Introduction
Endometrial cancer (EC) is a common gynecological
malignancy that endangers the lives of women worldwide (1,2).
Meanwhile, the incidence of EC is increasing, not only in developed
countries but also in developing ones, which renders EC an enormous
threat to public health (3,4). It
is proposed that obesity, diabetes, polycystic ovary syndrome and
Lynch syndrome are risk factors for EC; molecular abnormalities
such as PI3K/AKT and Wnt/β-catenin pathway mutations are also
highly associated with EC (5–8).
Currently, treatment strategies for EC mainly include surgical
resection, radiotherapy, hormone therapy and immunotherapy
(9,10); while efforts have never stopped in
the search for novel treatment targets of EC.
Integrins are a group of vital proteins that
regulate cell adhesion and signaling transduction, among which
integrin α7 (ITGA7) is involved in the pathogenesis and progression
of several types of cancer (11,12).
For example, ITGA7 knockdown suppresses cell proliferation, induces
apoptosis, reduces CD44 and CD133 expression levels as well as
decreases sensitivity to cisplatin in tongue squamous cell
carcinoma cell lines (13).
Another study revealed that ITGA7 modulates cell proliferation,
apoptosis and stemness through the PI3K/AKT pathway in
hepatocellular carcinoma (14).
Moreover, a previous study also revealed how important the
regulation of ITGA7 is for cell function in non-small cell lung
cancer (15). However, regarding
gynecological malignancy, only one previous study demonstrated that
ITGA7 is downregulated in high grade serous ovarian cancer tissues
compared with that of normal tissues (16). Based on the aforementioned
information, it was hypothesized that ITGA7 may also be involved in
the pathogenesis and progression of EC.
The present study aimed to investigate the effect of
ITGA7 knockdown on cell proliferation, apoptosis, invasion and its
potential downstream pathway in EC cell lines, then explore its
association with clinicopathological features in patients with
EC.
Materials and methods
Cell source and culture
conditions
Telomerase-immortalized human endometrial stromal
cells (THESCs) and human EC cell lines (including HEC-1A, RL95-2,
Ishikawa and KLE) were purchased from the American Type Culture
Collection or European Collection of Authenticated Cell Cultures.
THESCs, RL95-2 and KLE cells were cultured in DMEM/F-12 (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.). HEC-1A cells were
cultured in Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
containing 15% FBS (Gibco; Thermo Fisher Scientific, Inc.).
Ishikawa cells were cultured in MEM (MilliporeSigma) containing 5%
FBS (Gibco; Thermo Fisher Scientific, Inc.). The incubation
conditions were 5% CO2, 37°C for all the cells.
Detection of ITGA7 expression in EC
cell lines and THESCs
The ITGA7 mRNA and protein relative expression
levels in THESCs and EC cell lines (including HEC-1A, RL95-2,
Ishikawa and KLE) were analyzed using reverse
transcription-quantitative PCR (RT-qPCR) and western blot assays,
respectively, as described below.
Transfection of short interfering
(si)RNAs
ITGA7 siRNAs (si-ITGA7) and scrambled siRNA as a
negative control (si-NC) were purchased from Generay Biotech Co.,
Ltd. Ishiwaka, RL95-2 cells and THESCs were transfected with 50 nM
si-ITGA7 and 50 nM si-NC using HilyMAX reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), respectively, based on the manufacturer's
protocol. The siRNA sense sequences were as follows: Si-ITGA7-1,
5′-CAGCUACUUUGGCUUCUCUUU-3′; si-ITGA7-2,
5′-CAGCUACUUUGGCUUCUCUUU-3′; si-ITGA7-3,
5′-GGGUCUGUUUCAGCUACAUUU-3′;
si-NC,5′-GAAUUAAUUAAAGAUGGCCCGUUGUACU-3′.
RT-qPCR
THESCs, Ishiwaka and RL95-2 cells were harvested at
48 h after transfection. Total RNA of each group was extracted
using PureZOL RNA isolation reagent (Bio-Rad Laboratories, Inc.).
Qubit® 4 Flurometer (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for analyzing the RNA concentration.
Reverse transcription of RNA was performed using QuantiNova Reverse
Transcription Kit in accordance with the manufacturer's protocol
(Qiagen GmbH). qPCR was performed using QuantiNova SYBR®
Green PCR kit (Qiagen GmbH), and the following thermal cycles were
conducted: 95°C For 2 min, 1 cycle; 95°C for 5 sec; and 61°C for 30
sec, 40 cycles. Primers were obtained from Sangon Biotech Co., Ltd.
The primer sequences were listed as follows: ITGA7 Forward,
5′-GCCACTCTGCCTGTCCAATG-3′, and reverse,
5′-GGAGGTGCTAAGGATGAGGTAGA-3′. GAPDH forward,
5′-GAGTCCACTGGCGTCTTCAC-3′, and reverse,
5′-ATCTTGAGGCTGTTGTCATACTTCT-3′. ITGA7 mRNA expression was analyzed
using the 2−ΔΔCq calculation, with GAPDH as an internal
control (17).
Western blotting
At 48 h after transfection, Ishiwaka and RL95-2
cells were harvested and lysed in RIPA lysis buffer containing
Protease Inhibitor Cocktail at 1×107 cells per 200 µl
(MilliporeSigma) for protein extraction, based on the
manufacturer's protocol. A BCA protein concentration quantification
kit (Beyotime Institute of Biotechnology) was used for measuring
the protein concentration in each group. A total of 25 µg proteins
of each group were boiled at 98°C for 5 min and proteins were
separated using 10% NuPAGE Bis-Tris Gels (Thermo Fisher Scientific,
Inc.) and transferred into nitrocellulose membrane (Beijing
Solarbio Science and Technology Co., Ltd.). The membranes were
blocked with 5% BSA (Thermo Fisher Scientific, Inc.) for 1.5 h at
37°C, and then incubated with the ITGA7, AKT, p-AKT, PI3K, p-PI3K,
cleaved-caspase 3 (C-caspase 3) and GAPDH primary antibodies for
1.5 h at 37°C, respectively. Subsequently, the membranes were
incubated with the secondary antibodies for 50 min at 37°C.
Finally, the membranes were reacted with Pierce™ ECL Plus Western
Blotting Substrate Thermo Fisher Scientific, Inc. for
chemiluminescence. The source and dilution of antibodies are
presented in Table I.
 | Table I.Antibodies applied in western
blotting. |
Table I.
Antibodies applied in western
blotting.
| Antibody | Company | Cat. no. | Dilution |
|---|
| Primary
antibodies |
|
|
|
| ITGA7
mouse mAb | Santa Cruz
Biotechnology, Inc. | sc-81807 |
1:800 |
| AKT
rabbit mAb | Cell Signaling
Technology, Inc. | #4691 | 1:1,000 |
| p-AKT
rabbit mAb | Cell Signaling
Technology, Inc. | #4060 | 1:1,000 |
| PI3K
rabbit mAb | Cell Signaling
Technology, Inc. | #4257 | 1:1,000 |
| p-PI3K
rabbit mAb | Cell Signaling
Technology, Inc. | #17366 | 1:1,000 |
| Cleaved
caspase 3 rabbit mAb | Cell Signaling
Technology, Inc. | #9664 | 1:1,000 |
| GAPDH
mouse mAb | Abcam | ab9484 | 1:2,000 |
| Secondary
antibodies |
|
|
|
| Goat
anti-mouse IgG-HRP | Abcam | ab6789 | 1:4,000 |
| Goat
anti-rabbit IgG H&L (HRP) | Cell Signaling
Technology, Inc. | #7074 | 1:4,000 |
Cell proliferation assay
Cell proliferation of Ishiwaka and RL95-2 cells
after ITGA7 interference was performed using Cell Counting Kit-8
(CCK-8; Dojindo Laboratories, Inc.). In short, Ishiwaka and RL95-2
cells were seeded on a 96-well plate (4×103 cells in 100
µl medium) and incubated overnight. At 0, 24, 48 and 72 h after
transfection, 10 µl CCK-8 detection solution was added and
incubated for 2 h at 37°C, respectively. Optical density (OD) value
was measured using an Automated Enzyme Immunoassay Analyzer AIA-900
(Tosoh Corporation) at 450 nm.
Apoptosis assay
TUNEL apoptosis detection kit (Beyotime Institute of
Biotechnology) was used for analyzing Ishiwaka and RL95-2 apoptosis
after ITGA7 interference. In brief, at 48 h after transfection,
Ishiwaka and RL95-2 cells were fixed with 4% paraformaldehyde fix
solution (Beyotime Institute of Biotechnology) at 700 µl/well for
0.5 h. Triton X-100 solution (Beyotime Institute of Biotechnology)
was added to cells (200 µl) for 10 min. Subsequently, cells were
blocked in 5% BSA (Thermo Fisher Scientific) in TBST (0.05%
Tween-20) for 0.5 h. Cells were incubated with TUNEL apoptosis
detection solution (Beyotime Institute of Biotechnology) for 1 h.
Antifade mountant (Beyotime Institute of Biotechnology) was used to
reduce fluorescence quenching. All procedures of TUNEL assay were
carried out at room temperature. Fluorescent images of three random
fields were captured and analyzed using a fluorescence microscope
(Olympus Corporation).
Cell invasion assay
Transwell assay was used for analyzing Ishiwaka and
RL95-2 cell invasion after ITGA7 interference with Matrigel-plated
Transwell insert (Corning, Inc.). The Matrigel-plated Transwell
insert (Corning, Inc.) were precoated with Matrigel (BD Bioscience)
at 37°C for 1 h. Briefly, at 48 h after transfection,
4×104 Ishiwaka and RL95-2 cells in serum-free medium
(Gibco; Thermo Fisher Scientific, Inc.) were added into the upper
Matrigel-plated Transwell insert, and the lower wells contained 600
µl corresponding complete medium [MEM (MilliporeSigma) containing
5% FBS for Ishiwaka cells or DMEM/F-12 (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS for RL95-2 cells]. After 24 h
at 37°C, the non-migrated cells were gently removed. Subsequently,
migrated cells were fixed with 20% methanol for 20 min at room
temperature, followed by staining with crystal violet staining
solution (Beyotime Institute of Biotechnology) for 5 min at room
temperature. Images of stained cells were captured and cells were
counted by investigators using an inverted fluorescence microscope
(Olympus Corporation).
EC tissue sample detection
To further confirm the association of ITGA7 with EC
tumor features, 50 female patients (mean age, 63.9±10.1 years;
median age, 64.5 years; age range, 39–80 years) with primary EC who
underwent resection between November 2019 and June 2021 were
retrospectively analyzed after ethical approval by Union Hospital,
Tongji Medical College of Huazhong University of Science and
Technology (Wuhan, China). The inclusion criteria were: i)
Pathologically diagnosed as primary EC; ii) received tumor
resection; and iii) tumor tissue was accessible for
immunohistochemistry (IHC) assay. The exclusion criteria were: i)
History or complicated with other primary cancers; ii) received
neoadjuvant therapy; and iii) pregnant or lactating women. Written
informed consents were received from all the patients/guardians.
Myometrial invasion ≥1/2 or <1/2 indicated the depth of
myometrial invasion (10).
Paraffin-embedded tumor tissues and adjacent tissues (within 2-cm
next to tumor tissues) were acquired and IHC was used to detect
ITGA7 expression, and the ITGA7 IHC score was calculated. The IHC
assay and scoring method referred to a previous study (13). In brief, the tissues were fixed by
4% paraformaldehyde (Sangon Biotech Co., Ltd.) for 24 h at 4°C. The
tissues were then embedded in paraffine (Sangon Biotech Co., Ltd.)
and cut into 4-µm sections. The sections were then deparaffined in
xylene, rehydrated in descending alcohol series and antigen
retrieved in 98°C citric acid buffer for 3 min. The sections were
subsequently blocked by 5% BSA (Sangon Biotech Co., Ltd.) at 37°C
for 20 min after incubating in 3% H2O2 for 15
min. Next, the sections were incubated with ITGA7 antibody
(dilution rate, 1:150; cat. no. ab203254; Abcam) at 4°C overnight
followed by incubating with goat anti-rabbit IgG H&L (HRP;
dilution rate, 1,000; cat. no. #7074; Cell Signaling Technology,
Inc.) at 37°C for 1 h. The sections were stained by
3,3′-Diaminobenzidine (Sangon Biotech Co., Ltd.) for 10 min and
counterstained by hematoxylin (Sangon Biotech Co., Ltd.) for 2 min,
at last, at room temperature. The images with a magnification of
×200 were captured using a light microscope (Motic China Group Co.,
Ltd.) and analyzed by two independent pathologists. The total score
method was calculated by multiplying by intensity score and
percentage score of stained cells. The intensity score was as
follows: 0, No staining; 1, weak staining, light yellow; 2,
moderate staining, yellow brown; and 3, strong staining, brown. The
percentage score of stained cells was as below: 0, 0; 1, 1–25; 2,
26–50; 3, 51–75; and 4, 76–100% positive cells.
Statistical analysis
GraphPad Prism Software (version 7.0; GraphPad
Software, Inc.) was used for statistical analysis and graph
plotting in all assays. For cell experiments, data were presented
as mean with standard deviation (SD); multigroup comparison was
analyzed using one-way ANOVA followed by Dunnett's or Tukey's
multiple comparisons test. For clinical investigation, data were
presented as median with interquartile range (IQR); two-group
comparison was analyzed using Mann-Whitney U test or Wilcoxon
signed-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
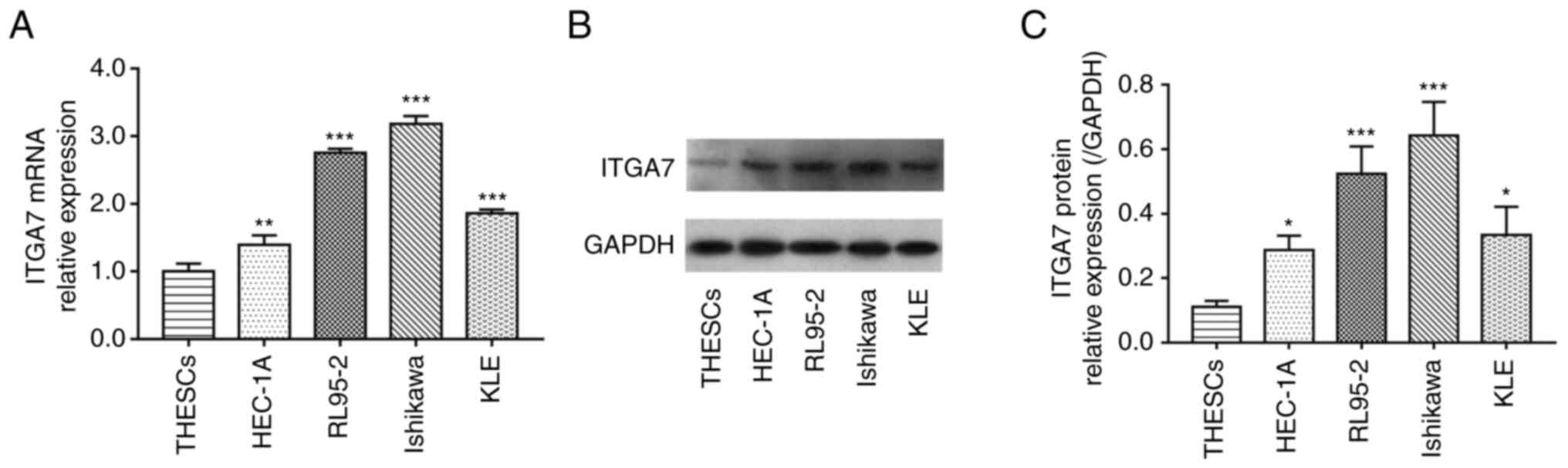
ITGA7 is overexpressed in EC cell
lines
The mRNA and protein levels of ITGA7 in THESCs and
EC cell lines were detected using RT-qPCR and western blotting,
respectively. Data demonstrated that ITGA7 mRNA expression was
significantly enhanced in EC cell lines (including HEC-1A, RL95-2,
Ishikawa and KLE) compared with THESCs (all P<0.01; Fig. 1A); meanwhile, the ITGA7 protein
expression was also significantly increased in EC cell lines
compared with THESCs (all P<0.05; Fig. 1B and C). RL95-2 and Ishikawa cell
lines were selected for further knockdown experiments due to the
fact that ITGA7 was most significantly increased in these two cell
lines. These data suggested that ITGA7 was highly expressed in EC
cells.
ITGA7 knockdown efficiency
Subsequently, in order to explore the effect of
ITGA7 on EC cell function and its downstream pathways, siRNA
transfection was conducted. In both Ishikawa and RL95-2 cell lines,
it was revealed that all three si-ITGA7 plasmids significantly
suppressed ITGA7 mRNA and protein levels compared with the si-NC
(all P<0.05; Fig. 2A-F).
Furthermore, the expression of ITGA7 in cells transfected with
si-ITGA7-2 had the lowest ITGA7 level, indicating that si-ITGA7-2
presented the most effective knockdown efficiency. Therefore, it
was used in further experiments.
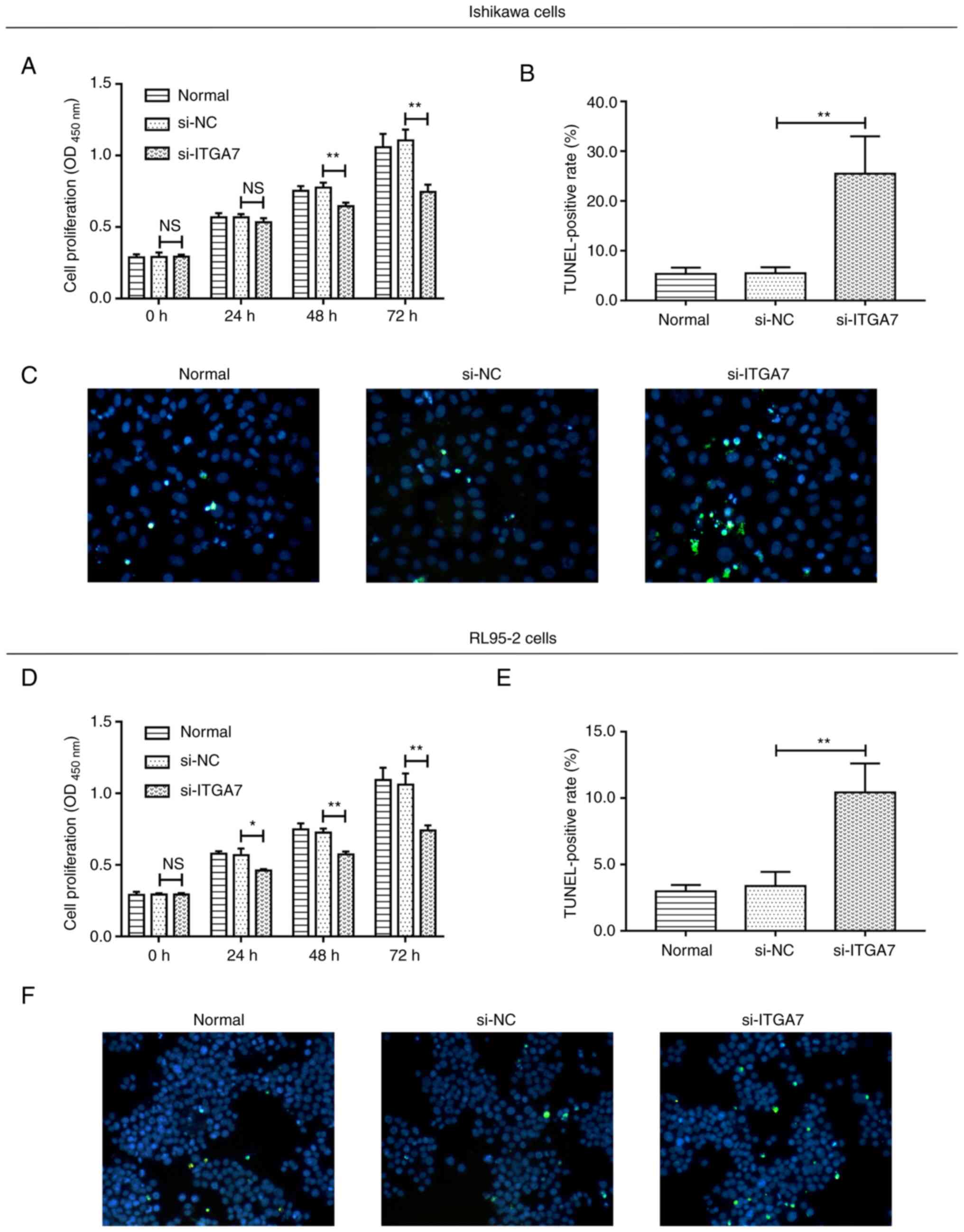
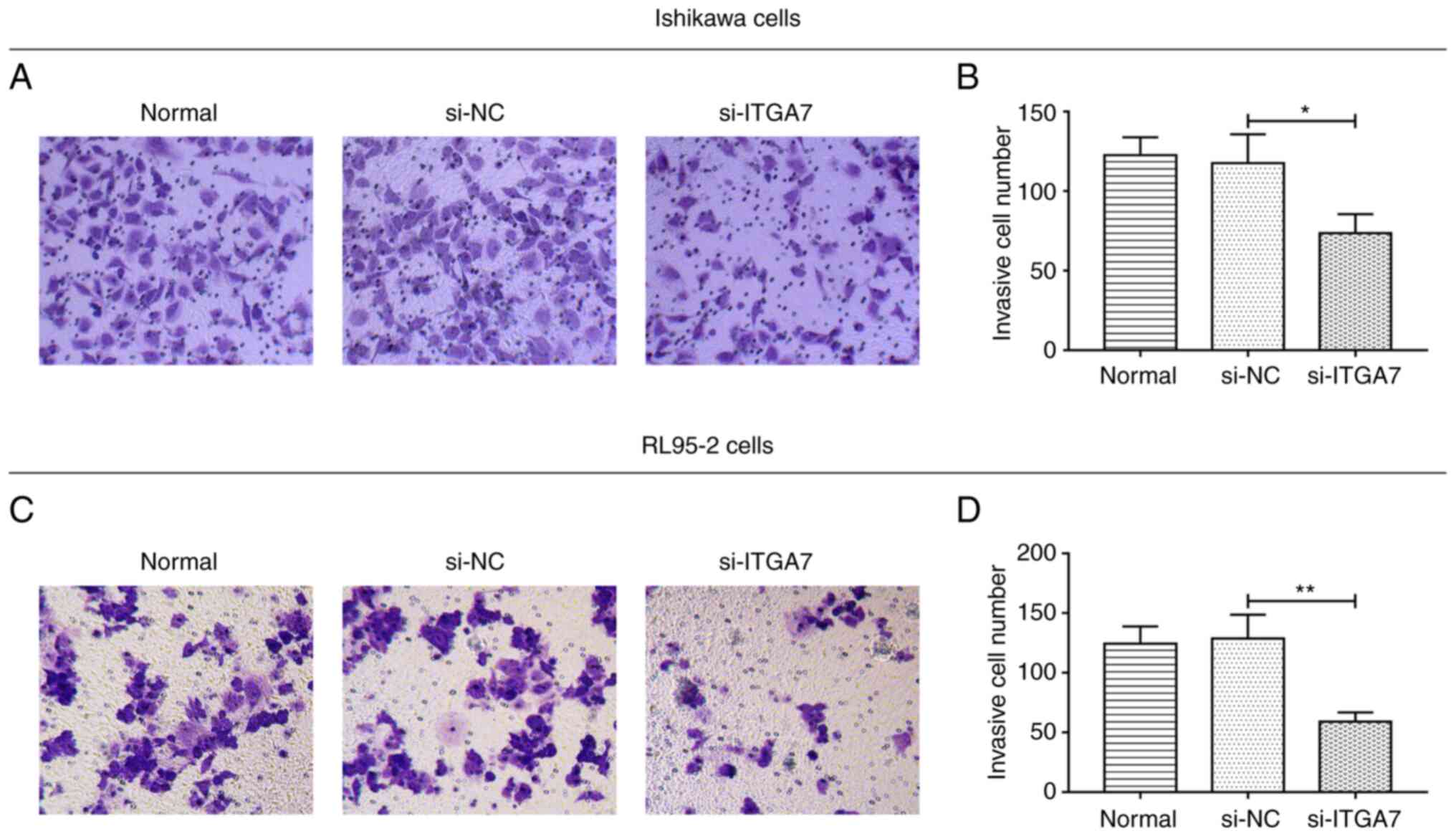
Si-ITGA7 suppresses proliferation and
invasion but promotes apoptosis in EC cell lines
The effect of ITGA7 knockdown on EC cell functions,
including proliferation, apoptosis and invasion, were investigated.
In Ishikawa cells, compared with the si-NC group, si-ITGA7
significantly suppressed cell proliferation at 48 and 72 h (both
P<0.01; Fig. 3A) and invasion
(P<0.05; Fig. 4A and B), while
it promoted apoptosis (P<0.01; Fig.
3B and C). Meanwhile, in RL95-2 cells, si-ITGA7 also
significantly inhibited cell proliferation at 24, 48 and 72 h (all
P<0.05; Fig. 3D) as well as
invasion (P<0.01; Fig. 4C and
D), but it significantly increased apoptosis (P<0.01;
Fig. 3E and F). C-caspase 3
determined by western blotting also confirmed that si-ITGA7
elevated apoptosis compared with si-NC in Ishikawa (grayscale
density, 0.556±0.072 vs. 0.386±0.049) and RL95-2 (grayscale
density, 0.552±0.049 vs. 0.344±0.050) cells (Fig. S1A and B). These data revealed that
knockdown of ITGA7 suppressed proliferation and invasion but
promoted apoptosis in EC cells.
Additionally, si-ITGA7 was transfected into THESCs,
in which transfection efficiency was demonstrated using RT-qPCR and
western blotting (Fig. S2A-C).
Subsequently, CCK-8, TUNEL and Transwell assays indicated that
si-ITGA7 had a decreased effect on proliferation, apoptosis and
invasion in THESCs, while these effects were minor and not
significant (Fig. S2D-G).
Si-ITGA7 inactivates PI3K/AKT
pathway
In addition, the implication of ITGA7 knockdown on
the PI3K/AKT pathway in EC cell lines was also assessed. In both
Ishikawa and RL95-2 cells, p-PI3K and p-AKT were reduced by
si-ITGA7; however, PI3K and AKT were not influenced by si-ITGA7
(Fig. 5A). Further grayscale
analyses demonstrated that the ratio of p-PI3K to PI3K and that of
p-AKT to AKT were both significantly reduced by si-ITGA7 compared
with the si-NC group in Ishikawa and RL95-2 cells (all P<0.01;
Fig. 5B). These data illustrated
that ITGA7 knockdown inactivated the PI3K/AKT pathway in EC
cells.
 | Figure 5.PI3K/AKT pathway after transfection in
Ishikawa and RL95-2 cells. (A) Representative images of PI3K,
p-PI3K, AKT, p-AKT detection by western blot analysis after
transfection. (B) Grayscale analyses of blots of PI3K, p-PI3K, AKT,
p-AKT levels after transfection. **P<0.01 and ***P<0.001.
ITGA7, integrin α7; NC, negative control; PI3K,
phosphatidylinositol 3-kinase; AKT, protein kinase B; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase; si-, short interfering;
p-, phosphorylated. |
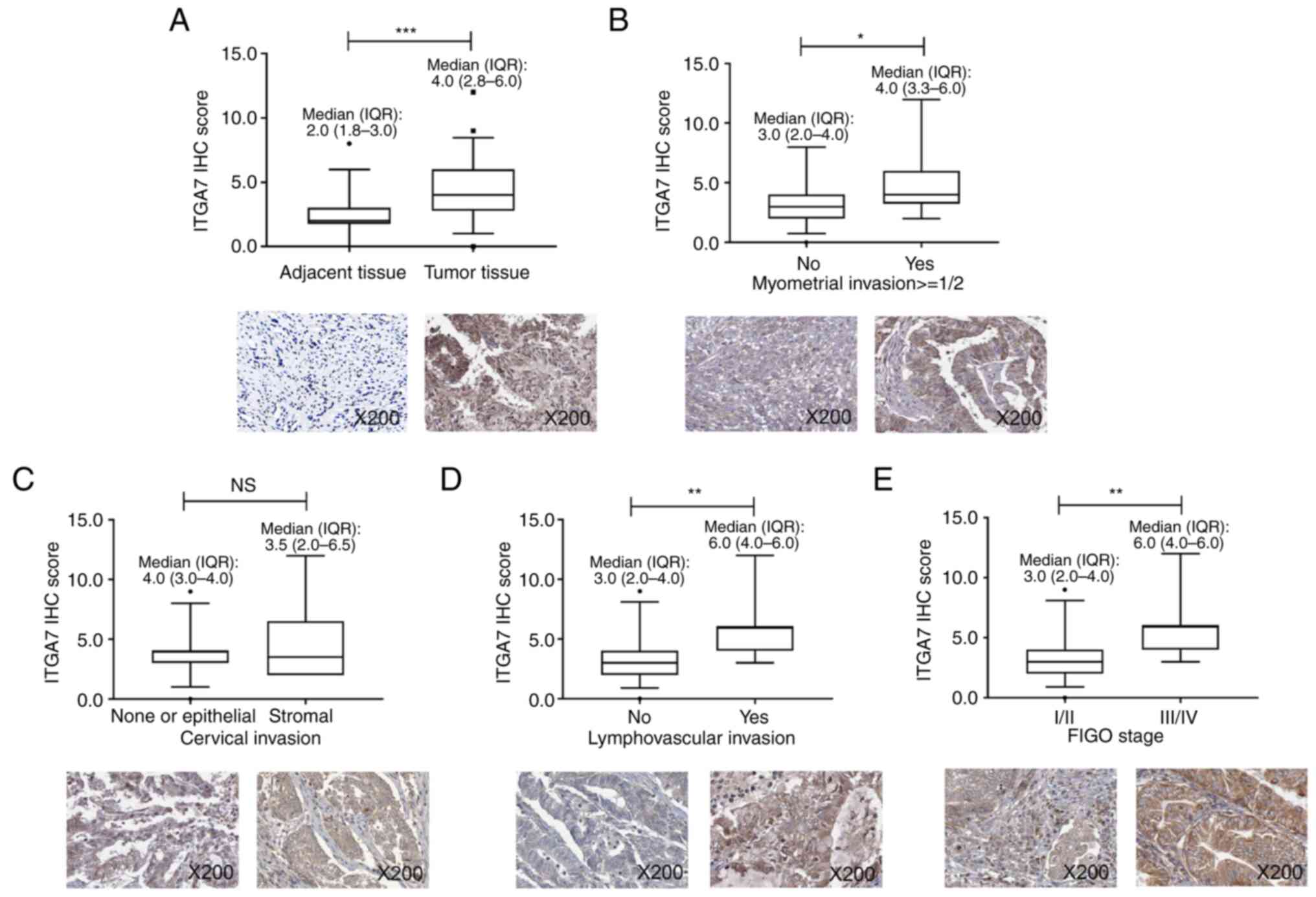
Reduced tumor ITGA7 is associated with
less advanced tumor features in patients with EC
In order to further verify the aforementioned data
in clinical setting, 50 patients with EC were enrolled. The
patients had a mean age of 63.9±10.1 years; 32% patients had
myometrial invasion >1/2, 20% patients had cervical invasion and
26% patients had lymphovascular invasion. Regarding the
International Federation of Gynecology and Obstetrics (FIGO) stage,
60, 14, 22 and 4% of patients were of stage I, II, III and IV,
respectively. ITGA7 levels in tumor and adjacent tissues from these
patients with EC were detected using IHC. Data demonstrated that
ITGA7 was elevated in tumor tissues compared with adjacent tissues
(P<0.001; Fig. 6A). Moreover, a
lower tumor ITGA7 IHC score was associated with the absence of
myometrial invasion ≥1/2 (P<0.05; Fig. 6B) and lymphovascular invasion
(P<0.01; Fig. 6D), as well as
FIGO stage I/II vs. III/IV (P<0.01; Fig. 6E), but not stromal invasion
(P>0.05; Fig. 6C). These data
suggested that lower level of tumor ITGA7 was associated with less
advanced tumor burden in patients with EC.
Discussion
ITGA7 is regarded as an important regulator for
carcinogenesis and progression, and its dysregulation has been
revealed in several types of cancer. For example, previous studies
have revealed that ITGA7 is increased in colorectal cancer cell
lines, hepatocellular carcinoma cell lines and non-small cell
cancer cell lines (14,15,18).
However, to the best of our knowledge, it has not yet been reported
whether ITGA7 is also upregulated in EC cell lines. Therefore, the
present study was conducted and revealed that ITGA7 expression was
significantly enhanced in EC cell lines (including HEC-1A, RL95-2,
Ishikawa and KLE) compared with THESCs. A possible explanation for
these data might be that high levels of ITGA7 could alter the
activation of several signaling pathways that are associated with
tumorigenesis, such as the PI3K/AKT, Wnt/β-catenin and Ras pathways
(18–20), to increase the malignant
proliferation of endometrial stromal cells and increase EC genesis.
Therefore, ITGA7 upregulation was noted in EC cell lines compared
with THESCs.
According to previous studies, ITGA7 modulates cell
function, including proliferation, apoptosis, stemness and
chemosensitivity, in several types of cancer, such as tongue
squamous cell carcinoma, hepatocellular carcinoma and non-small
cell lung cancer (13–15). The effect of ITGA7 on EC cell
function has yet to be elucidated. The present study discovered
that ITGA7 knockdown reduced cell proliferation and invasion, but
enhanced apoptosis in EC cell lines Ishikawa and RL95-2. Possible
explanations for these data may be: i) Low ITGA7 expression can
regulate the activation of several proliferation/apoptosis-related
pathways, such as the PI3K/AKT pathways (as shown by western
blotting) and Ras pathway [as in colorectal cancer (18)]. Therefore, ITGA7 knockdown
suppressed proliferation while promoting apoptosis in EC cell lines
Ishikawa and RL95-2. Or ii) low ITGA7 expression may suppress
several invasion-related mechanisms including the Wnt/β-catenin
pathway and epithelial-mesenchymal transition [as in hepatocellular
carcinoma (21)]. Therefore, ITGA7
knockdown repressed invasion in EC cell lines Ishikawa and
RL95-2.
The PI3K/AKT pathway is highly involved in cell
proliferation, survival and invasion (22). However, dysregulation of the
PI3K/AKT pathway is also a common phenomenon in human cancer
(23). Meanwhile, it is also
proposed that the PI3K/AKT pathway is one of the most common
pathways that is altered in EC: The molecular spectrum of EC
suggests that the mutation rates of PI3KCA, PIK3R1 and AKT1 are
59.7, 33 and 3.2 m respectively in all cases (22,24).
Previous studies illustrate that inhibiting the PI3K/AKT pathway
greatly suppresses the proliferation, migration and invasion of EC
cell lines (25,26). Moreover, it is reported that ITGA7
regulates PI3K/AKT (14).
Therefore, the effect of ITGA7 on the PI3K/AKT pathway in EC cell
lines was investigated in the present study. The data demonstrated
that ITGA7 knockdown inhibited the phosphorylation of PI3K and AKT.
These data were in line with a previous study, suggesting that
inhibiting ITGA7 also represses phosphorylation of the PI3K/AKT
pathway in hepatocellular carcinoma cell lines (14). Although the present study
discovered some notable findings, it is necessary to conduct in
vivo experiments in the future to further investigate the
effect of ITGA7 on EC progression using xenograft mice.
In order to further verify the in vitro
findings, the present study enrolled patients with EC and detected
ITGA7 in their tumor and adjacent tissues. Data demonstrated that
the expression levels of ITGA7 were higher in tumor tissues
compared with in adjacent tissues, which was in line with the
aforementioned data that ITGA7 was elevated in EC cell lines
compared with THESCs. Moreover, the present study revealed that
tumor ITGA7 was negatively associated with myometrial invasion
(≥1/2), lymphovascular invasion and FIGO stage. These data could be
explained by: i) ITGA7 Knockdown reduced invasion of EC cells, and
thus directly decreased myometrial and lymphovascular invasion; or
ii) ITGA7 knockdown might inactivate several pathways including the
PI3K/AKT and Wnt/β-catenin pathways (14,20)
to decrease proliferation and invasion of EC cells, which could
indirectly result in lower FIGO stage. ITGA7 was not associated
with cervical invasion, which could be explained by low statistical
power due to the small sample size (N=50). However, the prognostic
value of ITGA7 should be investigated in the future.
Collectively, ITGA7 knockdown represses
proliferation, invasion and the PI3K/AKT pathway while inducing
apoptosis in EC cell lines. Its insufficiency is associated with
less advanced tumor features in patients with EC. These indicate
that ITGA7 may be a potential target for the treatment of EC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JM contributed to the conception and design of the
study. ML contributed to performing the experiments. CL contributed
to data acquisition and analysis. TL and SG contributed to the
analysis and interpretation of data. JM and ML confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics approval of
Union Hospital, Tongji Medical College of Huazhong University of
Science and Technology. Written informed consents were received
from all the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lu KH and Broaddus RR: Endometrial cancer.
N Engl J Med. 383:2053–2064. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paleari L, Pesce S, Rutigliani M, Greppi
M, Obino V, Gorlero F, Vellone VG and Marcenaro E: New insights
into endometrial cancer. Cancers (Basel). 13:14962021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saleh M, Virarkar M, Bhosale P, El Sherif
S, Javadi S and Faria SC: Endometrial cancer, the current
international federation of gynecology and obstetrics staging
system, and the role of imaging. J Comput Assist Tomogr.
44:714–729. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Javadian P and Nezhat F: Endometrial
carcinoma and its precursors. Adv Exp Med Biol. 1242:59–72. 2020.
View Article : Google Scholar
|
|
5
|
Friedenreich CM, Ryder-Burbidge C and
McNeil J: Physical activity, obesity and sedentary behavior in
cancer etiology: Epidemiologic evidence and biologic mechanisms.
Mol Oncol. 15:790–800. 2021. View Article : Google Scholar
|
|
6
|
Ryan NA, McMahon RF, Ramchander NC, Seif
MW, Evans DG and Crosbie EJ: Lynch syndrome for the gynaecologist.
Obstet Gynaecol. 23:9–20. 2021. View Article : Google Scholar
|
|
7
|
Kyo S and Nakayama K: Endometrial cancer
as a metabolic disease with dysregulated PI3K signaling: Shedding
light on novel therapeutic strategies. Int J Mol Sci. 21:60732020.
View Article : Google Scholar
|
|
8
|
McMellen A, Woodruff ER, Corr BR, Bitler
BG and Moroney MR: Wnt Signaling in gynecologic malignancies. Int J
Mol Sci. 21:42722020. View Article : Google Scholar
|
|
9
|
Gómez-Raposo C, Merino Salvador M, Aguayo
Zamora C, Garcia de Santiago B and Casado Sáenz E: Immune
checkpoint inhibitors in endometrial cancer. Crit Rev Oncol
Hematol. 161:1033062021. View Article : Google Scholar
|
|
10
|
Concin N, Creutzberg CL, Vergote I, Cibula
D, Mirza MR, Marnitz S, Ledermann JA, Bosse T, Chargari C, Fagotti
A, et al: ESGO/ESTRO/ESP guidelines for the management of patients
with endometrial carcinoma. Virchows Arch. 478:153–190. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cooper J and Giancotti FG: Integrin
signaling in cancer: Mechanotransduction, stemness, epithelial
plasticity, and therapeutic resistance. Cancer Cell. 35:347–367.
2019. View Article : Google Scholar
|
|
12
|
Park HJ, Park JE, Lee H, Kim SJ, Yun JI,
Kim M, Park KH and Lee ST: Integrins functioning in uterine
endometrial stromal and epithelial cells in estrus. Reproduction.
153:351–360. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv Z, Yang Y and Yang C: Integrin α7
correlates with worse clinical features and prognosis, and its
knockdown inhibits cell proliferation and stemness in tongue
squamous cell carcinoma. Int J Oncol. 56:69–84. 2020.
|
|
14
|
Ge JC, Wang YX, Chen ZB and Chen DF:
Integrin alpha 7 correlates with poor clinical outcomes, and it
regulates cell proliferation, apoptosis and stemness via
PTK2-PI3K-Akt signaling pathway in hepatocellular carcinoma. Cell
Signal. 66:1094652020. View Article : Google Scholar
|
|
15
|
Xia D, Chen B and Yang X: Correlation of
integrin alpha 7 with clinicopathological characteristics and
survival profiles, as well as its regulatory role in cell
proliferation, apoptosis, and stemness in non-small-cell lung
cancer. J Clin Lab Anal. 33:e229732019. View Article : Google Scholar
|
|
16
|
Zhu T, Chen R, Wang J, Yue H, Lu X and Li
J: The prognostic value of ITGA and ITGB superfamily members in
patients with high grade serous ovarian cancer. Cancer Cell Int.
20:2572020. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Wang J, Zhang C, Lin C, Zhang J,
Zhang W, Zhang W, Lu Y, Zheng L and Li X: Circular RNA circITGA7
inhibits colorectal cancer growth and metastasis by modulating the
Ras pathway and upregulating transcription of its host gene ITGA7.
J Pathol. 246:166–179. 2018. View Article : Google Scholar
|
|
19
|
Tian T, Lai X, Xiang K, Han X, Yin S,
Cabrera RM, Steele JW, Lei Y, Cao X, Finnell RH, et al:
Hypermethylation of PI3K-AKT signalling pathway genes is associated
with human neural tube defects. Epigenetics. 17:133–146. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao HD, Mao Y and Ying YG: The
involvement of the laminin-integrin α7β1 signaling pathway in
mechanical ventilation-induced pulmonary fibrosis. J Thorac Dis.
9:3961–3972. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Z, Kong X and Wang Z: Integrin α7
knockdown suppresses cell proliferation, migration, invasion and
EMT in hepatocellular carcinoma. Exp Ther Med. 21:3092021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi X, Wang J, Lei Y, Cong C, Tan D and
Zhou X: Research progress on the PI3K/AKT signaling pathway in
gynecological cancer (review). Mol Med Rep. 19:4529–4535.
2019.PubMed/NCBI
|
|
23
|
du Rusquec P, Blonz C, Frenel JS and
Campone M: Targeting the PI3K/Akt/mTOR pathway in estrogen-receptor
positive HER2 negative advanced breast cancer. Ther Adv Med Oncol.
12:17588359209409392020. View Article : Google Scholar
|
|
24
|
Cancer Genome Atlas Research Network, .
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu H, Zhang L, Zhang X and Cui Z:
PI3K/AKT/mTOR pathway promotes progestin resistance in endometrial
cancer cells by inhibition of autophagy. Onco Targets Ther.
10:2865–2871. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ni M, Zhao Y and Wang X: Suppression of
synuclein gamma inhibits the movability of endometrial carcinoma
cells by PI3K/AKT/ERK signaling pathway. Genes Genomics.
43:633–641. 2021. View Article : Google Scholar : PubMed/NCBI
|