR. This experiment was repeated 10 times using hMSCs from
all 10 different donors.
Effects of hMSC on HLaC78 and Cal27
proliferation in co-culture and with conditioned media
The viability of HLaC78 and Cal27 cells co-cultured
with hMSCs, adipo-hMSCs or osteo-hMSCs was measured by counting the
cells using an electronic cell counter (CASY Cell Counter; OMNI
Life Science GmbH). Identical measurements were also performed
after the cultivation of the two HNSCC tumor cell lines with the
conditioned medium of hMSCs (hMSC-CM), adipo-hMSC (adipo-hMSC-CM)
and osteo-hMSC (osteo-hMSC-CM) at 37°C for 3 days. Conditioned
media were obtained after 3 days at 37°C of hMSC, adipo-hMSC and
osteo-hMSC incubation with DMEM-EM. Before the conditioning process
the differentiation media was removed.
Cytokine analysis using dot blot
assay
The Human Cytokine Array C3 dot blot assay (cat. no.
AAH-CYT-3-4; Raybiotech, Inc.) was used to measure hMSC, adipo-hMSC
and osteo-hMSC cytokine secretion after incubation with their
respective differentiation media for 3 weeks. After removing of the
differentiation media hMSCs, adipo-hMSCs and osteo-hMSCs were first
incubated with DMEM without any supplements. After a period of 48 h
at 37°C, the supernatants of the hMSCs from the 10 patients were
then collected and pooled. The cytokine profile was analyzed
according to the manufacturer's protocol. The chemiluminescence was
assessed using an X-ray film. Semi-quantitative detection of IL-6
concentration was performed by density measurements using the
ImageJ software (version 1.52a; National Institutes of Health) in
relation to the positive control dot density. According to the
manufacturer's declarations, the signal of the positive control
spots is associated with the amount of biotinylated antibody
printed onto the array.
Quantitative measurements of IL-6 by
ELISA
Supernatants of the Cal27 cell culture after
co-culture with hMSCs or incubation with the hMSC-CM for 3 days at
37°C were collected and analyzed for IL-6 levels using the ELISA
kit human IL-6 (cat. no. 950.030.096; Diaclone SAS). All
experiments were repeated with hMSCs from seven donors. The plates
were read out at 450 nm (Titertek Multiskan PLUS; Thermo Fisher
Scientific, Inc.). The standard curve was created by recombinant
IL-6.
STAT3 protein analysis in Cal27 and
HLaC78 cells by western blotting
Cal27 and HlaC78 cells were incubated with hMSC-CM
with or without 5 µg/ml anti-IL6 (cat. No. MAB2061; R&D
Systems, Inc.), adipo-hMSC-CM and osteo-hMSC-CM at 37°C for 2 days.
After washing the Cal27 and HlaC78 cells with PBS, they were
harvested using a cell scraper and dissolved in RIPA buffer (PBS
containing 1% NP40, 0.5% sodium deoxycholate and 0.1% SDS)
supplemented with 10 µg/ml phenylmethanesulfonyl fluoride. Protein
determination was performed by bicinchoninic acid method (Pierce
BCA Protein Assay Kit; cat. no. #23227; Thermo Fisher Scientific,
Inc.) Equal amounts (20 µg) of the total protein lysates were
separated in a 10% SDS-polyacrylamide gel, before they were
transferred onto polyvinylidene difluoride membranes. The membranes
were blocked for 1 h at room temperature with TBST (10 mM Tris, 150
mM NaCl and 0.05% Tween-20, pH 8.0) containing 5% non-fat dry milk.
The membranes were next incubated with the primary antibodies
against phosphorylated (p-) STAT3 (1:1,000; rabbit; cat. No. 9145;
Cell Signaling Technology, Inc.), STAT3 (1:2,000; rabbit; cat. No.
12640; Cell Signaling Technology, Inc.) and β-actin (1:2,000;
mouse; cat. No. MA5-15739; Thermo Fisher Scientific, Inc.)
overnight at 4°C. The membranes were then washed with TBST (Tween
0.1%) and incubated with a species-specific HRP-conjugated IgG
secondary antibody (1:10,000; cat. no. 7074; Cell Signaling
Technology, Inc.) for 1 h at room temperature. The bands were
visualized using a chemiluminescence system (iBright1500;
Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
Statistical analysis
All data were transferred into standard
spreadsheets. Differences between groups were examined for
significance, with one-way ANOVA performed using GraphPad Prism 6.0
statistics software (GraphPad Software, Inc.). For post hoc testing
Dunnett´s multiple comparisons test was used (Fig. 3), for multiple comparisons Tukey's
test was used (Fig. 5, Fig. 6, Fig.
7). All results were presented as mean ± SD. P<0.05 was
considered to indicate a statistically significant difference and
marked with an asterisk.
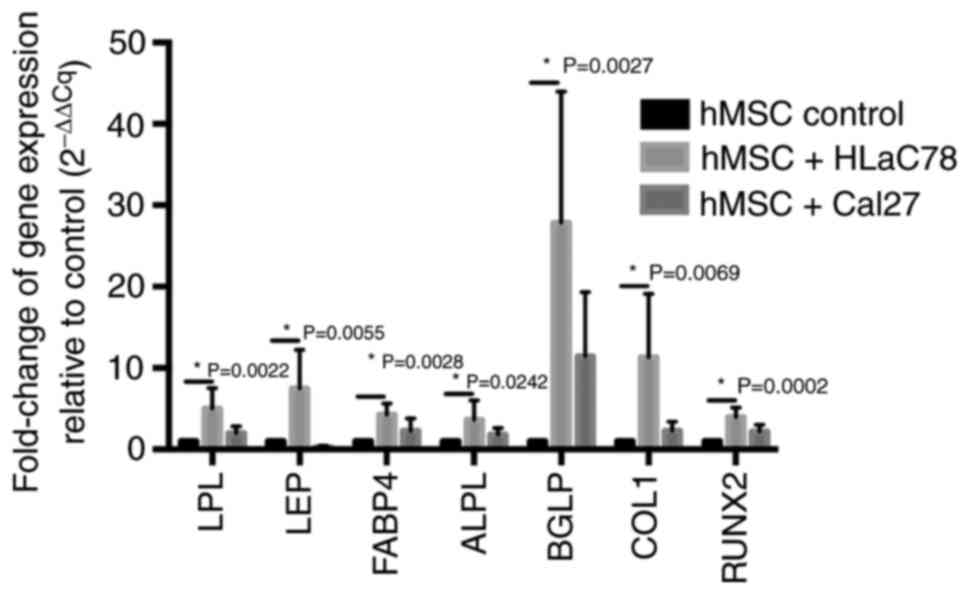 | Figure 3.Measurement of adipogenic and
osteogenic marker expression in hMSCs after co-culture with head
and neck squamous cell carcinoma cell lines Cal27 and HLaC78. Fold
change in the mRNA expression of adipogenic (LPL, LEP and FABP4)
and osteogenic (ALPL, BGLP, COL1 and RUNX2) differentiation markers
in hMSCs is elevated after 3 weeks of co-culture with Cal27 and
HLaC78 cells relative to hMSC control. hMSC control was cultured
under the same conditions with DMEM-EM. n=5 different hMSC donors.
hMSCs, human mesenchymal stem cells; LPL, lipoprotein lipase; LEP,
leptin; FABP4, fatty acid binding protein 4; ALPL, alkaline
phosphatase; BGLP, osteocalcin; COL1, collagen 1; RUNX2,
runt-related transcription factor 2. |
 | Figure 7.Cytokine assay of pooled hMSC
supernatants as assessed using dot blot assay and ELISA. The levels
of cytokine secretion after hMSC differentiation were measured.
Representative dot blot images of (A) hMSCs, (B) adipo-hMSCs and
(C) osteo-hMSCs. (D) Densitometric analysis of IL-6 (marked with
white boxes in A-C) relative to that of control dots. (E) Map of
cytokines analyzed using dot blot assay shown in (A-C). ELISA of
IL-6-concentration the culture supernatant of Cal27 cells (F) after
they were incubated with hMSC-CM, adipo-hMSC-CM and osteo-hMSC-CM
or (G) co-cultured with hMSCs, adipo-hMSCs and osteo-hMSCs. hMSCs,
human mesenchymal stem cells; CM, conditioned medium; nat-, native;
adipo-, adipogenic lineage; osteo-, osteogenic lineage; ENA-78,
C-X-C motif chemokine 5; GCSF, granulocyte-colony stimulating
factor; GM-CSF, granulocyte macrophage-colony stimulating factor;
GRO, C-X-C motif ligand 1; MCP, monocyte chemoattractant protein;
MCSF, macrophage-colony stimulating factor; MDC, macrophage-derived
chemokine; MIG, C-X-C motif ligand 9; MIP, macrophage inflammatory
protein; RANTES, regulated on activation, normal T expressed and
secreted; SCF, stem cell factor; SDF1, stromal cell-derived factor
1; TARC, thymus- and activation-regulated chemokine; IGF-1,
insulin-like growth factor-1; PDGF-BB, platelet-derived growth
factor. |
Results
hMSC morphology and differentiation
capability
The hMSCs exhibited a fibroblast-shaped morphology
when observed using microscopy (Fig.
1A). The Oil Red O, von Kossa and Alizarin Red staining
revealed characteristics of osteogenic and adipogenic
differentiation in osteo-hMSCs and adipo-hMSCs, respectively
(Fig. 1). The successful
osteogenic and adipogenic differentiation of hMSC was verified by
qPCR.
To evaluate the extent of hMSC differentiation
towards the osteogenic and adipogenic lineages, RT-qPCR was
performed. hMSCs incubated in DMEM-EM without differentiation
medium served as the control. After 1 week of incubation with
either osteogenic or adipogenic media, the expression of adipogenic
cell markers FABP4, LEP and LPL and osteogenic cell markers ALPL,
BGLP, Col 1 and RUNX-2 were measured. Compared with that in the
control group, adipogenic differentiation medium induced a
229.4-fold increase in FABP4 expression, a 275.4-fold increase of
LPL expression and a 206.6-fold increase of LEP expression
expression. In terms of osteogenic differentiation, there was a
4.2-fold increase in ALPL expression, a 3.7-fold increase in BGLP
expression, a 1.5-fold increase in Col 1 expression and a 1.4-fold
increase in RUNX2 expression (Table
I).
 | Table I.Reverse transcription-quantitative
PCR analysis of adipogenic or osteogenic differentiation marker
expression in hMSCs after incubation for 1 week with adipogenic or
osteogenic differentiation medium relative to untreated control
cellsa. |
Table I.
Reverse transcription-quantitative
PCR analysis of adipogenic or osteogenic differentiation marker
expression in hMSCs after incubation for 1 week with adipogenic or
osteogenic differentiation medium relative to untreated control
cellsa.
| hMSC type | FABP4 | LPL | LEP | ALPL | BGLP | Col 1 | RUNX2 |
|---|
| hMSC | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Adipo-hMSC | 229.9±295.4 | 275.4±370.1 | 206.6±204.3 | 2.6±2.5 | 0.4±0.46 | 1.6±0.4 | 1.5±0.8 |
|
Osteo-hMSC | 0.4±0.4 | 0.8±0.9 | 0.7±0.7 | 4.2±3.5 | 3.7±6.9 | 1.5±0.3 | 1.4±0.6 |
According to flow cytometry analysis, the hMSCs were
found to express surface markers CD90, CD73 and CD44 (Fig. 2). By contrast, hematopoietic
markers CD45 and CD34 could not be detected (Fig. 2).
Cal27 and HLaC78 promote the
osteogenic and adipogenic differentiation of hMSCs
Co-culturing hMSCs with Cal27 or HLaC78 increased
the expression of osteogenic and adipogenic lineages markers
measured by RT-qPCR, compared with that in control cells (Fig. 3). However there was only a slightly
increase of adipogenic markers after co-culturing hMSCs with Cal27.
Furthermore, compared with that in the control group, Oil Red O
staining of hMSCs co-cultured with Cal27 and HLaC78 cells showed
markedly higher lipid droplet production (Fig. 4A-C). According to von Kossa
staining, the quantity of calcium deposits was only increased
slightly after hMSC co-cultivation with Cal27, but was more notably
increased after co-cultivation with HLaC78 cells (Fig. 4D-F).
Effects of hMSC, adipo-hMSC and
osteo-hMSC on Cal27 and HLaC78 cell viability in co-culture
systems
After co-cultivation of hMSC, adipo-hMSC and
osteo-hMSC with Cal27 and HLaC78 cells, the number of HNSCC cells
were counted. The number of Cal27 cells was increased significantly
after co-cultivation with adipo-hMSC and osteo-hMSC compared with
that in the groups of Cal27 cells that were not co-cultured
(Fig. 5). Furthermore, there was
an increased Cal27 cell count after co-cultivation with adipo-hMSC
compared with hMSC (Fig. 5). In
addition, the count of viable HLaC78 cells was significantly higher
after co-cultivation with adipo-hMSC (Fig. 5). However, co-culturing with
undifferentiated hMSCs did not alter the number of Cal27 and HLaC78
cells compared with that in monoculture cells (Fig. 5). Furthermore, there was no
significant difference in the viability of HLaC78 cells after
co-cultivation with osteo-hMSCs (Fig.
5).
Effects of hMSC-CM, adipo-hMSC-CM and
osteo-hMSC-CM on Cal27 and HLaC78 cell viability
Following the treatment of Cal27 and HLaC cells with
media conditioned by hMSCs, increased cell viability was observed.
Adipo-hMSC-CM treatment significantly enhanced tumor cell viability
compared with that in cells treated with osteo-hMSC-CM (Fig. 6). There was no statistically
significant difference in the cell count compared to the incubation
with the control groups DMEM or hMSC-CM (Fig. 6).
Differences in cytokine secretion by
hMSC, adipo-hMSC and osteo-hMSC
Dot blot assay was used to investigate the profile
of cytokine secretion in hMSCs, adipo-hMSCs and osteo-hMSCs. Due to
the high expression level and their central role in tumour growth
stimulation and inflammation, IL-6 was chosen for further detailed
analysis. The secretion of IL-6 by adipo-hMSCs was markedly higher
compared with that by osteo-hMSCs, but lower compared with in hMSCs
(Fig. 7).
Differences in cytokine secretion of
Cal27 cells after co-culture
According to ELISA, IL-6 levels in the Cal27 cell
culture supernatants were markedly increased after incubation with
hMSC-CM and co-cultivation of Cal27 with hMSCs, compared with those
in the supernatant of control Cal27 cells incubated with DMEM
(Fig. 7F and G). In addition,
comparably high levels of IL-6 were detected after incubation of
Cal27 cells with hMSC-CM and in those co-cultured with osteo-hMSCs
compared to Cal27 cells incubated with DMEM (Fig. 7F and G).
STAT3 activation in Cal27 and HLaC78
cells
STAT3 activation at protein level was next evaluated
by western blotting. Cal27 and HLaC78 cells were cultured with
hMSC-CM, adipo-hMSC-CM and osteo-hMSC-CM. Furthermore to evaluate
the influence of IL-6 on STAT3-activation, anti-IL-6 was added to
hMSC-CM, adipo-hMSC-CM and osteo-hMSC-CM. The cultivation of Cal27
and HLaC78 cells with DMEM-EM served as the control. Markedly
enhanced activation of STAT3 was observed after the treatment of
Cal27 cells with CM compared with that in the control group
(Fig. 8). However this was not
observable for HLaC78 cells. The adipogenic or osteogenic
differentiation had no influence on the level of STAT3-activation.
The addition of anti-IL-6 reduced the STAT3 phosphorylation.
Discussion
Tumors are comprised of malignant cells surrounded
by a complex TME that contains different cell types, including
fibroblasts, endothelial cells, hMSCs, innate and adaptive immune
cells (23). A number of studies
previously reported an important role of hMSC in cancer pathology,
such as head and neck cancer (3,24).
hMSCs consist of a heterogenic cell population with a range of
properties, including migration towards wounds, immune modulation
and enhancement of wound repair (25–27).
Furthermore, hMSC have the reported ability to differentiate into
cancer-associated fibroblasts (28–30).
Mishra et al (30)
demonstrated the trans-differentiation of hMSCs after exposure to
breast cancer cell-conditioned media. In addition, another previous
study showed that AML cells can induce chemotactic effects on hMSCs
and osteogenic differentiation of these migrated cells (14). Osteogenic differentiation mediated
an important impact on AML cell proliferation by an enhanced
leukemia engraftment in a transgenic mouse model (14). However, to the best of our
knowledge, no evidence of HNSCC-induced hMSC differentiation exists
to date.
In the present study, differentiation of hMSC
towards both adipogenic and osteogenic lineages was shown after
co-culturing with Cal27 and HLaC78 cells. However, spontaneous
differentiation was previously described as an effect of hMSC aging
in long-term cultures (31).
Compared with that in control cells, without co-culturing with
tumor cells, a higher rate of differentiation of hMSC into
adipogenic and osteogenic cells in terms of morphology in addition
to the expression of their markers, which was revealed by RT-qPCR.
The effects of these differentiated hMSCs on tumor biology remains
poorly understood. Tu et al (32) found an inhibition of the cancer
cell survival by hMSCs in an osteosarcoma model though
TGF-β/Smad2/3 signaling. In this previous study, an increase of
VEGF- and IL-6-expession in hMSCs was observed (32). Furthermore, Paino et al
(33) previously investigated the
potential effects of SAOS2 and MCF7 cancer cell lines on hMSC
differentiation. Neither alterations in the expression hMSC surface
markers, including CD90, CD29 and vimentin, nor variations in the
expression of transcription factors Twist and Slug, could be
observed (33). However, an
upregulation in the expression of stemness genes, such as OCT3/4
and Nanog, was observed (33).
During the pathogenesis of breast cancer, adipocytes
serve an important role (34).
They are one of the main components of the breast microenvironment,
where they have the ability to provide pro-tumorigenic signals
(34). In the present study,
differentiation of hMSCs towards adipocytes led to an enhancement
of HNSCC cell viability. Furthermore, an enhanced activation of
STAT3 in Cal27 cells was found after cultivation with hMSC-CM,
adipo-hMSC-CM or osteo-hMSC-CM. The STAT3-activation was reduced
after adding anti-IL-6.
A potential reason for this pro-tumorigenic effect
of adipo-hMSC may be the paracrine secretion of IL-6. STAT3 is
activated particularly by the IL-6 family of cytokines, which
include IL-6, IL-8, IL-11 and Oncostatin (35). However, IL-6 is the most potent
activator of STAT3 (36). Adipose
tissue is a key source of IL-6, which produces 33% IL-6 found in
the plasma (37). However,
comparably low concentrations of IL-6 were found in the
adipo-hMSC-CM and in the supernatant of Cal27 cells co-cultured
with adipo-hMSCs. Therefore, differences in STAT3 activation and
cell viability could not be explained solely by effects mediated by
IL-6.
The effects of osteo-hMSCs on HNSCC cells were found
to be ambiguous. Cultivation of Cal27 cells with osteo-hMSCs
resulted in a positive effect on cell viability, evaluating the
bi-directional effects, based on the reciprocal influences of hMSCs
and tumour cells. However, no such effects could be detected on
HLaC78 cells. This raised the question of whether cell viability
was influenced by the cultivation of Cal27 and HLaC78 cells with
osteo-hMSC-CM. No statistically significant effects could be
detected after the cultivation of Cal27 and HLaC78 cells with
osteo-hMSC-CM. One possible explanation could be the low
concentrations of IL-6 in the osteo-hMSC-CM, compared with those in
hMSC-CM and adipo-hMSC-CM media as shown in the dot blot analysis.
However, these results remain ambiogious according to the IL-6
ELISA-measurements. Therefore, IL-6 alone is not sufficient to
explain the differences in STAT3 activation and cell viability, in
addition to differences in the osteogenic differentiated
lineages.
A large degree of variability was found in the
present study, with high standard deviations in almost all data.
Furthermore, there was ambiguous observations in only a slightly
increase of adipogenic markers in qPCR, but a clear uptake of lipid
droplets in the Oil Red O staining after co-culturing hMSCs with
Cal27, and in addition only a slightly increase of ossification in
the von Kossa staining and at the same time an increase of
osteogenic markers in qPCR for Cal27. One reason for the mismatch
of pPCR and morphology results could be that qPCR results represent
the mRNA level and the mRNA level does not in every case correlate
with the protein level. Another explanation for this finding could
be the biological behavior of hMSCs in vitro. Despite
characterizing hMSCs by their ability to adhere to plastic,
cellular morphology and expression of different cell surface
markers, these hMSCs remain highly heterogenic. This heterogeneity
can be influenced by age, sex or the immune status of donors in
addition to the culture conditions (38,39).
Since the donors of hMSCs exhibited high variability in age, sex
and immune status, this heterogeneity may have led to these
ambiguous results.
For future investigations the impact of the
chrondrogenic differentiation lineage of hMSCs should be focused
upon. Furthermore, the use of ≥ two different cell lines from a
specific cancer would be beneficial to focus any studies into the
molecular mechanism.
In conclusion, data from the present study suggest
that co-cultivation of hMSCs with Cal27 and HLaC78 cells can
promote hMSC differentiation into adipogenic and osteogenic
lineages. Furthermore, pro-tumorigenic effect of hMSCs
differentiated towards adipogenic lineage was observed. One
possible mechanism was the increased STAT3 activation in Cal27 and
HLaC78 cells incubated with adipo-hMSC-CM. Therefore, further
investigations into the underlying mechanisms are highly
warranted.
Acknowledgements
The authors would like to thank Mr. Michael Kessler
and Mrs. Silke Hummel (Department of Oto-Rhino-Laryngology,
Plastic, Aesthetic and Reconstructive Head and Neck Surgery,
University Hospital Würzburg, Würzburg, Germany) for their
technical support in their role as technician assistants.
Funding
The present study was funded by the Interdisciplinary Centre for
Clinical Science (IZKF) at the University of Würzburg (to TJM;
grant no. Z-2/78).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AS and SH designed the study. AS, TJM and MS
performed the experiments. MH, TG, RH and NK were involved in data
interpretation and critically reviewed the manuscript. TJM and AS
confirm the authenticity of all the raw data. All authors read and
approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Medical Faculty of the University of Würzburg
(approval no. 91/19). The present study was performed in accordance
with the World Medical Association Declaration of Helsinki. All
participants gave an informed written consent.
Patient consent for participation
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gupta S, Kong W, Peng Y, Miao Q and
Mackillop WJ: Temporal trends in the incidence and survival of
cancers of the upper aerodigestive tract in Ontario and the United
States. Int J Cancer. 125:2159–2165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bergfeld SA and DeClerck YA: Bone
marrow-derived mesenchymal stem cells and the tumor
microenvironment. Cancer Metastasis Rev. 29:249–261. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmitz S and Machiels JP: Targeting the
tumor environment in squamous cell carcinoma of the head and neck.
Curr Treat Options Oncol. 17:372016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ausoni S, Boscolo-Rizzo P, Singh B, Da
Mosto MC, Spinato G, Tirelli G, Spinato R and Azzarello G:
Targeting cellular and molecular drivers of head and neck squamous
cell carcinoma: Current options and emerging perspectives. Cancer
Metastasis Rev. 35:413–426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Friedman MS, Long MW and Hankenson KD:
Osteogenic differentiation of human mesenchymal stem cells is
regulated by bone morphogenetic protein-6. J Cell Biochem.
98:538–554. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Short B, Brouard N, Occhiodoro-Scott T,
Ramakrishnan A and Simmons PJ: Mesenchymal stem cells. Arch Med
Res. 34:565–571. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freitag J, Bates D, Wickham J, Shah K,
Huguenin L, Tenen A, Paterson K and Boyd R: Adipose-derived
mesenchymal stem cell therapy in the treatment of knee
osteoarthritis: A randomized controlled trial. Regen Med.
14:213–230. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baron R and Rawadi G: Targeting the
Wnt/beta-catenin pathway to regulate bone formation in the adult
skeleton. Endocrinology. 148:2635–2643. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tencerova M, Rendina-Ruedy E, Neess D,
Faergeman N, Figeac F, Ali D, Danielsen M, Haakonsson A, Rosen CJ
and Kassem M: Metabolic programming determines the
lineage-differentiation fate of murine bone marrow stromal
progenitor cells. Bone Res. 7:352019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Corre J, Mahtouk K, Attal M, Gadelorge M,
Huynh A, Fleury-Cappellesso S, Danho C, Laharrague P, Klein B, Rème
T and Bourin P: Bone marrow mesenchymal stem cells are abnormal in
multiple myeloma. Leukemia. 21:1079–1088. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fairfield H, Costa S, Falank C, Farrell M,
Murphy CS, D'Amico A, Driscoll H and Reagan MR: Multiple myeloma
cells alter adipogenesis, increase senescence-related and
inflammatory gene transcript expression, and alter metabolism in
preadipocytes. Front Oncol. 10:5846832020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Battula VL, Le PM, Sun JC, Nguyen K, Yuan
B, Zhou X, Sonnylal S, McQueen T, Ruvolo V, Michel KA, et al:
AML-induced osteogenic differentiation in mesenchymal stromal cells
supports leukemia growth. JCI Insight. 2:e900362017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Martinaud C, Desterke C, Konopacki J,
Pieri L, Torossian F, Golub R, Schmutz S, Anginot A, Guerton B,
Rochet N, et al: Osteogenic potential of mesenchymal stromal cells
contributes to primary myelofibrosis. Cancer Res. 75:4753–4765.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zenner HP, Lehner W and Herrmann IF:
Establishment of carcinoma cell lines from larynx and submandibular
gland. Arch Otorhinolaryngol. 225:269–277. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gioanni J, Fischel JL, Lambert JC, Demard
F, Mazeau C, Zanghellini E, Ettore F, Formento P, Chauvel P and
Lalanne CM: Two new human tumor cell lines derived from squamous
cell carcinomas of the tongue: Establishment, characterization and
response to cytotoxic treatment. Eur J Cancer Clin Oncol.
24:1445–1455. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee RH, Kim B, Choi I, Kim H, Choi HS, Suh
K, Bae YC and Jung JS: Characterization and expression analysis of
mesenchymal stem cells from human bone marrow and adipose tissue.
Cell Physiol Biochem. 14:311–324. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Scherzed A, Hackenberg S, Froelich K,
Kessler M, Koehler C, Hagen R, Radeloff A, Friehs G and Kleinsasser
N: BMSC enhance the survival of paclitaxel treated squamous cell
carcinoma cells in vitro. Cancer Biol Ther. 11:349–357. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Horwitz EM, Le Blanc K, Dominici M,
Mueller I, Slaper-Cortenbach I, Marini FC, Deans RJ, Krause DS and
Keating A; International Society for Cellular Therapy, :
Clarification of the nomenclature for MSC: The international
society for cellular therapy position statement. Cytotherapy.
7:393–295. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop D and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The international society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Poggi A and Giuliani M: Mesenchymal
stromal cells can regulate the immune response in the tumor
microenvironment. Vaccines (Basel). 4:412016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Scherzad A, Taeger J, Gehrke TE, Hagen R,
Kleinsasser N and Hackenberg S: Mesenchymal stem cells: Cancer
promoting effects or tumor suppression-A current overview.
Laryngorhinootologie. 97:678–687. 2018.PubMed/NCBI
|
|
25
|
Xia X, Chen W, Ma T, Xu G, Liu H, Liang C,
Bai X, Zhang Y, He Y and Liang T: Mesenchymal stem cells
administered after liver transplantation prevent acute
graft-versus-host disease in rats. Liver Transpl. 18:696–706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prockop DJ, Gregory CA and Spees JL: One
strategy for cell and gene therapy: Harnessing the power of adult
stem cells to repair tissues. Proc Natl Acad Sci USA. 100 (Suppl
1):S11917–S11923. 2003. View Article : Google Scholar
|
|
27
|
Cuiffo BG and Karnoub AE: Mesenchymal stem
cells in tumor development: Emerging roles and concepts. Cell Adh
Migr. 6:220–230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Q, Chai S, Wang W, Wan C, Zhang F,
Li Y and Wang F: Macrophages activate mesenchymal stem cells to
acquire cancer-associated fibroblast-like features resulting in
gastric epithelial cell lesions and malignant transformation in
vitro. Oncol Lett. 17:747–756. 2019.PubMed/NCBI
|
|
29
|
Spaeth EL, Dembinski JL, Sasser AK, Watson
K, Klopp A, Hall B, Andreeff M and Marini F: Mesenchymal stem cell
transition to tumor-associated fibroblasts contributes to
fibrovascular network expansion and tumor progression. PLoS One.
4:e49922009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mishra PJ, Mishra PJ, Humeniuk R, Medina
DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW and Banerjee D:
Carcinoma-associated fibroblast-like differentiation of human
mesenchymal stem cells. Cancer Res. 68:4331–4339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Z, Liu C, Xie Z, Song P, Zhao RC, Guo
L, Liu Z and Wu Y: Epigenetic dysregulation in mesenchymal stem
cell aging and spontaneous differentiation. PLoS One. 6:e205262011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tu B, Peng ZX, Fan QM, Du L, Yan W and
Tang TT: Osteosarcoma cells promote the production of pro-tumor
cytokines in mesenchymal stem cells by inhibiting their osteogenic
differentiation through the TGF-β/Smad2/3 pathway. Exp Cell Res.
320:164–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paino F, La Noce M, Di Nucci D, Nicoletti
GF, Salzillo R, De Rosa A, Ferraro GA, Papaccio G, Desiderio V and
Tirino V: Human adipose stem cell differentiation is highly
affected by cancer cells both in vitro and in vivo: Implication for
autologous fat grafting. Cell Death Dis. 8:e25682017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wolfson B, Eades G and Zhou Q: Adipocyte
activation of cancer stem cell signaling in breast cancer. World J
Biol Chem. 6:39–47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Banerjee K and Resat H: Constitutive
activation of STAT3 in breast cancer cells: A review. Int J Cancer.
138:2570–2578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Heinrich PC, Behrmann I, Haan S, Hermanns
HM, Muller-Newen G and Schaper F: Principles of interleukin
(IL)-6-type cytokine signalling and its regulation. Biochem J.
374:1–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Galic S, Oakhill JS and Steinberg GR:
Adipose tissue as an endocrine organ. Mol Cell Endocrinol.
316:129–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pacini S: Deterministic and stochastic
approaches in the clinical application of mesenchymal stromal cells
(MSCs). Front Cell Dev Biol. 2:502014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pevsner-Fischer M, Levin S and Zipori D:
The origins of mesenchymal stromal cell heterogeneity. Stem Cell
Rev. 7:560–568. 2011. View Article : Google Scholar : PubMed/NCBI
|






















