Introduction
Ovarian cancer is one of the most common gynecologic
malignancies and advanced ovarian cancer has a very low 5-year
survival rate, despite improvements in medical strategies in recent
years (1,2). The high mortality is partially due to
the lack of effective diagnostic markers for ovarian cancer at an
early stage and the prevalence of diffuse intra-abdominal
metastasis (3).
Long non-coding RNA plasmacytoma variant
translocation 1 (LncRNA PVT1) is a non-coding RNA of >200 bp,
which is located in the well-known cancer-associated chromosomal
region 8q24 (4). It has been
implicated in the biological processes of several cancers,
including ovarian epithelial cancer (4). In addition, PVT1 has been suggested to
be a marker of poor prognosis in several types of cancer, which can
promote malignancy by modulating various biological processes,
including epithelial-mesenchymal transition (EMT) (5,6). A
previous study demonstrated that PVT1 is upregulated in ovarian
cancer, and that a high level of PVT1 expression is associated with
a poor prognosis in patients with ovarian cancer (7). However, the biological role and
underlying mechanism of lncRNA PVT1 in ovarian cancer cells remain
unclear.
Connective tissue growth factor (CTGF) is a secreted
protein belonging to the cellular communication network (CCN)
family (8). It plays an important
role in the remolding of the extracellular matrix and the
development of connective tissues such as those constituting the
skeleton (8,9). The dysregulation of CTGF is implicated
in the development of pathological conditions such as diabetic
retinopathy and the progression of cancer (10,11).
However, the regulatory mechanisms controlling the expression of
CTGF under pathological conditions require clarification.
In the present study, the functional role of lncRNA
PVT1 in the proliferation, migration, invasion and EMT of the SKOV3
and CAOV3 human ovarian cancer cell lines was explored. In
addition, the potential involvement of CTGF in the biological role
of lncRNA PVT1 as a regulator of the malignant phenotype of ovarian
cancer cells was also investigated.
Materials and methods
Cell culture and transfection
The SKOV3 and CAOV3 human ovarian cancer cell lines
were obtained from Qilu Medical College of Shandong University. The
cells were maintained in DMEM (Hyclone; Cytiva) supplemented with
10% fetal bovine serum (Hyclone; Cytiva), 100 U/ml penicillin and
100 mg/ml streptomycin (Biosharp Life Sciences) in a humidified
incubator at 37°C with 5% CO2. Recombinant human CTGF
(rhCTGF; PeproTech, Inc.; 500 mg/l) was added to the culture medium
to examine its role in the cell phenotype.
For transfection, SKOV-3 and CAOV3 cells were seeded
in 24-well plates at a density of 2×105 cells/well.
Then, 50 nM small interfering RNA (siRNA) targeting PVT1 (si-PVT1)
or negative control siRNA (si-NC) was respectively transfected into
cells at ambient temperature using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Cells were subjected to further
experiments 48 h after transfection. The sequence of the
PVT1-specific siRNA was 5′-GGCACCTTCCAGTGGATTT-3′. The si-NC was
scrambled siRNA with the following sequences: Sense,
UGCUGACUCCAAAGCUCUGdTdT and anti-sense, CAGAGCUUUGGAGUCAGCAdTdT.
The si-PVT1 and si-NC were purchased from Guangzhou Ruibo
Biotechnology Co., Ltd.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RT was performed to generate cDNA using a ReverTra Ace™ qPCR
RT kit (Toyobo Life Science) at 42°C for 1 h on a PCR machine. The
relative expression level of each gene was measured using
Thunderbird SYBR qPCR Mix (Toyobo Life Science) on a 7500 Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The following thermocycling conditions were applied: 40 cycles of
denaturation at 95°C for 15 sec, primer annealing at 58°C for 20
sec and extension at 72°C for 20 sec. The relative expression
levels of the target genes were normalized to GAPDH using the
2−∆∆Cq method (12). The
primer sequences used are shown in Table I.
 | Table I.Quantitative polymerase chain reaction
primer sequences. |
Table I.
Quantitative polymerase chain reaction
primer sequences.
| Genes |
| Primer sequences
(5′-3′) |
|---|
| PVT1 | F |
TCTGGGGAATAACGCTGGTG |
|
| R |
CTTCACCAGGAAGAGTCGGG |
| CTGF | F |
ACCGACTGGAAGACACGTTTG |
|
| R |
CCAGGTCAGCTTCGCAAGG |
| E-cadherin | F |
TGAAAACAGCAAAGGGCTTGGA |
|
| R |
GCAGTGTCTCTCCAAATCCGA |
| Vimentin | F |
CTGGATTCACTCCCTCTGGT |
|
| R |
CGTGATGCTGAGAAGTTTCG |
| GAPDH | F |
GGAGCGAGATCCCTCCAAAAT |
|
| R |
GGCTGTTGTCATACTTCTCATGG |
Cell Counting Kit-8 (CCK-8)
proliferation assay
The transfected cells (5×103 cells/well)
were seeded in 96-well plates. The cells were cultured for 0, 24,
48, 72, 96 and 120 h, respectively. After culture, 10 µl CCK-8
solution (Dojindo Laboratories, Inc.) was added to each well and
the cells were further incubated for 1 h. The absorbance at 450 nm
was measured using a microplate reader (Thermo Fisher Scientific,
Inc.).
Colony formation assay
The transfected cells (5×102 cells/well)
were seeded in 6-well plates and co-cultured with or without rhCTGF
for 14 days. The cell medium was refreshed every 3 days. On day 14,
the cells were fixed with 4% paraformaldehyde at room temperature
for 10 min and then stained with 0.5% crystal violet (Beyotime
Institute of Biotechnology) for 20 min at ambient temperature.
Subsequently, the number of colonies (with >50 cells considered
to be a colony) was counted manually under a Leica AM6000
microscope (Leica Microsystems GmbH).
Wound-healing assay
Transfected SKOV-3 and CAOV3 cells with or without
rhCTGF were seeded into 6-well plates at a density of
5.0×105 cells/well. Cells were serum-starved for 18 h.
When the degree of confluence reached ~90%, a scratch wound was
created in the cell layer using a sterile 200-µl pipette tip in the
central region of each well. The wounded cells were incubated at
37°C for 48 h. Cell images were then captured using an inverted
light microscope (Leica AM6000). The distance that the cells
migrated was analyzed using ImageJ software (version 1.8.0)
(National Institutes of Health). The migration rate was calculated
as ratio of the wound distance at 48 h to the wound distance at 0
h.
Transwell migration and invasion
assays
The migration and invasion ability of the ovarian
cancer cells was detected using a 24-well Transwell chamber
(Costar; Corning, Inc.) with an 8.0-µm pore size. The transfected
cells were suspended in serum-free DMEM and inoculated in the upper
chamber at a density of 5.0×105 cells/well.
Matrigel-coated chambers (BD Biosciences) were used for the
invasion assay, while chambers without Matrigel coating were used
for the migration assay. The chamber coating was performed at 37°C
for 30 min. Medium containing 10% FBS was added to the lower
chamber with or without rhCTGF. After 48 h of incubation at 37°C,
the migratory or invading cells on the membrane were fixed with
methanol and stained with 0.1% crystal violet (Beyotime Institute
of Biotechnology) at ambient temperature for 20 min. The number of
migrating and invading cells was counted in 5 random fields using a
Leica AM6000 microscope.
Western blotting
The protein levels of CTGF, E-cadherin and vimentin
were examined by western blotting. Total protein was extracted
ovarian cancer cells using RIPA lysis buffer containing protease
inhibitor cocktail (Thermo Fisher Scientific, Inc.). Cells
suspended in the RIPA buffer were lysed on ice for 10 min and then
centrifuged at 13,200 × g for 10 min. The supernatant containing
total protein lysate was quantified using a BCA Protein assay kit
(Beyotime Institute of Biotechnology,). Then, 10 µg protein sample
per lane was loaded for separation on 12% gels by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The
separated proteins on the SDS-PAGE gel were transferred onto
polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc.).
After blocking with 5% skimmed milk for 1 h at ambient temperature,
the membrane was incubated with the following primary antibodies at
4°C overnight: Anti-CTGF (ab5097; dilution 1:1,000; Abcam,),
E-cadherin (ab231303; dilution 1:1,500; Abcam), vimentin (ab92547;
dilution 1:1,000; Abcam) and anti-GAPDH (BM1985; dilution 1:2,500;
Boster Biological Technology) The membranes were washed three times
with TBS with 2.5% Tween 20 (TBST) buffer and then incubated with
HRP-conjugated secondary antibody (#7074; dilution 1:3,000; Cell
Signaling Technology, Inc.) at room temperature for 1 h. After
further washes with TBST buffer, the protein signals were developed
using Super ECL Plus Detection Reagent (Tanon Science and
Technology Co., Ltd.) and images captured using a gel imager system
(Bio-Rad Laboratories, Inc.). The western blot experiment was
performed once for each condition.
Statistical analysis
SPSS 25.0 (IBM Corp.), ImageJ and GraphPad Prism 8
(GraphPad Software, Inc.) were used for data analysis. Data are
presented as the mean ± standard deviation of three independent
experiments. Unpaired Student's t-test was used for comparisons
between two groups. Comparisons among multiple groups were analyzed
using one-way analysis of variance (ANOVA) with Tukey's post hoc
test for subsequent pairwise comparisons. Comparisons of data at
multiple time points were examined using two-way ANOVA, with
Bonferroni's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Silencing PVT1 suppresses the
proliferation, migration and invasion ability of ovarian cancer
cells
To investigate the biological roles of lncRNA PVT1
in the phenotype of ovarian cancer cells, si-PVT1 and si-NC were
transfected into SKOV3 and CAOV3 cells. The transfection of si-PVT1
significantly reduced the expression of PVT1 in SKOV3 and CAOV3
cells compared with that in cells transfected with si-NC (Fig. 1A). Comparison of the transfected
cells in CCK-8, colony formation, wound healing and Transwell
assays revealed that the proliferation, migration and invasion
abilities of both cell lines were significantly impaired following
PVT1 silencing (Fig. 1B-E). These
results suggest that lncRNA PVT1 contributes to the malignant
phenotype of ovarian cancer cells.
PVT1 knockdown reduces CTGF expression
and regulates the levels of EMT-related genes
To investigate whether PVT1 modulate the EMT
process, the level of CTGF and EMT-associated genes, namely
E-cadherin and vimentin, were examined using RT-qPCR and western
blotting. In SKOV3 and CAOV3 cells, PVT1 silencing suppressed the
expression of CTGF and vimentin but increased the expression of
E-cadherin at the mRNA and protein levels (Fig. 2). These results indicate that the
downregulation of PVT1 inhibits the expression of CTGF and
suppresses the EMT process.
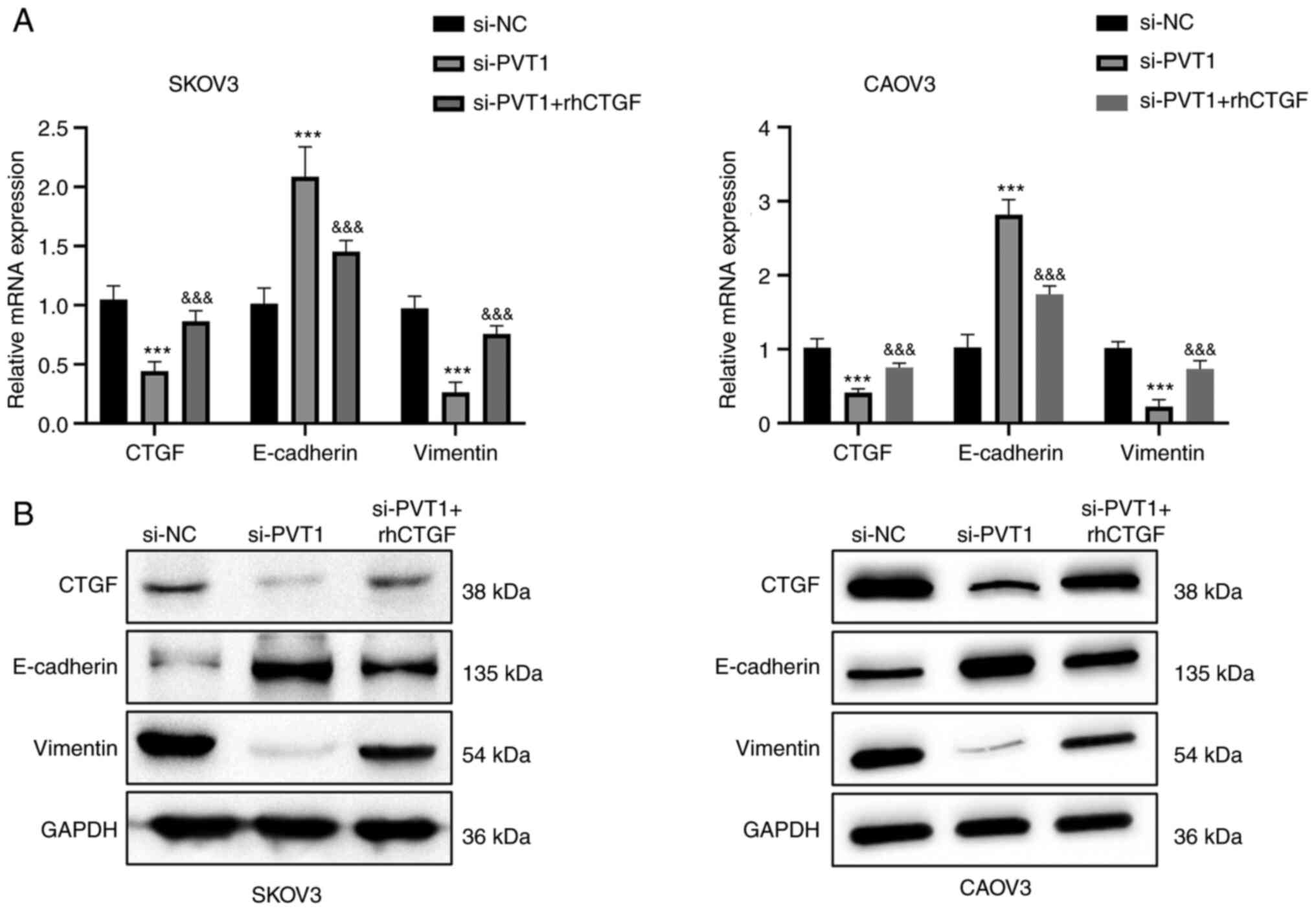
rhCTGF partially abrogates the
inhibitory effects of PVT1 knockdown
As CTGF expression was found to be decreased by PVT1
knockdown, whether CTGF is a downstream mediator of PVT1 function
was investigated via the addition of rhCTGF to the cell culture.
Consistent with the previous results, the proliferation, migration
and invasion abilities of SKOV3 and CAOV3 cells were repressed by
PVT1 siRNA. However, the inhibitory effect was partially attenuated
by treatment with rhCTGF (Fig. 3).
In addition, the rhCTGF treatment also attenuated the reduction in
the expression levels of CTGF and vimentin induced by PVT1
knockdown, and reduced the PVT1 knockdown-induced increase in the
expression level of E-cadherin (Fig.
4). Together, these results suggest that CTGF mediates the
biological effects of lncRNA PVT1 in ovarian cancer cells.
Discussion
The lncRNA PVT1 has been recognized as an oncogenic
non-coding RNA and is a potential prognostic marker for various
cancers. It has been implicated in the development and progression
of various tumors, including ovarian cancer (13,14).
However, its functional roles and regulatory mechanisms in ovarian
cancer remain unclear. Previous studies suggest that lncRNA PVT1 is
involved in the regulation of EMT processes in pancreatic, prostate
and breast cancer (6,15,16).
Chen et al (17)
demonstrated that PVT1 contributes to regulation of the EMT process
by silencing microRNA-214 in ovarian cancer. In addition, other
studies have demonstrated that PVT1 regulates the expression of
CTGF (18,19). The present study clarified the roles
of lncRNA PVT1 and CTGF and their relationship in ovarian
cancer.
CTGF is an extracellular factor of the CCN family,
members of which are implicated in remodeling the extracellular
matrix and signal transduction (11,20).
The upregulation of CTGF has been reported to promote cancer
initiation, progression and metastasis via the augmention of cell
proliferation, migration, invasion, drug resistance and the EMT
process (11,20). In addition, Yang et al
(21) reported that CTGF is a key
factor dictating the malignancy and EMT processes in ovarian cancer
cells. The EMT process has been proposed to be a key event in the
invasion and metastatic dissemination of cancer cells, during which
cells gradually gain enhanced mobility and invasive potential; the
ability of ovarian cancer cells to invade and metastasize is
enhanced through EMT (22,23).
In the present study, it was demonstrated that the
proliferation, migration and invasion abilities of SKOV3 and CAOV3
cells were inhibited following PVT1 knockdown, which was consistent
with previous studies (24–26). In addition, the present study showed
that CTGF and the mesenchymal marker protein vimentin were
downregulated in cells with PVT1 knockdown, while the epithelial
marker E-cadherin was upregulated. Importantly, these effects were
partially attenuated when the cells were treated with rhCTGF. These
data indicate that lncRNA PVT1 promotes the proliferation and
migratory abilities of ovarian cancer cells via the regulation of
CTGF expression, which may also induce EMT in the progression of
ovarian cancer.
However, the present study is limited by the lack of
clinical samples to validate the regulation of CTGF by lncRNA PVT1
in ovarian cancer tissues. The regulation of CTGF by lncRNA PVT1
during the progression of ovarian cancer also merits evaluation in
a xenograft mouse model. In addition, the mechanisms underlying the
dysregulation effect of lncRNA PVT1 require further study in
ovarian cancer tissues and cell lines.
In summary, lncRNA PVT1 serves as an oncogenic
factor by facilitating the proliferation, migration, invasiveness
and EMT process in ovarian cancer cells via the regulation of CTGF.
Therefore, lncRNA PVT1 and CTGF may be considered as therapeutic
targets to limit the progression of ovarian cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by the Scientific Research Project of
Weifang Health Commission (grant no. 2019W02432), the Science and
Technology Development Program of Weifang City (grant no.
2019YX015) and the General Project of Shandong Provincial Natural
Science Foundation (grant no. ZR2020MH242).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LD, HW, YG, SW and WW contributed to manuscript
drafting, writing and revision, as well as study concept and
design. LD, HW, YG and SW were responsible for the collection,
assembly and interpretation of the data and figure drawing. All
authors read and approved the final manuscript. LD and WW confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Shetty M: Imaging and differential
diagnosis of ovarian cancer. Semin Ultrasound CT MR. 40:302–318.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eisenhauer EA: Real-world evidence in the
treatment of ovarian cancer. Ann Oncol. 28 (Suppl_8):viii61–viii65.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang JY, Lu AQ and Chen LJ: LncRNAs in
ovarian cancer. Clin Chim Acta. 490:17–27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ogunwobi OO and Segura MF: Editorial: PVT1
in Cancer. Front Oncol. 10:5887862020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Tan Y, Wang H, Xu M and Xu L:
Long Non-Coding RNA plasmacytoma variant translocation 1 (PVT1)
enhances proliferation, migration, and epithelial-mesenchymal
transition (EMT) of pituitary adenoma cells by activating
β-Catenin, c-Myc, and Cyclin D1 expression. Med Sci Monit.
25:7652–7659. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Q, Yu Y, Sun Z and Pan Y: Long
non-coding RNA PVT1 promotes cell proliferation and invasion
through regulating miR-133a in ovarian cancer. Biomed Pharmacother.
106:61–67. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kubota S and Takigawa M: Cellular and
molecular actions of CCN2/CTGF and its role under physiological and
pathological conditions. Clin Sci (Lond). 128:181–196. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arnott JA, Lambi AG, Mundy C, Hendesi H,
Pixley RA, Owen TA, Safadi FF and Popoff SN: The role of connective
tissue growth factor (CTGF/CCN2) in skeletogenesis. Crit Rev
Eukaryot Gene Expr. 21:43–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Klaassen I, van Geest RJ, Kuiper EJ, van
Noorden CJ and Schlingemann RO: The role of CTGF in diabetic
retinopathy. Exp Eye Res. 133:37–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen YW, Zhou YD, Chen HZ, Luan X and
Zhang WD: Targeting CTGF in Cancer: An emerging therapeutic
opportunity. Trends Cancer. 7:511–524. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho SW, Xu J, Sun R, Mumbach MR, Carter
AC, Chen YG, Yost KE, Kim J, He J, Nevins SA, et al: Promoter of
lncRNA Gene PVT1 is a tumor-suppressor DNA boundary element. Cell.
173:1398–1412.e22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu C, Jin J, Liang D, Gao Z, Zhang Y, Guo
T and He Y: Long Noncoding RNA PVT1 as a novel predictor of
metastasis, clinicopathological characteristics and prognosis in
human cancers: A meta-analysis. Pathol Oncol Res. 25:837–847. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu BQ, Jiang Y, Zhu F, Sun DL and He XZ:
Long noncoding RNA PVT1 promotes EMT and cell proliferation and
migration through downregulating p21 in pancreatic cancer cells.
Technol Cancer Res Treat. 16:819–827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang Z, Cui J and Song Y: Long noncoding
RNA PVT1 promotes EMT via mediating microRNA-186 targeting of
Twist1 in prostate cancer. Gene. 654:36–42. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Y, Du H, Bao L and Liu W: LncRNA PVT1
promotes ovarian cancer progression by silencing miR-214. Cancer
Biol Med. 15:238–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng J, Hu L, Cheng J, Xu J, Zhong Z,
Yang Y and Yuan Z: lncRNA PVT1 promotes the angiogenesis of
vascular endothelial cell by targeting miR-26b to activate
CTGF/ANGPT2. Int J Mol Med. 42:489–496. 2018.PubMed/NCBI
|
|
19
|
Ding LB, Li Y, Liu GY, Li TH, Li F, Guan J
and Wang HJ: Long non-coding RNA PVT1, a molecular sponge of
miR-26b, is involved in the progression of hyperglycemia-induced
collagen degradation in human chondrocytes by targeting CTGF/TGF-β
signal ways. Innate Immun. 26:204–214. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang CG, Lv L, Liu FR, Wang ZN, Na D, Li
F, Li JB, Sun Z and Xu HM: Connective tissue growth factor is a
positive regulator of epithelial-mesenchymal transition and
promotes the adhesion with gastric cancer cells in human peritoneal
mesothelial cells. Cytokine. 61:173–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang L, Hou J, Cui XH, Suo LN and Lv YW:
MiR-133b regulates the expression of CTGF in epithelial-mesenchymal
transition of ovarian cancer. Eur Rev Med Pharmacol Sci.
21:5602–5609. 2017.PubMed/NCBI
|
|
22
|
Prieto-García E, Díaz-García CV,
García-Ruiz I and Agulló-Ortuño MT: Epithelial-to-mesenchymal
transition in tumor progression. Med Oncol. 34:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding Y, Fang Q, Li Y and Wang Y:
Amplification of lncRNA PVT1 promotes ovarian cancer proliferation
by binding to miR-140. Mamm Genome. 30:217–225. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu C, Dai C, Guo Y, Qin R and Liu J: Long
non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role
in the regulatory mechanism of ovarian cancer. Biosci Rep.
40:BSR202008002020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yi K, Hou M, Yuan J, Yang L, Zeng X, Xi M
and Chen J: LncRNA PVT1 epigenetically stabilizes and
post-transcriptionally regulates FOXM1 by acting as a microRNA
sponge and thus promotes malignant behaviors of ovarian cancer
cells. Am J Transl Res. 12:2860–2874. 2020.PubMed/NCBI
|