Introduction
Endometrial cancer (EC) is the most common
gynecological malignancy in the developed countries (1). Its rate of incidence per 100,000
people in Europe was 32 and in the Czech Republic was 39 in the
year 2020 (https://ecis.jrc.ec.europa.eu/). Most EC cases are
diagnosed post-menopausally (with a peak incidence between 65–69
years) and in early stages with relatively favorable prognosis
(2). EC mortality is approximately
four times lower than EC incidence (<20%; www.svod.cz). However, the mortality may vary based on
geography and race (3).
Many non-genetic factors modify EC risk. While
excess of endogenous estrogens, obesity, insulin resistance, and
tamoxifen use increase EC risk, oral contraceptives and sufficient
physical activity have protective effects (4).
The risk of EC development is also affected by
genetic factors. Germline pathogenic variants (PV) in known
EC-predisposition genes are considered the most clinically
important [reviewed in (5)].
Germline variants in EC patients were studied by several next
generation sequencing (NGS) based studies, dominantly using limited
gene panels (21–84 genes) (6–15).
These studies reported variable prevalence of germline variants in
EC patients ranging from 4.5 to 23%. Majority of hereditary EC
cases are associated with Lynch syndrome (LS; also known as
hereditary nonpolyposis colorectal cancer), which is caused by
germline PV in mismatch repair genes (MMR; MLH1, MSH2, MSH6,
PMS2, and structural alterations of 3′ end of EPCAM)
(16). Guidelines for clinical
follow-up of carriers of germline PV in LS genes include specific
management of increased EC risk. Modest increase of EC risk has
been suggested in BRCA1 and BRCA2 PV carriers (most
notably the serous-like EC subtype), and other hereditary breast
and ovarian cancer genes (HBOC; ATM, BARD1, BRCA1, BRCA2, BRIP1,
CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, STK11, TP53)
(17). Other noteworthy candidate
EC-predisposition genes include e.g. POLD1 and POLE
(18). Germline missense PV
affecting proofreading capabilities of POLE/POLD1 are
associated with increased EC risk as a part of polymerase
proofreading-associated polyposis, but the importance of germline
POLD1/POLE truncating variants remains rather elusive
(18). Importantly, the genetic
basis of most EC cases has not been explained yet as the diagnosis
itself is not a criterion for germline genetic testing unless
fulfilling LS criteria (5).
We aimed to evaluate germline genetic background of
527 patients with uterine tumors to identify genes associated with
EC risk in our population, and to evaluate clinicopathological
features in germline PV carriers.
Materials and methods
Patients
For this retrospective cohort study, we collected
527 patients with uterine malignancies diagnosed at nine Czech
health care centers (General University Hospital in Prague, Masaryk
Memorial Cancer Institute, AGEL Laboratories, Gennet, GHC Genetics,
University Hospital Pilsen, Pronatal, Palacky University Olomouc)
and the Bank of Clinical Samples (First Faculty of Medicine). The
full list of all participating institutions is provided in the
Table SI. Patients were enrolled
between 2011–2021 and were Caucasians of the Czech origin. The
clinicopathological characteristics (Table I) revealed that endometrial cancers
(EC; 89.7%) were the dominant type of collected uterine
malignances, therefore the whole cohort of patients with uterine
malignancies will be hereafter referred to as ‘EC patients’.
Deficient MMR, microsatellite instability and MLH1
hypermethylation statuses were not available. We divided patients
according to national indication criteria for germline genetic
testing of LS and/or HBOC patients:
 | Table I.Clinicopathological characteristics
of 527 patients with EC. |
Table I.
Clinicopathological characteristics
of 527 patients with EC.
| Variables | All patients with
EC (N=527) | LS only
(N=151) | HBOC only
(N=16) | LS + HBOC
(N=82) | Non-indicated
(N=278) |
|---|
| Age at EC
diagnosis |
|
|
|
|
|
| Mean,
years | 59.1 | 50.8 | 59.0 | 51.3 | 65.8 |
| Median,
years | 60.5 | 47.8 | 57.0 | 49.0 | 65.3 |
| Range,
years | 24-92 | 24-91 | 51-73 | 29-82 | 50-92 |
| <50
years, n (%) | 120 (23.2) | 79 (53.4) | 0 | 41 (51.3) | 0 |
| ≥50
years, n (%) | 397 (76.8) | 69 (46.6) | 15 (100.0) | 39 (48.8) | 274 (100.0) |
| N.A.,
n | 10 | 3 | 1 | 2 | 4 |
| Histology of
uterine malignances, n (%) |
|
|
|
|
|
|
Endometrial carcinoma | 349 (89.7) | 76 (85.4) | 8 (72.7) | 48 (100.0) | 217 (90.0) |
|
Endometrioid
adenocarcinoma | 284 (73.0) | 65 (73.0) | 7 (63.6) | 44 (91.7) | 168 (69.7) |
|
Serous | 35 (9.0) | 4 (4.5) | 1 (9.1) | 3 (6.3) | 27 (11.2) |
| Clear
cell | 7 (1.8) | 2 (2.2) | 0 | 0 | 5 (2.1) |
|
Undifferentiated | 3 (0.8) | 0 | 0 | 0 | 3 (1.2) |
| Mixed
(endometroid/serous) | 3 (0.8) | 0 | 0 | 0 | 3 (1.2) |
| Mixed
(endometroid/serous/clear cell) | 1 (0.3) | 1 (1.1) | 0 | 0 | 0 |
| Mixed
(endometroid/clear cell) | 4 (1.0) | 3 (3.4) | 0 | 0 | 1 (0.4) |
|
EIN | 8 (2.1) | 1 (1.1) | 0 | 1 (2.1) | 6 (2.5) |
|
Unspecified | 4 (1.0) | 0 | 0 | 0 | 4 (1.7) |
|
Sarcoma | 40 (10.3) | 13 (14.6) | 3 (27.3) | 0 | 24 (10.0) |
|
Leiomyosarcoma | 32 (8.2) | 9 (10.1) | 2 (18.2) | 0 | 21 (8.7) |
|
Undifferentiated | 2 (0.5) | 0 | 0 | 0 | 2 (0.8) |
|
Endometrial stromal
sarcoma | 3 (0.8) | 2 (2.2) | 0 | 0 | 1 (0.4) |
|
Unspecified | 3 (0.8) | 2 (2.2) | 1 (9.1) | 0 | 0 |
| Unknown
malignant tumor of corpus uteri | 138 | 62 | 5 | 34 | 37 |
| FIGO grade, n
(%) |
|
|
|
|
|
| 1 | 123 (35.9) | 35 (48.6) | 4 (40.0) | 16 (45.7) | 68 (30.1) |
| 2 | 100 (29.2) | 15 (20.8) | 3 (30.0) | 12 (34.3) | 70 (31.0) |
| 3 | 120 (35.0) | 22 (30.6) | 3 (30.0) | 7 (20.0) | 88 (38.9) |
|
N.A. | 184 | 79 | 6 | 47 | 52 |
| FIGO stage, n
(%) |
|
|
|
|
|
| 0 | 8 (2.8) | 1 (2.1) | 0 | 1 (4.2) | 6 (2.8) |
| I | 176 (60.9) | 33 (68.8) | 4 (66.7) | 17 (70.8) | 122 (57.8) |
| II | 38 (13.1) | 5 (10.4) | 1 (16.7) | 2 (8.3) | 30 (14.2) |
|
III | 48 (16.6) | 8 (16.7) | 1 (16.7) | 2 (8.3) | 37 (17.5) |
| IV | 19 (6.6) | 1 (2.1) | 0 | 2 (8.3) | 16 (7.6) |
|
N.A. | 238 | 103 | 10 | 58 | 67 |
| Multiple primary
tumors in personal history, n (%) |
|
|
|
|
|
|
Present | 214 (40.6) | 69 (45.7) | 16 (100.0) | 82 (100.0) | 47 (16.9) |
|
Absent | 313 (59.4) | 82 (54.3) | 0 | 0 | 231 (83.1) |
| Multiple primary
tumors in personal history, n (%) |
|
|
|
|
|
|
CRC | 31 (5.9) | 31 (20.5) | 0 | 0 | 0 |
| OC | 59 (11.2) | 0 | 1 (6.3) | 58 (70.7) | 0 |
| BC | 80 (15.2) | 14 (9.3) | 15 (93.8) | 13 (15.9) | 38 (13.7) |
| Triple
primary EC+(BC/OC/CRC) | 13 (2.5) | 2 (1.3) | 0 | 11 (13.4) | 0 |
|
Other | 31 (5.9) | 22 (14.6) | 0 | 0 | 9 (3.2) |
|
None | 313 (59.4) | 82 (54.3) | 0 | 0 | 231 (83.1) |
| Family cancer
history (first/second degree), n (%) |
|
|
|
|
|
|
Positive | 353 (69.8) | 120 (81.6) | 13 (100.0) | 56 (73.7) | 164 (60.7) |
|
Negative | 153 (30.2) | 27 (18.4) | 0 | 20 (26.3) | 106 (39.3) |
|
Unknown | 21 | 4 | 3 | 6 | 8 |
| Tumors in family
history, n (%) |
|
|
|
|
|
| EC | 35 (6.9) | 14 (9.5) | 1 (7.7) | 6 (7.9) | 14 (5.2) |
|
CRC | 88 (17.4) | 39 (26.5) | 4 (30.8) | 15 (19.7) | 30 (11.1) |
| OC | 15 (3.0) | 7 (4.8) | 1 (7.7) | 5 (6.6) | 2 (0.7) |
| BC | 60 (11.9) | 14 (9.5) | 3 (23.1) | 9 (11.8) | 34 (12.6) |
|
Multiple (EC/OC/CRC) | 10 (2.0) | 10 (6.8) | 0 | 0 | 0 |
|
Other | 145 (28.7) | 36 (24.5) | 4 (30.8) | 21 (27.6) | 84 (31.1) |
|
None | 153 (30.2) | 27 (18.4) | 0 | 20 (26.3) | 106 (39.3) |
|
Unknown | 21 | 4 | 3 | 6 | 8 |
Breast cancer or ovarian cancer (C50/C56)-national
indication criteria for germline genetic testing [HBOC criteria;
cancer diagnoses (C##) correspond to the International
Classification of Diseases 10; available at https://icd.who.int/browse10/2019/en#/C00-C97].
Personal history: i) patient is diagnosed with C50 <45 years or
<50 years, if family history is unknown; ii) patient has
bilateral C50 with the age of diagnosis of the first one <50
years and of both <60 years; iii) patient is diagnosed with
triple negative C50 ≤60 years; iv) patient is a male diagnosed with
C50; v) patient is diagnosed with either C56, C57 or C48; vi)
patient has a duplicity od C50 and C25 regardless of age. Family
history: i) patient and two relatives are diagnosed with C50; ii)
patient and one relative are diagnosed with C50 <50 years or
both C50 <60 years (patient included); iii) patient and a direct
relative (parent, sibling, child, alternatively mother or father's
sister) are diagnosed with either ovarian cancer, fallopian tube or
primary peritoneal tumor, triple negative C50/medullar C50, male
relative diagnosed with C50, pancreatic cancer, prostate cancer
with Gleason score ≥7 or primary metastatic C61.
Colorectal cancer or EC-national indication criteria
for germline genetic testing (LS criteria): i) Age of diagnosis
<50 years; ii) proven microsatellite instability <60 years;
iii) patient has a concurrent diagnosis linked to LS (colorectal
cancer, stomach cancer, pancreatic cancer, ovarian cancer, small
intestine cancer, ureter cancer, renal pelvis cancer, bile tract
cancer, glioblastoma); iv) patient and one first degree relative
have diagnoses linked to LS <50 years; v) patient and two second
degree relatives have diagnoses linked to LS regardless of the age
of diagnosis; and vi) patients with colorectal cancer and more than
ten adenomas/polyps.
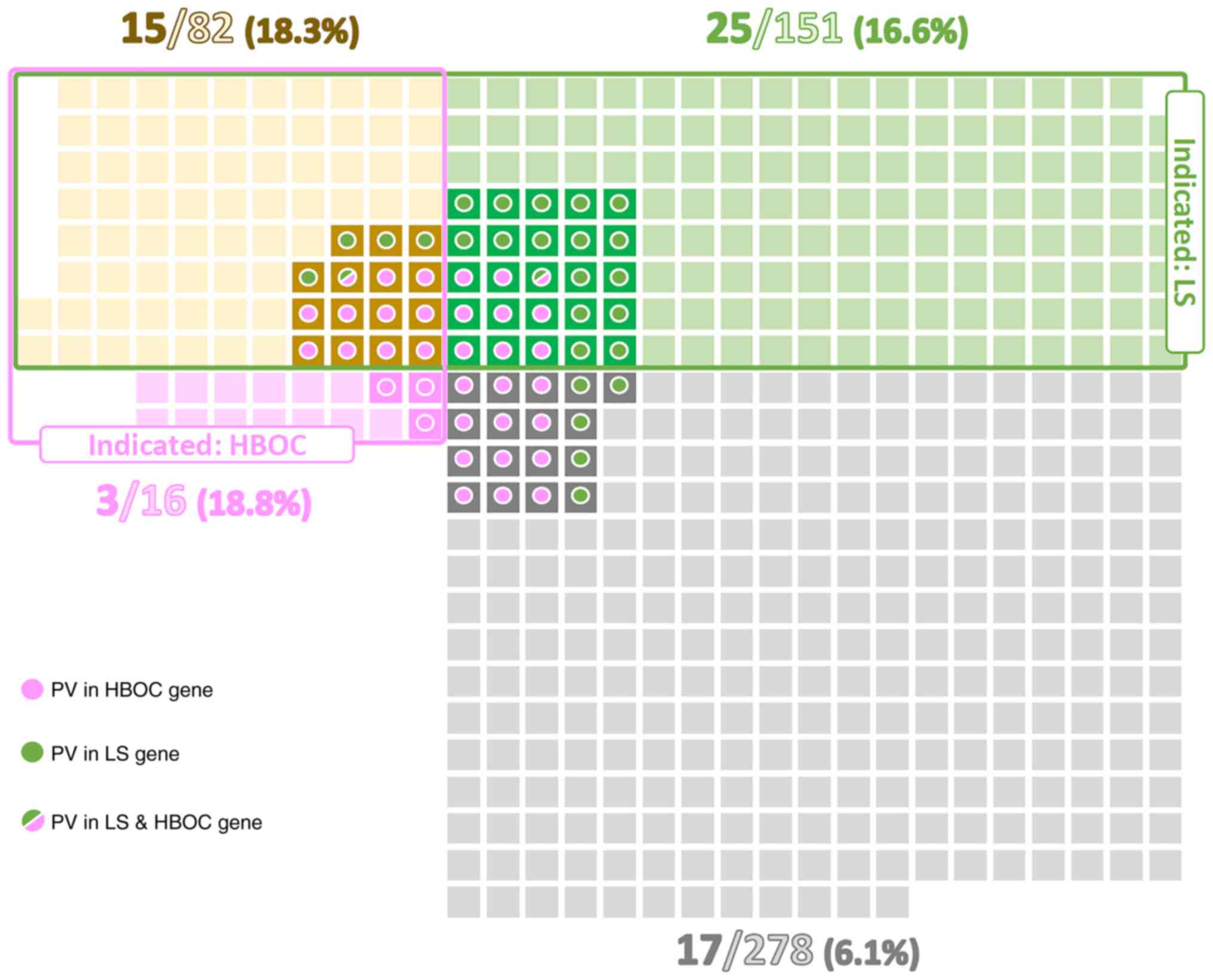
Of all patients 151/527 (28.7%) met only LS genetic
testing criteria, 16/527 (3.0%) met only HBOC criteria, and 82/527
(15.6%) met both these criteria. A total of 278/527 (52.7%)
patients would not be indicated for germline genetic testing.
The study was approved by Ethics Committees of
participating institutions. Written consent for the research
analysis was obtained from all participants. Clinicopathological
information was collected during genetic counselling or retrieved
from patients' record.
Two sets of population-matched controls (PMC) were
used for comparisons with analyzed EC patients. First, used as a
reference for genetic variant prioritization, included 777
non-cancer volunteers aged >60 years that were analyzed
identically with EC patients as described previously (19). Second group, used in case-control
analyses, included 1662 PMC analyzed as described previously
(20). Briefly, the unselected
controls (1,170 males and 492 females; median age 57 years, range
18–88 years) were unrelated individuals analyzed by whole-exome
sequencing by National Center for Medical Genomics (https://ncmg.cz/) for various noncancer
conditions.
Genetic testing using panel NGS
Genomic DNA was isolated from peripheral blood
collected at the time of enrollment in each respective center. DNA
samples were analyzed by NGS using a custom-designed CZECANCA panel
as described previously (21) with
minor modifications reflecting recent technological updates. These
modifications included a new probe synthesis HyperDesign (Roche)
improving target coverage for all 226 genes (the sequence capture
panel development is shown in detail on the panel web page:
http://www.czecanca.cz/eng/panel.html, and full list
of genes targeted in this project is described in Table SII). Further modifications included
usage of cheaper and faster enzymatic fragmentation replacing
ultrasound DNA fragmentation, preparation of DNA libraries using
recently introduced KAPA HyperPlus Library Preparation kit (Roche;
according to the manufacturer's instruction) and Illumina
NextSeq500 sequencing. Resulting NGS data were processed by an
in-house bioinformatics pipeline as we described previously
(21). Briefly, SAM files were
generated from FASTQ using NovoAlign v2.08.03 (http://www.novocraft.com/products/novoalign/) and
transformed into BAM by Picard tools v1.129 (https://broadinstitute.github.io/picard/). The Genome
Analysis Toolkit v3.8.1 (https://software.broadinstitute.org/gatk/) (22) was used to prepare variant-call
format, annotated by SnpEff v4.3 (http://pcingola.github.io/SnpEff/). Identification of
medium size indels was performed by Pindel v0.2.5a7 (http://gmt.genome.wustl.edu/packages/pindel/) and copy
number variations (CNV) were detected using CNV kit v0.7.4
(https://pypi.python.org/pypi/CNVkit).
All 226 analyzed genes were divided into 19 known
EC-predisposition genes described by NCCN guidelines or reviewed by
Spurdle et al (5) and 207
other ‘candidate’ genes. Five genes associated with LS (MLH1,
MSH2, MSH6, PMS2, EPCAM) and 14 genes associated with HBOC
(ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, NF1, PALB2, PTEN,
RAD51C, RAD51D, STK11, TP53) were considered as the
EC-predisposition genes. The remaining 207 candidate
cancer-susceptibility genes included those that have been
episodically associated with EC predisposition (incl. APC,
MUTYH, NBN, POLD1, POLE; Table
SII) (5).
Variant prioritization
Genetic variants found in patients were filtered,
excluding variants: i) with low sequencing quality (q<150); ii)
with a high minor allele frequency (MAF >0.001) in population
databases (gnomAD https://gnomad.broadinstitute.org/, Exome Sequencing
Project https://evs.gs.washington.edu/EVS/, 1000 Genomes
Project http://www.internationalgenome.org/) (23–25)
unless classified as pathogenic/likely pathogenic (P/LP) in the
ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) (26); iii) present with frequency higher
than 0.5% in a group of 777 PMC, except for variants with P/LP
ClinVar classification; iv) in untranslated region, intronic
outside of consensus splice sites, synonymous and
insertion/deletions not resulting in a frameshift unless classified
as pathogenic/likely pathogenic (P/LP) in the ClinVar database; v)
classified as benign/likely benign in ClinVar with at least
two-star rating; vi) low risk variants in BRCA2
(c.9976A>T; p.Lys3326Ter) and in CHEK2 (c.470T>C;
p.Ile157Thr).
Resulting set of variants was evaluated according to
the ACMG (American College of Medical Genetics) recommendations
(27). Variants mentioned in
ClinVar as a single submitter or with a conflicting interpretation
of pathogenicity were categorized as variants of uncertain
significance (VUS). Whole gene duplication and truncating variants
localized in the last exon were considered VUS, unless they were
classified as P/LP in ClinVar. All PV were inspected in Integrative
Genomics Viewer or confirmed using Sanger sequencing or multiplex
ligation-dependent probe amplification analysis (MRC Holland).
Confirmed PV were submitted to ClinVar database.
Statistical analysis
The frequencies of PV in EC patients were compared
with the frequencies of PV in a group of 1662 unselected PMC. Odds
ratios (OR) with 95% confidence intervals (CI) were calculated for
EC patients carrying found germline PV using 2×2 contingency table.
The χ2 or Fisher's exact tests were used for the
calculation of P-values (considered significant when P<0.05).
Differences in age at diagnosis were analyzed by one-way ANOVA
followed by Tukey-Kramer's test. Statistical analysis was performed
using the R language v4.1.
Results
Germline PV in patients with uterine
malignances
We performed germline genetic testing in 527 Czech
EC patients including 249 individuals fulfilling LS, HBOC, LS +
HBOC indication criteria and 278 individuals not fulfilling any
criteria for germline genetic testing. Germline PV were
significantly more frequent in patients (118/527; 22.4%) than in
population-matched controls (290/1662; 17.4%; P=0.011).
Germline PV in EC-predisposition
genes
PV were found in 12 (out of 19 tested)
EC-predisposition genes (Table
II). Frequency of these variants was more than four-times
higher in EC patients (60/527; 11.4%; Table SIII) than in PMC (46/1662; 2.8%;
P=9.7×10−16). PV in LS genes were found in 27/527 (5.1%)
patients (half of them were MSH6 PV carriers) and in 4/1662
(0.25%) controls and they represented the strongest genetic risk
factor for EC development (OR=22.4, P=1.8×10−17).
Interestingly, no PMS2 PV were observed among patients.
BRCA1, BRCA2 and CHEK2 were the most frequently
mutated HBOC genes, their PVs conferred significantly increased EC
risk for female carriers. However, this risk was lower in
comparison to LS genes (ranging from high EC risk in BRCA2
to moderate EC risk in BRCA1 and CHEK2,
respectively). PV in the remaining 11 HBOC genes were not
identified or did not differ significantly from PMC (Table II). Two carriers harbored
coincidental mutations in MLH1/BRCA1 and
MSH2/ATM, respectively.
 | Table II.Frequencies of germline PV in 19
ec-predisposition genes. |
Table II.
Frequencies of germline PV in 19
ec-predisposition genes.
|
|
| Indication for
germline genetic testing |
|
| All patients with
EC vs. PMC |
|---|
|
|
|
|
|
|
|---|
| Gene group | Germline PV | LS, n (%)
(N=151) | HBOC, n (%)
(N=16) | LS+HBOC, n (%)
(N=82) | Non-indicated, n
(%) (N=278) | All patients with
EC, n (%) (N=527) | PMC, n (%)
(N=1662) |
|
|---|
| OR (95% CI) | P-value |
|---|
| LS |
MLH1a | 3 (2.0) | 0 | 2a (2.4) | 1 (0.4) | 6a (1.1) | 1 (0.1) | 19.1 |
1.3×10−4 |
|
|
|
|
|
|
|
|
| (2.3-159.1) |
|
|
|
MSH2b | 6b (4.0) | 0 | 2 (2.4) | 0 | 8b (1.5) | 0 | N.A. |
|
|
| MSH6 | 8 (5.3) | 0 | 1 (1.2) | 4 (1.4) | 13 (2.4) | 0 | N.A. |
|
|
| PMS2 | 0 | 0 | 0 | 0 | 0 | 3 (0.2) | N.A. |
|
|
| EPCAM | 0 | 0 | 0 | 0 | 0 | 0 | N.A. |
|
|
| All LS | 17 | 0 | 5 (6.1) | 5 (1.8) | 27 (5.1) | 4 (0.2) | 22.4 |
1.8×10−17 |
|
| genes | (11.3) |
|
|
|
|
| (7.8-64.3) |
|
| HBOC | ATMb | 1b (0.7) | 0 | 1 (1.2) | 3 (1.1) | 5b (1.0) | 7 (0.4) | 2.3 | 0.2 |
|
|
|
|
|
|
|
|
| (0.2-7.2) |
|
|
| BARD1 | 0 | 0 | 1 (1.2) | 0 | 1 (0.2) | 0 | N.A. |
|
|
|
BRCA1a | 2 (1.3) | 2 (12.5) | 6a (7.3) | 1 (0.4) | 11a (2.1) | 9 (0.5) | 3.9 |
1.0×10−3 |
|
|
|
|
|
|
|
|
| (1.6-9.5) |
|
|
| BRCA2 | 1 (0.7) | 1 (6.3) | 0 | 5 (1.8) | 7 (1.3) | 3 (0.2) | 7.4 |
2.0×10−3 |
|
|
|
|
|
|
|
|
| (1.9-28.9) |
|
|
| BRIP1 | 0 | 0 | 0 | 1 (0.4) | 1 (0.2) | 3 (0.2) | 1.1 | >0.9 |
|
|
|
|
|
|
|
|
| (0.1-10.1) |
|
|
| CDH1 | 0 | 0 | 0 | 0 | 0 | 0 | N.A. |
|
|
| CHEK2 | 3 (2.0) | 0 | 1 (1.2) | 2 (0.7) | 6 (1.1) | 6 (0.4) | 3.2 |
4.0×10−2 |
|
|
|
|
|
|
|
|
| (1.0-9.9) |
|
|
| NF1 | 0 | 0 | 0 | 0 | 0 | 1 (0.1) | N.A. |
|
|
| PALB2 | 1 (0.7) | 0 | 0 | 0 | 1 (0.2) | 8 (0.5) | 0.4 | 0.4 |
|
|
|
|
|
|
|
|
| (0.1-3.1) |
|
|
| PTEN | 1 (0.7) | 0 | 0 | 0 | 1 (0.2) | 1 (0.1) | 3.2 | 0.4 |
|
|
|
|
|
|
|
|
| (0.2-50.5) |
|
|
| RAD51C | 0 | 0 | 2 (2.4) | 0 | 2 (0.4) | 2 (0.1) | 3.2 | 0.2 |
|
|
|
|
|
|
|
|
| (0.4-22.5) |
|
|
| RAD51D | 0 | 0 | 0 | 0 | 0 | 0 | N.A. |
|
|
| STK11 | 0 | 0 | 0 | 0 | 0 | 0 | N.A. |
|
|
| TP53 | 0 | 0 | 0 | 0 | 0 | 2 (0.1) | N.A. |
|
|
| All | 9 (6.0) | 3 (18.8) | 11 (13.4) | 12 (4.3) | 35 (6.6) | 42 (2.5) | 2.7 |
7.9×10−5 |
|
| HBOC |
|
|
|
|
|
| (1.7-4.3) |
|
| All genes | All PV | 26 | 3 | 16 | 17 | 62 | 46 |
|
|
| All genes | All | 25b | 3 (18.8) | 15a (18.3) | 17 (6.1) | 60a,b (11.4) | 46 (2.8) |
|
|
|
| carriers | (16.6) |
|
|
|
|
|
|
|
Indication criteria for identification
of PV carriers
Among EC patients indicated for germline genetic
testing according to the above-mentioned criteria, the proportions
of PV carriers fulfilling criteria for LS, HBOC, and both
conditions were similar (16.6, 18.8, and 18.3%, respectively).
These proportions were approximately three-times higher than in EC
patients not fulfilling any criteria for germline genetic testing
(6.1%; Fig. 1). As expected, the
highest proportion of PV in LS genes (11.3%) was detected in a
subgroup of patients fulfilling criteria only for LS testing.
Similarly, patients meeting solely the HBOC testing criteria had
the highest frequency (18.8%) of PV in HBOC genes. Even though the
overall percentage of PV carriers differed between subgroups of
patients meeting both LS + HBOC genetic testing criteria (18.3%)
and not fulfilling any criteria (6.1%), the ratio of carriers of PV
in LS:HBOC genes in these two subgroups was similar (5:11 vs. 5:12;
Table II, Fig. 1). On the other hand, highly
penetrant genes (MLH1, MSH2, BRCA1) were predominantly
affected in the subgroup fulfilling both criteria, whereas the
subgroup of non-indicated patients was characterized by PV in less
penetrant genes (MHS6, ATM).
Moreover, among non-indicated patients we found 2
PVs in HBOC genes in subset of 41 patients with double primary EC
and breast cancer (BC; 1×ATM, 1×BRCA1; 2/41; 4.9%)
and 3 PVs in HBOC genes in subset of 31 patients with EC and BC in
family cancer history (2×BRCA2, 1×CHEK2; 3/31;
9.7%).
Germline PV in other candidate cancer
predisposition genes
The overall prevalence of PV in remaining candidate
genes (identified in 48 out of 207 genes) was significantly higher
in EC patients (66/527; 12.5%) compared to controls (139/1662;
8.4%; P=0.004; Table SIV). Eight
EC patients (and no PMC) carried a coincidental PV in
EC-predisposition and candidate genes. Excluding all 60 carriers of
PV in EC-predisposition genes, the frequency of PV carriers in
other candidate genes was still significantly higher in 467 EC
patients (N=58; 12.4%) in comparison to 1616 PMC (N=139; 8.6%;
P=0.01). The most frequent PV were found in MUTYH
(monoallelic PV in 5/467, 1.1%) and FANCA (4/467; 0.8%).
Their frequencies, however, did not differ from that in PMC
(MUTYH−18/1616, 1.1%; FANCA−10/1616, 0.6%).
Interestingly, three patients carried germline
truncating variant in the genes coding for DNA polymerases (two in
POLE and one in POLD1) that have been linked to
EC-predisposition previously (5).
In contrast, only one POLE and no POLD1 mutation was
detected in PMC. Thus, the overall frequency of PV in DNA
polymerases was significantly higher in EC-predisposition gene
negative patients (3/467; 0.6%) than in PMC (1/1616; 0.06%;
OR=10.44; 95% CI 1.08-100.51; P=0.012).
Regarding subgroups of patients based on indication
criteria for genetic testing, the frequency of PV in candidate
predisposition genes (after excluding the carriers of PV in
EC-predisposition genes) was significantly higher in patients
fulfilling both indication criteria for LS + HBOC (14/67; 20.9%) in
comparison to subgroup of patients fulfilling no genetic testing
criteria (28/261; 10.7%; P=0.04, Table
SIV). The frequencies of PV in patients meeting indication
criteria for LS only and HBOC only did not differ significantly
(14/126; 11.1 and 2/13; 15.4%, respectively).
Clinicopathological characteristics in
germline PV carriers
The median age at EC onset was significantly lower
only in patients with PV in LS genes compared to non-carriers (51.0
vs. 61.4 years, P=0.01, Fig.
2A).
Concerning the histology subtypes (Fig. 2C), the overall frequency of PV in
EC-predisposition genes was similar in patients with endometrial
carcinoma to those with sarcoma subtypes (39/349, 11.2% and 4/40,
10.0%; respectively); however, no carrier of PV in LS gene was
diagnosed with sarcoma. Interestingly, two out of eight patients
diagnosed with precancerous EIN (endometrial intraepithelial
neoplasia) carried a PV in BRCA1. Unfortunately, the
histologic subtypes of endometrial carcinomas other than
endometrioid were rarely represented, thus the frequencies of PVs
in these subgroups cannot be calculated and compared.
Analysis of patients with second primary tumors
(Fig. 2B) revealed that the highest
frequency of PV in EC-predisposition genes was found in patients
with 3 primary tumors and in patients with second primary
colorectal cancer (CRC). The proportion between carriers of PV in
LS and HBOC genes respected the corresponding indication criteria:
the carriers of LS gene variants were enriched in patients with EC
+ CRC and 3 primary tumors. Accordingly, all 13 patients with 3
primary tumors developed either CRC (N=5) and/or ovarian cancer
(OC; N=10). The carriers of PV in HBOC genes were more frequent in
patients with EC + OC and EC + BC.
When considering family cancer history (Fig. 2D), the highest frequency (reaching
40%) of PV in EC-predisposition genes were found in small subgroups
of patients with family history of multiple primary tumors and
family history of ovarian tumors. Not surprisingly, predominant
tumor types in a family were in concordance with the elevated
frequencies of PV in LS or HBOC genes.
The prevalence of carriers of PV in candidate
predisposition genes did not differ from that of non-carriers in
any of the clinicopathological categories.
The information about immunohistochemistry and
microsatellite instability in EC tumor specimens was
unavailable.
Discussion
Pathogenic germline alterations in LS genes are
considered the most significant genetic risk factor for EC
predisposition (5). In our study,
the carriers of PV in LS genes represented 5.1% of all analyzed EC
patients. This frequency is approximately in the middle of
frequencies reported by other studies (Fig. 3). Variable frequencies result from
inconsistent patients' enrollment criteria. Studies reporting the
highest frequency (Tian et al (7), Karpel et al (13), Susswein et al (14), Heald et al (15) with 22.7, 9.4, 8.4, and 8.2% of LS PV
carriers, respectively) analyzed high-risk EC patients enriched in
individuals with familial LS criteria or in patients with positive
MMR gene immunohistochemistry [Tian et al (7)]. In contrast, the lowest frequency of
PV in LS genes was reported by studies with unselected EC cases,
including Huang et al (28)
(1.1%), a study of EC samples from The Cancer Genome Atlas (TCGA).
We have found similar differences as we identified 22/233 (9.4%)
vs. 5/294 (1.7%) carriers of PV in LS genes in LS-indicated vs. LS
non-indicated patients, respectively (Fig. 1). Interestingly, despite differences
in frequencies of PV in EC patients, the risk of EC development in
LS PV carriers was similar in our and LaDuca et al (29) study (OR 22.4 and 20.1, respectively;
Fig. 3), the only study among those
previously published that quantified the EC risk associated with PV
in LS genes.
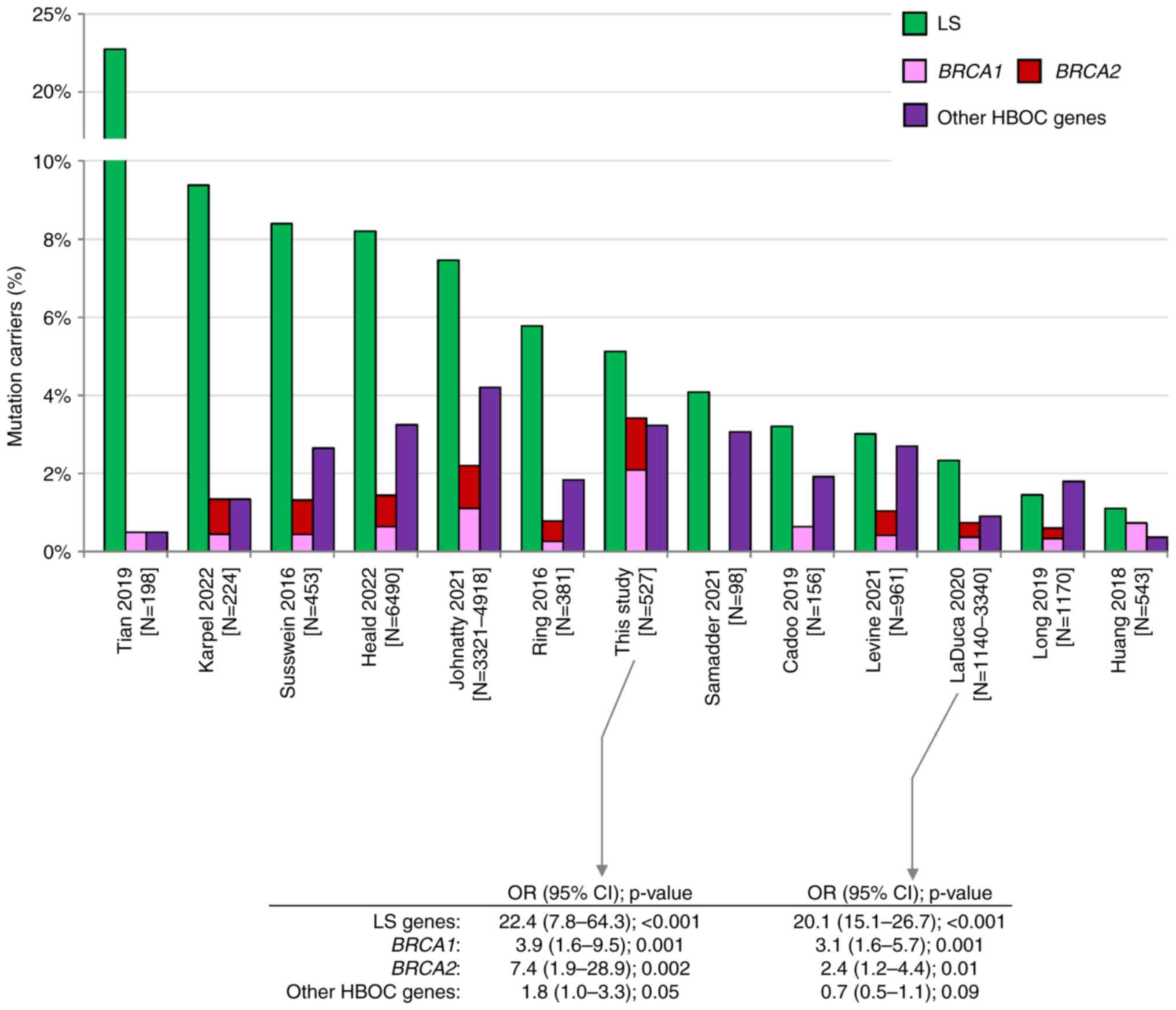 | Figure 3.Comparison among previously published
studies describing germline PV in patients with endometrial cancer
(6–15,28,29).
Green, pink, red and purple bars represent the prevalence of PV in
LS genes, BRCA1, BRCA2 and other HBOC genes (ATM, BARD1,
BRIP1, CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, STK11 and
TP53), respectively. CI, confidence interval; HBOC,
hereditary breast and ovarian cancer; LS, Lynch syndrome; N,
number; OR, odds ratio; PV, pathogenic variant. |
Even though only less than 20% of analyzed EC
patients (98/527, 18.6%) met the HBOC germline genetic testing
criteria, the overall frequencies of PV carriers in
BRCA1/BRCA2 were unusually high in contrast to other studies
(Fig. 3). We identified 11 PVs in
BRCA1 (2.1%) and 7 PVs in BRCA2 (1.3%). Compared to
frequencies of PVs in controls we calculated the risks OR=3.9 for
BRCA1 and OR=7.4 for BRCA2 (Table II). The risk of EC development
associated with BRCA1 and BRCA2 mutations was
substantially lower than in LS carriers, and similar to EC risk
reported previously by LaDuca et al (29). Our results suggest that PV in
BRCA1/BRCA2 are associated with at least moderate EC
risk. Among 16 EC patients meeting only the HBOC criteria, three
harbored BRCA1/BRCA2 mutation. This was also
documented by results of a small study by Vietri et al
(30), who identified PV in
BRCA1/BRCA2 in 9/21 hereditary EC patients fulfilling
HBOC testing criteria. In the group of 82 patients meeting both LS
and HBOC testing criteria, BRCA1 PV were more frequent than
PV in LS genes. Moreover, up to 5 and 10% of PVs in HBOC genes were
identified in non-indicated EC patients with BC in personal or
family cancer history, respectively. This further implies that the
diagnosis of EC should be considered as a part of indication
criteria for HBOC germline genetic testing irrespective to EC
histology subtype. Among PV carriers in other HBOC genes, PV in
CHEK2 and ATM were the most frequent. Importantly, PV
in CHEK2 were associated with moderately increased risk
(OR=3.2, P=0.04). Mutations in CHEK2 were associated with
predisposition to EC in several studies previously (31).
Our analysis of other candidate genes showed that
only PVs in POLD1 and POLE (three truncating
variants, one in POLD1, two in POLE) were
significantly associated with EC risk. Germline truncating variants
in DNA polymerase genes in our EC patients conferred about 10-times
increased risk of EC development. Germline missense PV in both DNA
polymerase genes affecting exonuclease domains were previously
linked to EC predisposition (5) and
their specific somatic missense PV represent important predictive
markers for favorable prognosis and/or immune checkpoint therapy in
EC patients (32–34). However, the exact risk as well as
the overall role of germline truncating variants needs to be
further validated in larger cohorts due to the low frequency of
POLD1 and POLE mutation carriers.
Analysis of clinicopathological characteristics
confirmed an earlier age at disease onset in carriers of LS gene
mutations in comparison to non-carriers as referred in other
studies (6,7,9,10). The
age at EC onset varied even among the carriers of PV in particular
LS genes: the carriers of PV in MSH6 had later age at onset
(56 years) compared to the MLH1/MSH2 PV carriers (48
years), as previously described by Tian et al (7). Interestingly, the age at EC onset in
carriers of PV in HBOC genes did not differ from non-carriers.
As expected, other differences in
clinicopathological characteristics largely corresponded to
subgroups of patients classified according to the germline genetic
testing criteria. PV in LS genes were most frequently identified in
patients with ≥3 primary tumors or second primary CRC in personal
cancer history, or multiple primary tumors/CRC in family cancer
history. Similarly, carriers of PV in HBOC genes recruited in
majority from individuals with BC/OC in personal or family cancer
history. On the other hand, clinicopathological characteristics did
not differ in carriers of candidate EC-predisposition genes and
non-carriers.
Generally and as expected, we have identified the
majority of PV in the groups of patients fulfilling genetic testing
criteria for LS or HBOC with majority of PV in genes related to a
corresponding cancer syndrome. Overall, 43/60 PV (71.7%) carriers
were indicated for germline genetic testing. Importantly, remaining
17 PV carriers, who would not be indicated for genetic testing
using current indication criteria, still represent a significant
proportion (28.3%) of cases carrying a germline PV in the LS
(MLH1, MSH6) or the HBOC (ATM, BRCA1, BRCA2, BRIP1,
CHEK2) genes. Of these, two had double primary tumors and an
additional 10 had a positive family cancer history. The frequency
of PV carriers among EC patients with double primary tumors was
15.4% (33/214). While we have found eight PV carriers in HBOC genes
and four carriers in LS genes (including a patient with
co-occurrence of BRCA1 and MLH1 PV and diagnosed with
EC, OC, and BC) in the group of 69 patients with EC and OC (11.6%),
we have identified eight carriers of LS genes mutation and only one
additional carrier of the CHEK2 gene mutation (a patient
with EC, CRC, and melanoma) in the group of 34 patients with EC and
CRC (26.5%). This suggests that the presence of double primary
tumors could potentially represent a sole indication criterion for
germline genetic testing, as indicated by previous studies
(19,20,35,36).
Strengths of our study include homogeneity of
studied population consisting of Caucasians, Slavs of the Czech
origin and inclusion of PMC that allowed calculation of
overall/gene-level risks for EC development. Study limitations
include retrospective design and unavailability of EC tumors
immunohistochemistry, microsatellite instability and mutation
status of POLE, which prevented us from correlating presence
of germline mutations with different molecular subtypes of EC.
Moreover, as approximately half of the analyzed EC patients
(292/527, 55.4%) were recruited from the CZECANCA consortium
(focused on analyses of genetic cancer predisposition), we cannot
exclude a potential bias toward enriched prevalence of PV carriers.
To minimize this bias, we divided all enrolled patients according
to the testing criteria and analyzed them independently.
In conclusion, over 11% of EC patients carried a
germline PV in genes associated with established germline cancer
predisposition. EC patients fulfilling LS criteria had five-times
higher chance to carry a LS gene PV than EC patients not fulfilling
criteria for germline genetic testing. Presence of PV in LS gene
increases the EC risk 20-fold when compared with non-carriers.
However, 28.3% of PV carriers in clinically relevant genes would
not be indicated for germline genetic testing using current
indication criteria. Therefore, we believe that EC as a second
primary tumor in proband or occurrence of EC in a family cancer
history should be considered within the indication criteria for
germline genetic testing. This is of particular importance for
countries where reflex testing is not routinely performed in EC
patients.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Stanislav
Kmoch and Dr Viktor Stranecky (National Center for Medical
Genomics, Prague, Czech Republic) (LM2018132) for performing
whole-exome sequencing of PMC (project
CZ.02.1.01/0.0/0.0/18_046/0015515).
Funding
This work was supported by the Ministry of Health of the Czech
Republic (grant no. NU20-03-00285), institutional support of the
Ministry of Health of the Czech Republic (grant nos. DRO VFN 64165
and DRO MMCI 00209805), the Ministry of Education, Youth, and
Sports of the Czech Republic (grant nos. BBMRI-CZ LM2023033,
LM2023067 and LX22NPO5102), the Charles University (grant no.
GAUK902120), and the Charles University institutional programs
(grant nos. SVV260516 and COOPERATIO, research area DIAG).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to restrictions
imposed by national regulatory authorities but are available from
the corresponding author on reasonable request.
Authors' contributions
JK, SJ, MB, LC, MC, PD, KH, MH, SC, MaKo, MoKo, VK,
EM, BO, AP and ST performed the experiments. MV, LD, LF, OH, MaKa,
JN, MS, JiS, VS, IS and JaS provided additional data, and
participated in data analysis and interpretation. PZ, PN and MZ
participated in data analysis and interpretation, as well as
revision of the manuscript. PK, ZK, TZ and MJ designed the study
and were involved in drafting and editing of the manuscript. JK and
MJ confirm the authenticity of all the raw data. All authors agreed
to be accountable for all aspects of the work. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent. The
study was conducted in line with the Helsinki Declaration and was
approved by The Ethics Committee of General University Hospital in
Prague, Prague, Czech Republic under reference number 11/20 Grant
VFN IGP.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ACMG
|
American College of Medical
Genetics
|
|
BC
|
breast cancer
|
|
CI
|
confidence interval
|
|
CNV
|
copy number variation
|
|
CRC
|
colorectal cancer
|
|
EC
|
endometrial cancer
|
|
EIN
|
endometrial intraepithelial
neoplasia
|
|
FIGO
|
The International Federation of
Gynecology and Obstetrics
|
|
GGT
|
germline genetic testing
|
|
HBOC
|
hereditary breast and ovarian
cancer
|
|
LS
|
Lynch syndrome
|
|
MMR
|
mismatch repair
|
|
N
|
number
|
|
N.A.
|
not available
|
|
NCCN
|
National Comprehensive Cancer
Network
|
|
NGS
|
next generation sequencing
|
|
OC
|
ovarian cancer
|
|
OR
|
odds ratio
|
|
P/LP
|
pathogenic/likely pathogenic
|
|
PMC
|
population-matched controls
|
|
PV
|
pathogenic variant
|
|
VUS
|
variant of uncertain significance
|
References
|
1
|
Lortet-Tieulent J, Ferlay J, Bray F and
Jemal A: International patterns and trends in endometrial cancer
incidence, 1978–2013. J Natl Cancer Inst. 110:354–361. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arend RC, Jones BA, Martinez A and
Goodfellow P: Endometrial cancer: Molecular markers and management
of advanced stage disease. Gynecol Oncol. 150:569–580. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raglan O, Kalliala I, Markozannes G,
Cividini S, Gunter MJ, Nautiyal J, Gabra H, Paraskevaidis E,
Martin-Hirsch P, Tsilidis KK and Kyrgiou M: Risk factors for
endometrial cancer: An umbrella review of the literature. Int J
Cancer. 145:1719–1730. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spurdle AB, Bowman MA, Shamsani J and Kirk
J: Endometrial cancer gene panels: Clinical diagnostic vs research
germline DNA testing. Mod Pathol. 30:1048–1068. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Levine MD, Pearlman R, Hampel H, Cosgrove
C, Cohn D, Chassen A, Suarez A, Barrington DA, McElroy JP, Waggoner
S, et al: Up-front multigene panel testing for cancer
susceptibility in patients with newly diagnosed endometrial cancer:
A multicenter prospective study. JCO Precis Oncol. 5:1588–1602.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tian W, Bi R, Ren Y, He H, Shi S, Shan B,
Yang W, Wang Q and Wang H: Screening for hereditary cancers in
patients with endometrial cancer reveals a high frequency of
germline mutations in cancer predisposition genes. Int J Cancer.
145:1290–1298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cadoo KA, Mandelker DL, Mukherjee S,
Stewart C, DeLair D, Ravichandran V, Srinivasan P, Hurley D, Kemel
Y, Arnold AG, et al: Understanding inherited risk in unselected
newly diagnosed patients with endometrial cancer. JCO Precis Oncol.
3:PO.18.00338. 2019.
|
|
9
|
Long B, Lilyquist J, Weaver A, Hu C,
Gnanaolivu R, Lee KY, Hart SN, Polley EC, Bakkum-Gamez JN, Couch FJ
and Dowdy SC: Cancer susceptibility gene mutations in type I and II
endometrial cancer. Gynecol Oncol. 152:20–25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ring KL, Bruegl AS, Allen BA, Elkin EP,
Singh N, Hartman AR, Daniels MS and Broaddus RR: Germline
multi-gene hereditary cancer panel testing in an unselected
endometrial cancer cohort. Mod Pathol. 29:1381–1389. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Samadder NJ, Riegert-Johnson D, Boardman
L, Rhodes D, Wick M, Okuno S, Kunze KL, Golafshar M, Uson PLS Jr,
Mountjoy L, et al: Comparison of universal genetic testing vs
guideline-directed targeted testing for patients with hereditary
cancer syndrome. JAMA Oncol. 7:230–237. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Johnatty SE, Pesaran T, Dolinsky J, Yussuf
A, LaDuca H, James PA, O'Mara TA and Spurdle AB: Case-case analysis
addressing ascertainment bias for multigene panel testing
implicates BRCA1 and PALB2 in endometrial cancer. Hum Mutat.
42:1265–1278. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karpel HC, Chern JY, Smith JM, Smith AJ
and Pothuri B: Utility of germline multi-gene panel testing in
patients with endometrial cancer. Gynecol Oncol. 165:546–551. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Susswein LR, Marshall ML, Nusbaum R, Vogel
Postula KJ, Weissman SM, Yackowski L, Vaccari EM, Bissonnette J,
Booker JK, Cremona ML, et al: Pathogenic and likely pathogenic
variant prevalence among the first 10,000 patients referred for
next-generation cancer panel testing. Genet Med. 18:823–832. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heald B, Mokhtary S, Nielsen SM, Rojahn S,
Yang S, Michalski ST and Esplin ED: Unexpected actionable genetic
variants revealed by multigene panel testing of patients with
uterine cancer. Gynecol Oncol. 166:344–350. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Daniels MS and Lu KH: Genetic
predisposition in gynecologic cancers. Semin Oncol. 43:543–547.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Jonge MM, de Kroon CD, Jenner DJ,
Oosting J, de Hullu JA, Mourits MJE, Gómez Garcia EB, Ausems MGEM,
Margriet Collée J, van Engelen K, et al: Endometrial cancer risk in
women with germline BRCA1 or BRCA2 mutations: Multicenter cohort
study. J Natl Cancer Inst. 113:1203–1211. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mur P, Garcia-Mulero S, Del Valle J,
Magraner-Pardo L, Vidal A, Pineda M, Cinnirella G, Martin-Ramos E,
Pons T, López-Doriga A, et al: Role of POLE and POLD1 in familial
cancer. Genet Med. 22:2089–2100. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wieme G, Kral J, Rosseel T, Zemankova P,
Parton B, Vocka M, Van Heetvelde M, Kleiblova P, Blaumeiser B,
Soukupova J, et al: Prevalence of germline pathogenic variants in
cancer predisposing genes in Czech and Belgian pancreatic cancer
patients. Cancers (Basel). 13:44302021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lhotova K, Stolarova L, Zemankova P, Vocka
M, Janatova M, Borecka M, Cerna M, Jelinkova S, Kral J, Volkova Z,
et al: Multigene panel germline testing of 1333 Czech patients with
ovarian cancer. Cancers (Basel). 12:9562020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Soukupova J, Zemankova P, Lhotova K,
Janatova M, Borecka M, Stolarova L, Lhota F, Foretova L, Machackova
E, Stranecky V, et al: Validation of CZECANCA (CZEch CAncer paNel
for clinical application) for targeted NGS-based analysis of
hereditary cancer syndromes. PLoS One. 13:e01957612018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
1000 Genomes Project Consortium, .
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker
RE, Kang HM, Marth GT and McVean GA: An integrated map of genetic
variation from 1,092 human genomes. Nature. 491:56–65. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karczewski KJ, Francioli LC, Tiao G,
Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A,
Birnbaum DP, et al: The mutational constraint spectrum quantified
from variation in 141,456 humans. Nature. 581:434–443. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Exome Variant Server NGESPE, ; Available
online:, . http://evs.gs.washington.edu/EVS/15–March. 2022
|
|
26
|
Landrum MJ, Lee JM, Riley GR, Jang W,
Rubinstein WS, Church DM and Maglott DR: ClinVar: Public archive of
relationships among sequence variation and human phenotype. Nucleic
Acids Res. 42:(Database Issue). D980–D985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miller DT, Lee K, Chung WK, Gordon AS,
Herman GE, Klein TE, Stewart DR, Amendola LM, Adelman K, Bale SJ,
et al: ACMG SF v3.0 list for reporting of secondary findings in
clinical exome and genome sequencing: A policy statement of the
American college of medical genetics and genomics (ACMG). Genet
Med. 23:1381–1390. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang
J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, et al:
Pathogenic germline variants in 10,389 adult cancers. Cell.
173:355–370.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
LaDuca H, Polley EC, Yussuf A, Hoang L,
Gutierrez S, Hart SN, Yadav S, Hu C, Na J, Goldgar DE, et al: A
clinical guide to hereditary cancer panel testing: Evaluation of
gene-specific cancer associations and sensitivity of genetic
testing criteria in a cohort of 165,000 high-risk patients. Genet
Med. 22:407–415. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vietri MT, D'Elia G, Caliendo G,
Casamassimi A, Federico A, Passariello L, Cioffi M and Molinari AM:
Prevalence of mutations in BRCA and MMR genes in patients affected
with hereditary endometrial cancer. Med Oncol. 38:132021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stolarova L, Kleiblova P, Janatova M,
Soukupova J, Zemankova P, Macurek L and Kleibl Z: CHEK2 germline
variants in cancer predisposition: Stalemate rather than checkmate.
Cells. 9:26752020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yen TT, Wang TL, Fader AN, Shih IM and
Gaillard S: Molecular classification and emerging targeted therapy
in endometrial cancer. Int J Gynecol Pathol. 39:26–35. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
León-Castillo A, Britton H, McConechy MK,
McAlpine JN, Nout R, Kommoss S, Brucker SY, Carlson JW, Epstein E,
Rau TT, et al: Interpretation of somatic POLE mutations in
endometrial carcinoma. J Pathol. 250:323–335. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Green AK, Feinberg J and Makker V: A
review of immune checkpoint blockade therapy in endometrial cancer.
Am Soc Clin Oncol Educ Book. 40:1–7. 2020.PubMed/NCBI
|
|
35
|
Bychkovsky BL, Lo MT, Yussuf A, Horton C,
Richardson M, LaDuca H, Garber JE and Rana HQ: Prevalence and
spectrum of pathogenic variants among patients with multiple
primary cancers evaluated by clinical characteristics. Cancer.
128:1275–1283. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stolarova L, Jelinkova S, Storchova R,
Machackova E, Zemankova P, Vocka M, Kodet O, Kral J, Cerna M,
Volkova Z, et al: Identification of germline mutations in melanoma
patients with early onset, double primary tumors, or family cancer
history by NGS analysis of 217 genes. Biomedicines. 8:4042020.
View Article : Google Scholar : PubMed/NCBI
|