Introduction
Colorectal cancer (CRC) is a prevalent
gastrointestinal malignancy. Recent data show that it has the
second highest mortality and third highest incidence worldwide
(1). Despite improvements in early
diagnosis and treatment techniques, developing countries are still
experiencing an increase in the incidence rate and mortality of CRC
(2). Additionally, patients with
metastatic CRC have a poor prognosis with a median 5-year survival
rate of 18.5% in the United States and 27.7% in Europe (1). Therefore, it is imperative to identify
effective biomarkers and therapeutic targets to improve patient
prognosis.
Epithelial-mesenchymal transition (EMT) is a key
process in embryonic development. Studies have shown that it
contributes to tumor progression. EMT causes epithelial cells to
acquire fibroblast-like characteristics, decreases intercellular
adhesion and increases motility (3,4).
Snail, a member of the zinc finger transcription factor Snail
family, induces EMT by downregulating EMT-associated genes,
including E-cadherin (E-cad), claudin, occludin, protein associated
with mouse musculus veli-7, membrane-associated guanylate kinase
homolog (MAGUK) p55 family member and Pals1-associated tight
junction (PATJ) crumbs cell polarity complex component (4,5).
E-cad, a cadherin protein family member, is a component of adhesion
junctions and the primary organizer of the epithelial phenotype
(6,7). Studies have shown that the loss of
E-cad expression is associated with tumor progression and
metastasis and induced expression of E-cad in cancer cells can
prevent tumor progression and invasion (5,8).
The present study investigated the role of Snail and
E-cad in CRC and demonstrated that they can individually predict
CRC prognosis, with their joint prediction having a greater
combined effects that can more accurately predict patient
prognosis.
Materials and methods
Patients and cancer tissue
samples
Data of 470 patients with CRC were collected from
Yixing People's Hospital affiliated to Yangzhou University in the
present study. The patients underwent radical colon cancer surgery
at the Department of Oncology of Yixing People's Hospital between
January 2006 and December 2010 and were followed up for at least 5
years. The mean age of 470 patients is 63, these are 282 males and
188 females. The clinicopathological features are shown in our
previously published article (9).
Overall survival (OS) was the primary end point, which was
calculated from the date of surgery to the date of death or final
follow-up.
The Ethics Committee of Yixing Hospital approved the
present study, which was performed according to the principles of
the Declaration of Helsinki. The human and animal experiments were
approved by the Ethics Committee of Yixing Hospital Affiliated to
Yangzhou University (approval no. YXYLL-2015-42). All patients
provided written informed consent for use of their tissues.
Construction of tissue microarray
(TMA) and immunohistochemistry
Tumor tissues were selected from paraffin blocks and
confirmed by hematoxylin and eosin staining. TMA construction was
performed using cancer tissues and corresponding adjacent tissue (5
cm from cancer tissue). Each point on the TMA chip had a diameter
of 1.5 mm to accommodate both tumor and non-tumor tissue. TMA chips
were placed in a 55°C incubator for 10 min and cooled at room
temperature. These chips were placed in a cryostat and 4 µm thick
slices were produced. The slices were placed in water at 45°C for 2
min, baked at 58°C for 18 h and stored at −20°C for future use.
The immunostaining was performed as described
previously (10). Rabbit monoclonal
antibodies, including anti-C-terminus of Hsc70-interacting protein
(CHIP; 1:100; no. 1132; Cell Signaling Technology, Inc.) and Gal1
(cat. no. ab108389; 1:100; Abcam), were incubated at 4°C overnight.
The staining score of the tissue controls were pre-evaluated to
ensure the quality control of immunostaining for each microarray
slide.
Evaluation of immunostaining
Two pathologists who were blinded to the clinical
data scored the staining of Snail or E-cad in the tissue. The
presence of Snail or E-cad in cancer and adjacent tissue was
evaluated using the semi-quantitative immunoreactivity score (IRS)
reported previously (11).
Intensity of immunostaining was categorized as 0–3 (0, negative; 1,
weak; 2, moderate; 3, strong). Proportion of immunoreactive cells
was categorized as 1, (0–25%), 2 (26–50%), 3 (51–75%) and 4
(76–100%). The product of these scores was used to calculate IRS
ranging from 0 to 12. To determine the optimum cutoff value of
Snail or E-cad IRS for 1-, 3- and 5-year OS rate, receiver operator
characteristic (ROC) analysis was used. The optimum cutoff point
for CHIP IRS was 4 since it had the best predictive value for
survival.
Cell lines and animals
HCT 116 and HT 29 CRC cells were obtained from
Procell Life Science & Technology Co., Ltd. These cells were
cultured in RPMI-1640 medium supplemented with 10% FBS (Beyotime
Biotechnology) and 1% penicillin/streptomycin. All cells were
incubated at 37°C with 5% CO2. These cells were
authenticated by short tandem repeat profiling.
Female BALB/c nude mice were obtained from the
Comparative Medicine Laboratory Animal Center [license no. scxk
(SU) 2012–0004] of Yangzhou University. The mice (age, 6–8 weeks)
were kept in specific pathogen-free conditions and cared for
according to the National Institutes of Health Guide for the Care
and Use of Laboratory Animals.
A total of ~2×106 HCT 116 stable cells
and control cells (0.2 ml/mouse; 5 mice/group) were implanted
subcutaneously into the flank of each mouse. After 21 days, the
mice were sacrificed. All nude mice were euthanized by cervical
dislocation and all animal experiments were conducted under the
animal use license of Yangzhou University (no. SYXK2022-0044).
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from CRC
tissue and cells. cDNA was synthesized using a PrimeScript™ RT kit
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
instructions. SYBR Green qPCR analysis (Applied Biosystems) was
performed using the Applied Biosystems 7500 real-time PCR system
(Roche Applied Science). The method of quantification was
2−ΔΔCq (12). The
sequences of the primers were as follows (5′□3′): E-cad forward,
CGAGAGCTACACGTTCACGG and reverse, GGGTGTCGAGGGAAAAATAGG; Snail
forward, CCTCGCTGCCAATGCTCATCTG and reverse, CTCTGCCACCCTGGGACTCTC
and GAPDH forward, ACGGATTTGGTCGTATTGGG and reverse,
CGCTCCTGGAAGATGGTGAT (all Sangon Biotech Co., Ltd.).
Western blotting
Cells or tissues were lysed with cold lysis buffer
supplemented with a protease inhibitor (Beyotime Biotechnology) on
ice for 30 min. The total protein concentration was measured using
the Bicinchoninic Acid Protein assay kit (Thermo Fisher Scientific,
Inc.). Protein (80 µg/lane) was separated by SDS-PAGE on 10% gels.
Subsequently, protein was transferred to the PVDF membrane, which
was incubated with antibodies. The protocol was executed in the
aforementioned manner. Rabbit monoclonal anti-E-cad (cat. no.
ab40772; 1:1,000;Abcam), rabbit monoclonal anti-Snail (cat. no.
ab216347; 1:1,000; Abcam) and mouse monoclonal anti-β-actin (cat.
no. AF0003; 1:2,000; Beyotime Biotechnology) were used as the
primary antibodies. ImageJ software (v 1.44; National Institutes of
Health) was used to normalize expression to the expression of
β-actin and the band strength of each protein was
semi-quantified.
Wound scratch assay
HCT 116 cells (~5×105) were added into
each well of a marked six-well plate to ensure that the plate was
fully covered. After 24 h, a sterile pipette tip was used to
scratch a horizontal line perpendicular to the bottom of the plate
and the cells were washed with PBS three times. Serum-free
RPMI-1640 medium (Beyotime Biotechnology) was added and cells were
cultured in a 37°C 5% CO2 incubator. The process of
tumor cell migration was observed and photographed at 50×
magnification 0, 24 and 48 h after wounding.
High content cell migration
experiment
The cells were plated in 96-well plates (2,000
cells/well). When the cells grew to 80% confluence, the fresh
RPMI-1640 medium supplemented with 10% FBS was refreshed. Under the
conditions of 37°C and 5% CO2, the migration of cells
was observed using an Operetta CLS high connotation cell imaging
system (PerkinElmer). The cells were scanned and images were
captured every 10 min for 12 h. The cell migration curve was
constructed.
Lentiviral (LV) infection
LV vector (Shanghai GenePharma Co., Ltd.) was used
to knockdown or increase with Snail or E-cad expression. LV-Snail,
LV-Snail-control (ctrl), LV-Snail-short hairpin RNA (shRNA) and
LV-Snail-shRNA-ctrl; LV-E-cad, LV-E-cad-ctrl, LV-E-cad-shRNA and
LV-E-cad-shRNA-ctrl were transfected into HT 29 and HCT 116 cells
at a MOI of 20 with 10 µg/ml Polybrene (Shanghai GeneChem Co.,
Ltd.). The cells were maintained with normal RPMI-1640 culture
medium at 37°C with 5% CO2 for 24 h after lentiviral
infection 8 h. After 24 h, the cells were incubated in RPMI-1640
with 2 µg/ml puromycin. The sequences of the shRNAs and
shRNA-control were as follows (5′→3′): Snail: CCACTCAGATGTCAAGAAGTA
and ctrl: TTCTCCGAACGTGTCACGTTT. E-cad: CAUGGAUAACCAGAAUAAATT and
UUCUCCGAACGUGUCACGUTT.
Statistical analysis
The association between Snail and E-cad expression
and clinicopathological data was evaluated using Fisher's exact
test. IRS expression of Snail and E-cad in tumor and corresponding
non-tumor tissue was compared using Wilcoxon signed-rank test
(grouping). Kaplan-Meier (log-rank test) survival analysis was used
to determine differences in OS (13). Univariate or multivariate Cox
regression analysis was used to estimate the hazard ratio (HR) and
95% CI. STATA software (v10.1; StataCorp LP) was used to analyze
all experimental data. Data were analyzed by one-way ANOVA followed
by Tukey's post hoc test. Mann-Whitney U was used as non-parametric
test to compare unpaired data. P<0.05 was considered to indicate
a statistically significant difference. All experiments were
repeated in triplicate.
Results
Snail and E-cad expression in CRC vs.
non-cancerous tissue
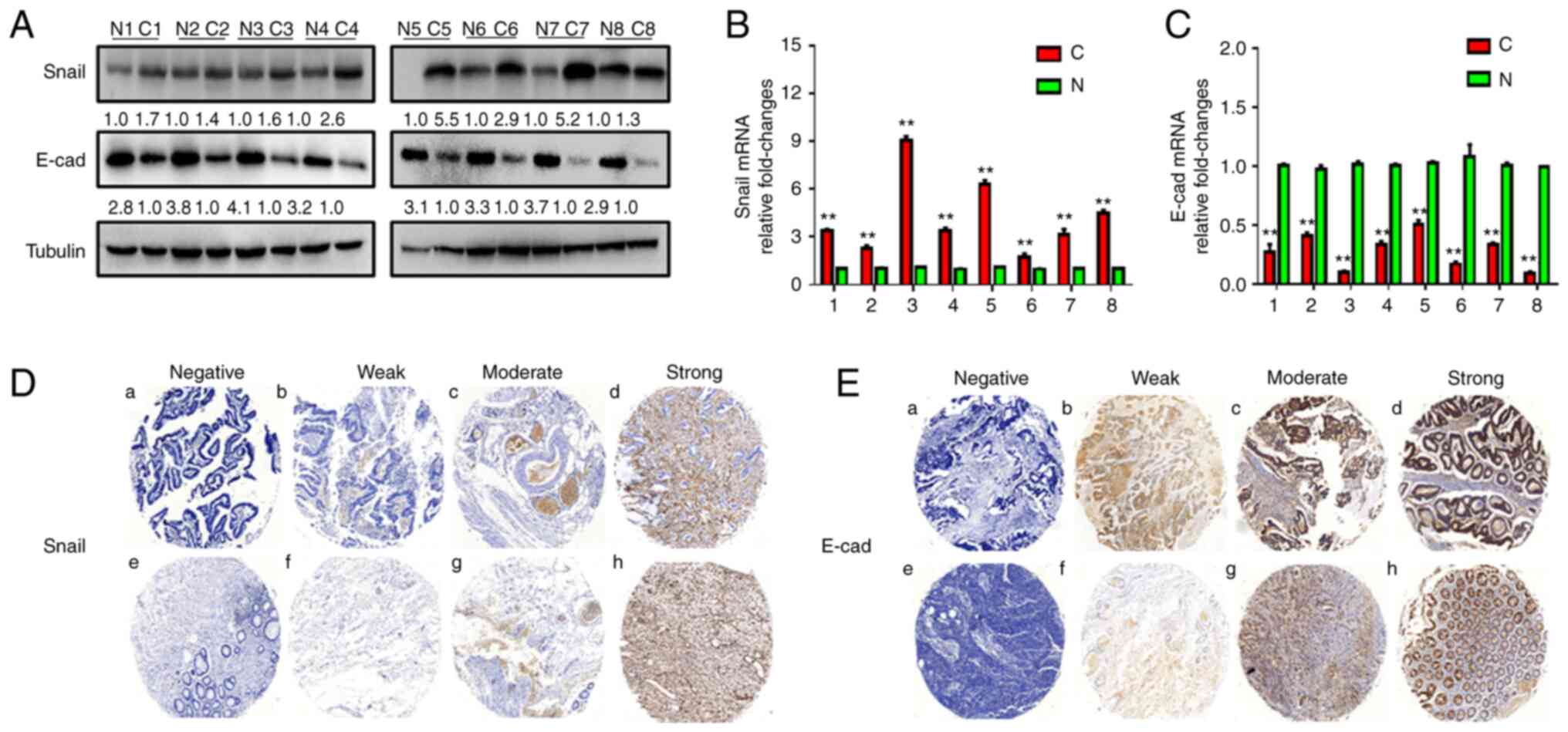
The present study used eight pairs of primary CRC
and adjacent normal tissues to detect the protein expression levels
of Snail and E-cad by western blotting. The protein expression
levels of E-cad were lower in tumor compared with adjacent normal
tissues, while expression levels of Snail were higher in tumor
tissue (Fig. 1A). RT-qPCR was used
to detect the mRNA levels of Snail and E-cad, which were higher and
lower, respectively, in tumor compared with corresponding normal
tissues (Fig. 1B and C).
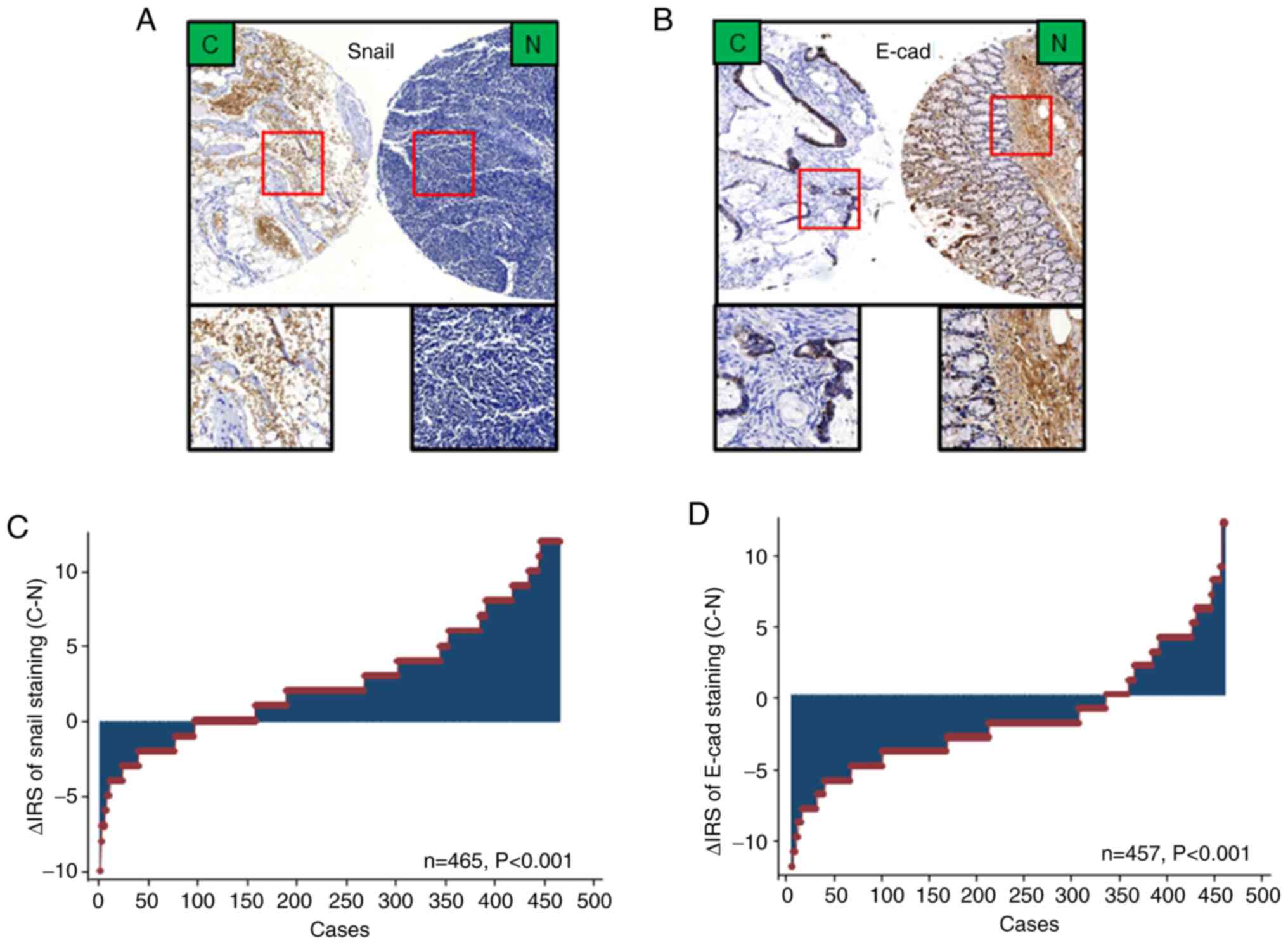
To confirm Snail and E-cad expression in CRC tissue,
immunohistochemical staining was performed (Figs. 1D and E and 2A and B). Expression levels of Snail were
markedly upregulated in cancer compared with adjacent normal
tissues. Similarly, the expression levels of E-cad were
downregulated in tumor compared with adjacent non-tumor tissue
(Fig. 2C and D).
Association between Snail and E-cad
expression and clinicopathological data in patients with CRC
The present study analyzed the association between
Snail and E-cad expression and clinicopathological characteristics
of 465 patients with CRC. Snail expression was significantly
associated with the depth of invasion, lymph node metastasis, TNM
stage and distal metastasis (Table
I; P<0.05). Similarly, expression levels of E-cad were
significantly associated with pathological classification, lymph
node metastasis, TNM stage and distal metastasis (Table II; P<0.05).
 | Table I.Association between expression levels
of Snail and clinicopathological features of patients with
colorectal cancer (n=469). |
Table I.
Association between expression levels
of Snail and clinicopathological features of patients with
colorectal cancer (n=469).
| Characteristic | Low Snail, n=275
(58.6%) | High Snail, n=194
(41.4%) |
P-valuea |
|---|
| Age, years |
|
| 0.077 |
|
≤65 | 164 (61.7) | 102 (38.3) |
|
|
>65 | 111 (54.7) | 92 (45.3) |
|
| Sex |
|
| 0.400 |
|
Male | 166 (59.3) | 114 (40.7) |
|
|
Female | 109 (57.7) | 80 (42.3) |
|
| Pathological
classificationb |
|
| 0.165 |
| I | 2 (50.0) | 2 (50.0) |
|
| II | 253 (59.8) | 170 (40.2) |
|
|
III | 16 (44.4) | 20 (55.6) |
|
| Depth of
invasionb |
|
| <0.001 |
|
T1/T2 | 83 (81.4) | 19 (18.6) |
|
|
T3/T4 | 187 (51.7) | 175 (48.3) |
|
| Lymph node
metastasisb |
|
| <0.001 |
| N0 | 210 (76.4) | 65 (23.6) |
|
|
N1/N2 | 61 (32.1) | 129 (67.9) |
|
| TNM
stageb |
|
| <0.001 |
| I | 74 (85.1) | 13 (14.9) |
|
| II | 131 (73.2) | 48 (26.8) |
|
|
III | 60 (33.3) | 120 (66.7) |
|
| IV | 5 (29.4) | 12 (70.6) |
|
| Tumor diameter,
cmb |
|
| 0.254 |
| ≤5 | 224 (59.4) | 153 (40.6) |
|
|
>5 | 50 (54.9) | 41 (45.1) |
|
| Distant
metastasis |
|
| 0.004 |
| M0 | 270 (60.0) | 180 (40.0) |
|
| M1 | 5 (26.3) | 14 (73.7) |
|
 | Table II.Association between expression levels
of E-cad and clinicopathological features in patients with
colorectal cancer (n=465). |
Table II.
Association between expression levels
of E-cad and clinicopathological features in patients with
colorectal cancer (n=465).
| Characteristic | Low E-cad, n=244
(52.5%) | High E-cad, n=221
(47.5%) |
P-valuea |
|---|
| Age, years |
|
| 0.111 |
|
≤65 | 131 (49.8) | 132 (50.2) |
|
|
>65 | 113 (55.9) | 89 (44.1) |
|
| Sex |
|
| 0.453 |
|
Male | 147 (52.9) | 131 (47.1) |
|
|
Female | 97 (51.9) | 90 (48.1) |
|
| Pathological
classificationb |
|
| 0.001 |
| I | 3 (60.0) | 2 (40.0) |
|
| II | 210 (50.1) | 209 (49.9) |
|
|
III | 29 (80.6) | 7 (19.4) |
|
| Depth of
invasionb |
|
| 0.098 |
|
T1/T2 | 47 (46.5) | 54 (53.5) |
|
|
T3/T4 | 196 (54.4) | 164 (45.6) |
|
| Lymph node
metastasisb |
|
| <0.001 |
| N0 | 112 (40.9) | 162 (59.1) |
|
|
N1/N2 | 131 (69.7) | 57 (30.3) |
|
| TNM
stageb |
|
| <0.001 |
| I | 37 (43.0) | 49 (57.0) |
|
| II | 68 (38.0) | 111 (62.0) |
|
|
III | 124 (69.7) | 54 (30.3) |
|
| IV | 14 (82.4) | 3 (17.6) |
|
| Tumor diameter,
cmb |
|
| 0.041 |
| ≤5 | 188 (50.3) | 186 (49.7) |
|
|
>5 | 55 (61.1) | 35 (38.9) |
|
| Distant
metastasis |
|
| 0.004 |
| M0 | 228 (51.1) | 218 (48.9) |
|
| M1 | 16 (84.2) | 3 (15.8) |
|
High Snail and low E-cad expression is
associated with shorter survival time in patients with CRC
Kaplan-Meier analysis showed that high Snail
expression or low E-cad expression in cancer tissue was
significantly associated with poorer 5-year survival rates in
patients with CRC (both P<0.001; log-rank test; Fig. 3C and D). Furthermore, univariate and
multivariate Cox regression analyses revealed that Snail or E-cad
were independent prognostic factors for patients with CRC. The
results of the univariate Cox regression analysis demonstrated that
Snail and E-cad expression were associated with OS in patients with
CRC (Table III). Additionally,
multivariate Cox regression analysis revealed that Snail and E-cad
expression was an independent prognostic factor in patients with
CRC (Table IV; Snail: HR, 0.181;
95% CI, 0.128-0.255; P<0.001; E-cad: HR, 0.212; 95% CI,
0.148-0.303; P<0.001).
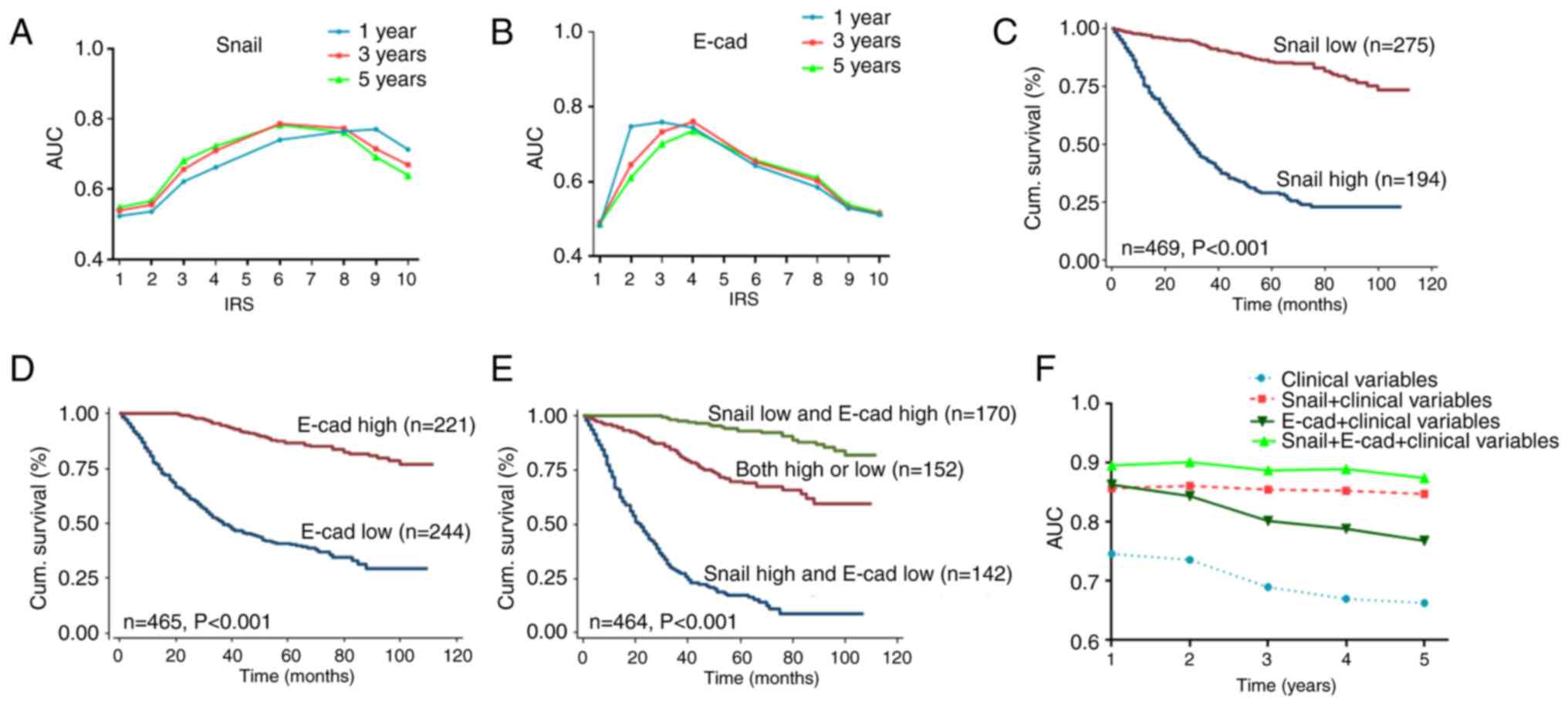 | Figure 3.Snail and E-cad expression predict
prognosis of CRC. AUC for (A) Snail or (B) E-cad plotted against
cut-off values for IRS for 1-, 3- and 5-year OS. Kaplan-Meier
curves of (C) Snail, (D) E-cad and (E) Snail + E-cad expression in
training cohort for OS. (F) Time-dependent receiver operating
characteristic analysis for clinical risk score (TNM stage,
histological type and tumor diameter), alone or in combination with
Snail, E-cad or Snail + E-cad. AUC, area under the curve; IRS,
immunoreactivity score; OS, overall survival; E-cad, E-cadherin;
CRC, colorectal cancer; cum, cumulative. |
 | Table III.Univariate Cox regression analysis of
Snail and E-cad expression predicting survival in patients with
colorectal cancer (n=470). |
Table III.
Univariate Cox regression analysis of
Snail and E-cad expression predicting survival in patients with
colorectal cancer (n=470).
| Expression, low vs.
high | HR (95% CI) | P-value |
|---|
| Snail | 0.143
(0.104-0.197) | <0.001 |
| E-cad | 0.166
(0.117-0.235) | <0.001 |
 | Table IV.Multivariate Cox regression analysis
of Snail, E-cad, Snail + E-cad expression and clinicopathological
variables predicting survival in patients with colorectal
cancer. |
Table IV.
Multivariate Cox regression analysis
of Snail, E-cad, Snail + E-cad expression and clinicopathological
variables predicting survival in patients with colorectal
cancer.
| A, Snail |
|---|
|
|---|
| Variable | HR (95% CI) | P-value |
|---|
| Age, ≤65 vs. >65
years | 1.707
(1.278-2.279) | <0.001 |
| Sex, male vs.
female | 0.887
(0.665-1.183) | 0.413 |
| Pathological
classification, I/II vs. III | 2.130
(1.331-3.411) | 0.002 |
| TNM stage, I/II vs.
III/IV | 1.797
(1.309-2.466) | <0.001 |
| Tumor diameter, ≤5
vs. >5 cm | 1.041
(0.727-1.493) | 0.825 |
| Expression, low vs.
high | 0.181
(0.128-0.255) | <0.001 |
|
| B,
E-cad |
|
|
Variable | HR (95%
CI) | P-value |
|
| Age, ≤65 vs. >65
years | 1.734
(1.304-2.307) | <0.001 |
| Sex, male vs.
female | 0.995
(0.745-1.328) | 0.972 |
| Pathological
classification, I/II vs. III | 1.620
(1.018-2.578) | 0.042 |
| TNM stage, I/II vs.
III/IV | 2.487
(1.839-3.363) | <0.001 |
| Tumor diameter, ≤5
vs. >5 cm | 1.033
(0.719-1.483) | 0.861 |
| Expression, low vs.
high | 0.212
(0.148-0.303) | <0.001 |
|
| C,
Snail/E-cad |
|
|
Variable | HR (95%
CI) | P-value |
|
| Age, ≤65 vs. >65
years | 1.781
(1.120-2.831) | <0.001 |
| Sex, male vs.
female | 1.056
(0.663-1.683) | 0.818 |
| Pathological
classification, I/II vs. III | 0.900
(0.324-2.501) | 0.840 |
| TNM stage, I/II vs.
III/IV | 1.862
(1.148-3.020) | 0.012 |
| Tumor diameter, ≤5
vs. >5 cm | 1.077
(0.596-1.947) | 0.806 |
| Expression |
|
|
| Both
low vs. one low | 0.272
(0.163-0.455) | <0.001 |
| Both
low vs. both high | 0.226
(0.172-0.298) | <0.001 |
Combined Snail and E-cad expression
has greater predictive ability of OS in patients with CRC
The survival rate of the Snail low expression and
E-cad high expression groups was higher than that of other groups
(P<0.001, log-rank test; Fig.
3E). To verify whether Snail combined with E-cad had a great
predictive effect on the prognosis of patients with CRC, the
clinical risk score (TNM stage, histological type and tumor
diameter), Snail expression, E-cad expression and Snail + E-cad
expression were used for time-dependent ROC analysis. The results
suggested that for patients with CRC, the clinical risk score
combined with Snail and E-cad expression had a greater contribution
than any of these markers alone (Fig.
3F).
Snail promotes CRC cell migration by
decreasing E-cad
Previous studies have shown an association between
Snail and E-cad expression and lymph node metastasis, TNM stage and
distant metastasis in CRC (4,5). To
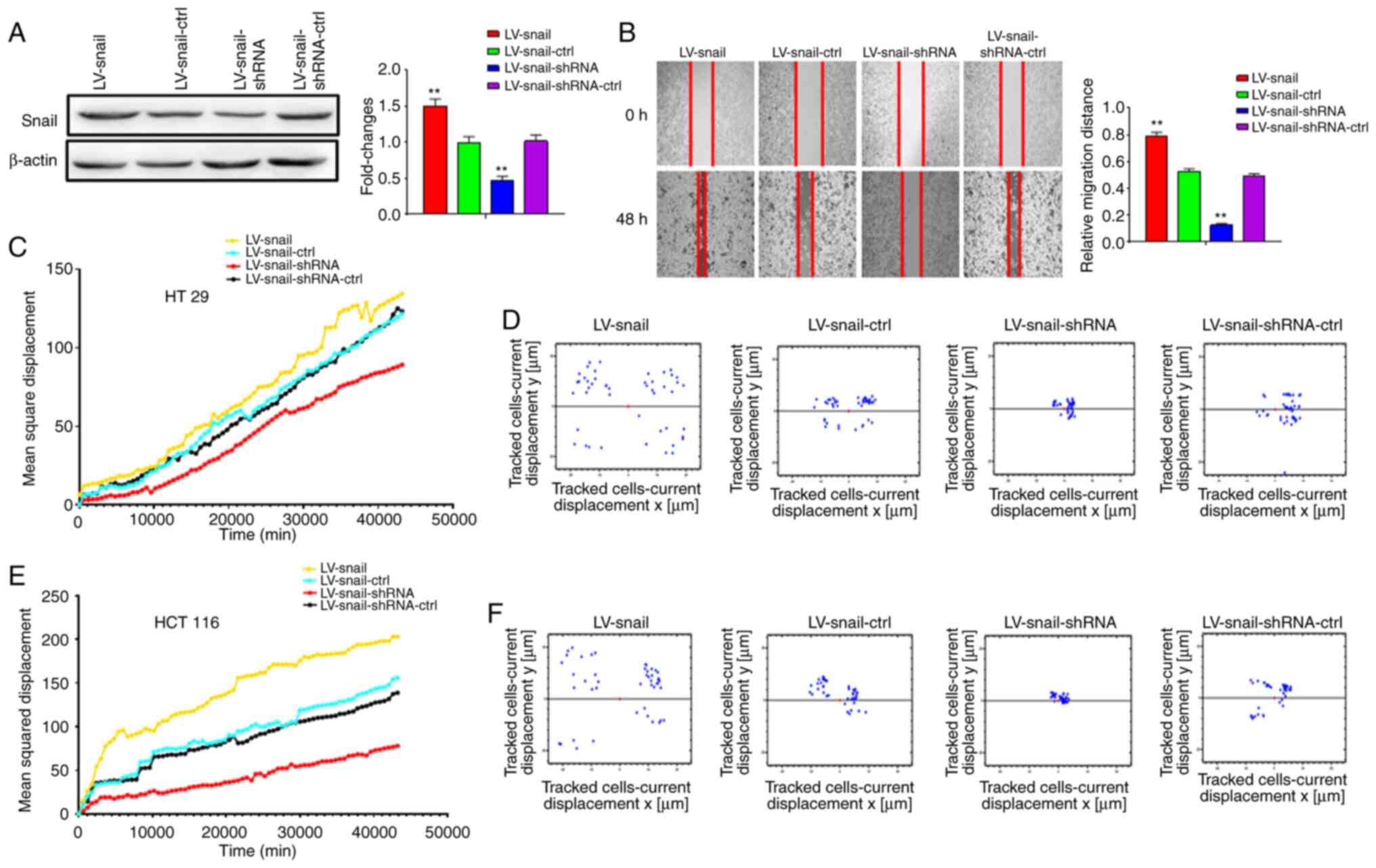
investigate the effects of Snail and E-cad on CRC cells, lentivirus
was used to generate stable cell lines of HCT 116 and HT 29
(Fig. 4A). There following groups
were established under normal culture conditions: Overexpression
LV-Snail, overexpression LV-E-cad, knockdown LV-Snail-shRNA,
knockdown LV-E-cad-shRNA and corresponding control groups. Wound
healing and high content cell migration assay were used to detect
changes in cell migration. The migration of LV-Snail cells was
significantly increased compared with the corresponding control
group, while the migration of LV-Snail-shRNA cells was decreased
(Fig. 4B-F).
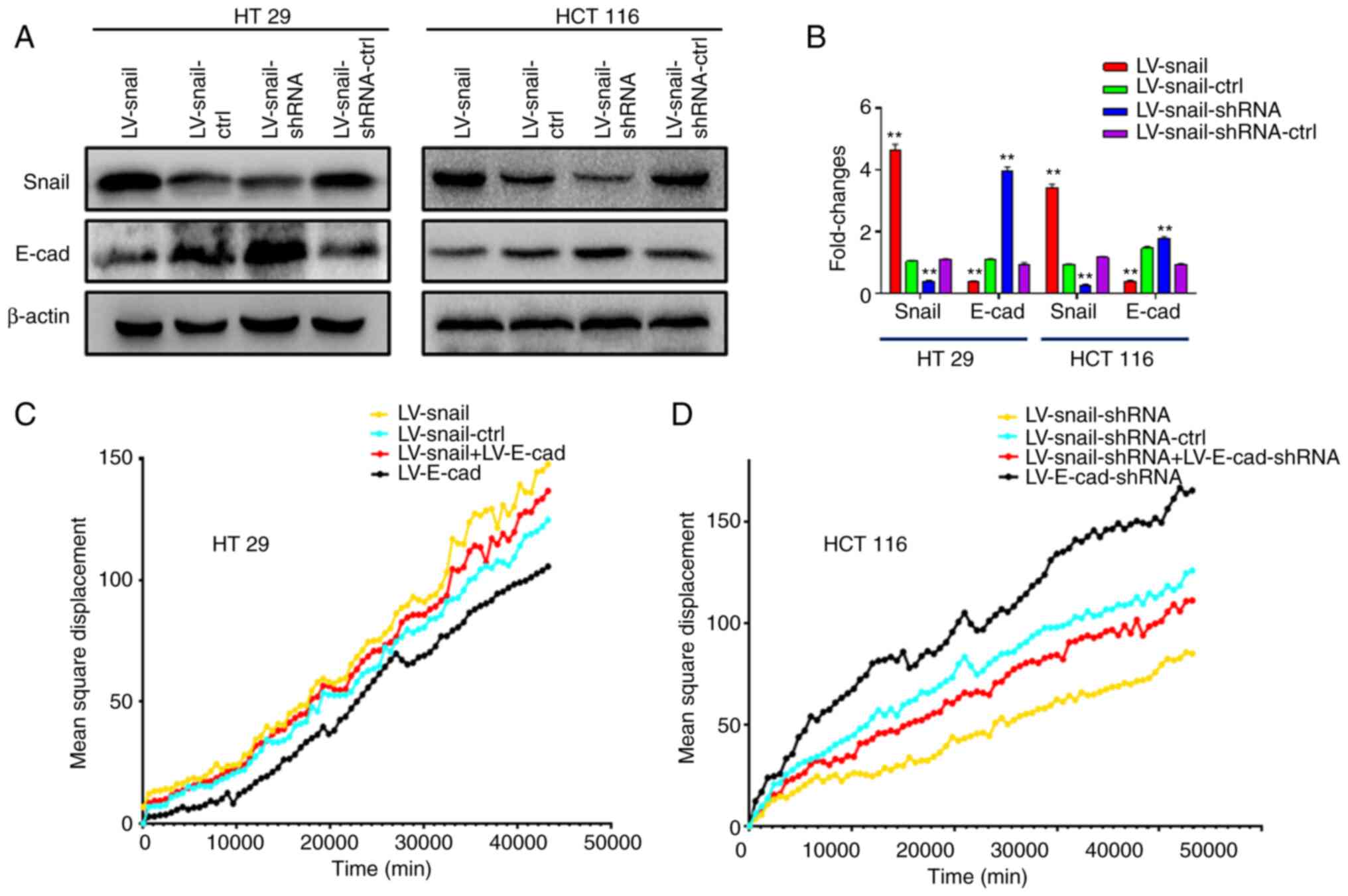
Subsequently, the present study detected expression
levels of Snail and E-cad in each group after lentivirus infection
by western blotting. The data showed that E-cad expression was
decreased in each group following infection with LV-Snail
lentivirus compared with respective control groups. Following
infection with LV-Snail-shRNA lentivirus, the expression levels of
E-cad were elevated (Fig. 5A and
B). The present study altered expression levels of E-cad by
secondary lentivirus infection. Cell migration was decreased after
re-infection with LV-E-cad lentivirus to increase the expression
levels of E-cad in LV-Snail CRC cells. By contrast, LV-E-cad-shRNA
cell migration was enhanced following infection with LV-E-cad-shRNA
lentivirus (Fig. 5C and D).
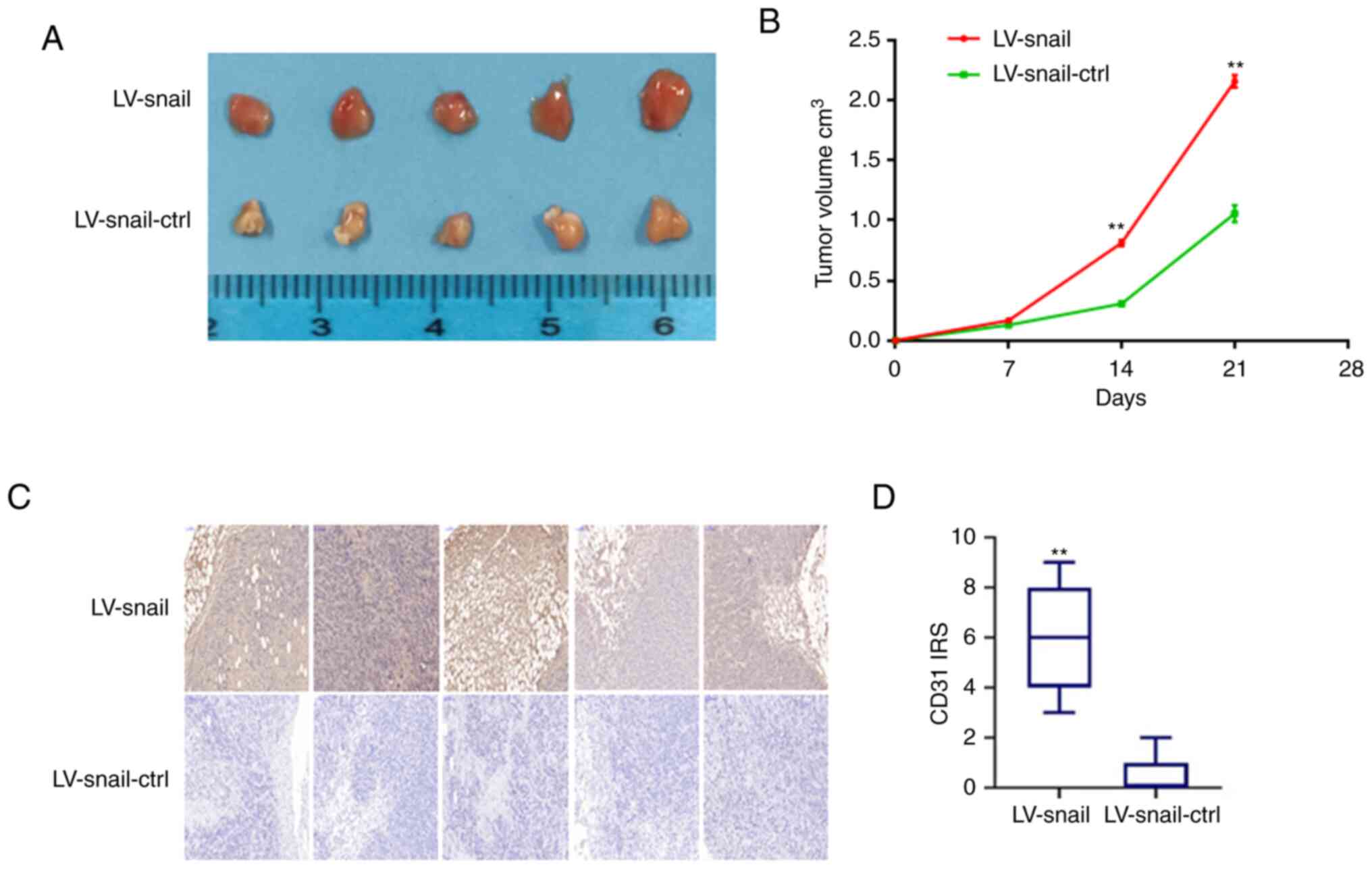
Snail promotes CRC cell proliferation
in vivo
To study the effect of Snail on the proliferation of
CRC cells in vivo, stable LV-Snail and LV-Snail-ctrl HCT 116
cells were subcutaneously injected into nude mice. The mice were
sacrificed after 21 days. There was much more vascular-rich cancer
tissue in LV-Snail group than in the control group (Fig. 6A). The tumor volume of each group
was measured every week. The diameter of the largest tumor tissue
was ~6 mm. The tumor volume was larger in LV-Snail group than in
the control group (Fig. 6B;
P<0.01). Expression levels of CD31 were detected in tissue by
IHC. The data revealed that the expression levels of CD31 in the
transplanted tumors in the LV-Snail group were higher than those in
the control group (Fig. 6C and D;
P<0.01), which suggested that Snail served a notable role in
promoting the angiogenesis of CRC cells in vivo.
Discussion
CRC is one of the most common types of cancer of the
digestive system. In 2020, there were ~1.9 million new cases of CRC
worldwide, resulting in >900,000 deaths (1). Although the incidence and mortality
rate of CRC have decreased steadily in previous years, there is an
upward trend in the incidence and mortality rates of individuals
<50 years old (14–17). Tumor metastasis is a complicated
process involving tumor cells invading the microenvironment,
entering the blood, migration, angiogenesis and proliferation.
Postoperative recurrence and metastasis are the primary reasons for
the low survival rate of patients with CRC (18). Therefore, it is imperative to find
molecular markers that can predict the prognosis of patients with
CRC.
EMT is a process in which epithelial cells lose
epithelioid features and switch to invasive mesenchymal cells,
manifested by decreased expression levels of epithelial genes, such
as E-cad and occludin, and increased expression of mesenchymal
genes, such as N-cad and Vimentin (19). EMT is mainly involved in embryo
development, wound healing, cancer cell metastasis and drug
resistance (20,21).
E-cad protein primarily exists in epithelial cells
and regulates cell adhesion in tissue. Reduction of E-cad
expression usually indicates the beginning of EMT. Studies have
demonstrated that expression of E-cad can inhibit tumor progression
and invasion, and thus E-cad is considered a classical tumor
inhibitor (4,5,8).
Snail is a member of the zinc finger transcription
factor Snail family. This family encodes transcriptional inhibitors
and shares a conserved C-terminal domain containing 4–6
C2H2 type zinc fingers that bind to the E-box
motif (5′-CANNTG-3′) of the target gene promoter (22). Snail is a primary inducer of EMT and
has been associated with recurrence, metastasis and poor prognosis
of breast cancer (23,24). Additionally, Snail is involved in
acquisition of tumor stem cell features and inhibits estrogen
receptor signaling (25,26), thus decreasing recurrence-free
survival in patients with low-grade breast cancer (27). To the best of our knowledge,
however, research on the role of Snail in CRC is currently
lacking.
The present study suggested that Snail could promote
the proliferation and migration of CRC cells in vitro.
Furthermore, Snail was found to be a poor prognostic marker for
patients with CRC. Based on our CRC database analysis, it was
concluded that Snail and E-cad were independent prognostic markers.
Next, the present study attempted to analyze whether these two
indicators had a greater combined effect in predicting CRC
prognosis. Notably, Snail and E-cad had a greater combined effect
based on Kaplan-Meier survival and ROC curve analysis of clinical
variables.
The present study was only a retrospective study of
a single center; a multi-center study should be performed in the
future to expand the sample size. In future, prospective studies
should be performed and database analysis conclusions should be
verified using cell phenotype experiments and animal models of
transplanted and metastatic tumors in vivo.
In conclusion, the present study demonstrated that
Snail and E-cad were prognostic molecular biomarkers in patients
with CRC. Snail promoted proliferation of CRC cells in vivo
and in vitro. Notably, the present study identified a
greater combined effect of Snail and E-cad in predicting prognosis.
Further research into the role of these proteins may improve the
survival of patients with CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Top Talent Support Program
for Young and Middle-aged People of Wuxi Health Committee (grant.
no. WX2022); Wuxi City Health Planning Commission project (grant.
no. Q202168); the Natural Science Foundation of Jiangsu Province
(grant. no. BK20191149) and Scientific Research Project of Maternal
and Child Health Care Association of Jiangsu Province (grant. no.
FYX202119)
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the SUB13034094 repository,
[https://www.ncbi.nlm.nih.gov/biosample/34085159 to https://www.ncbi.nlm.nih.gov/biosample/34085290.
Authors' contributions
WMW, YZ and JS conceived and designed the study and
methodology. WMW, JJ, ZZ, YFW and KM performed the experiments and
acquired the data. JJ, ZZ, YFW, KM, JPJ and XZ analyzed and
interpreted the data. WMW, JJ, ZZ and JPJ wrote, reviewed and
revised the manuscript. YZ and JS provided study supervision. All
authors have read and approved the final manuscript. WMW and YZ
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The Ethics Committee of Yixing Hospital approved the
present study, which was performed according to the principles of
the Declaration of Helsinki. The human and animal experiments were
approved by the Ethics Committee of Yixing Hospital Affiliated to
Yangzhou University (approval no. YXYLL-2015-42).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aigner K, Dampier B, Descovich L, Mikula
M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist
P, et al: The transcription factor ZEB1 (deltaEF1) promotes tumour
cell dedifferentiation by repressing master regulators of
epithelial polarity. Oncogene. 26:6979–6988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Whiteman EL, Liu CJ, Fearon ER and
Margolis B: The transcription factor snail represses Crumbs3
expression and disrupts apico-basal polarity complexes. Oncogene.
27:3875–3879. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gumbiner B, Stevenson B and Grimaldi A:
The role of the cell adhesion molecule uvomorulin in the formation
and maintenance of the epithelial junctional complex. J Cell Biol.
107:1575–1587. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gumbiner BM: Regulation of
cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol.
6:622–634. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Navarro P, Gómez M, Pizarro A, Gamallo C,
Quintanilla M and Cano A: A role for the E-cadherin cell-cell
adhesion molecule during tumor progression of mouse epidermal
carcinogenesis. J Cell Biol. 115:517–533. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Li D, Xiang L, Lv M, Tao L, Ni T,
Deng J, Gu X, Masatara S, Liu Y and Zhou Y: TIMP-2 inhibits
metastasis and predicts prognosis of colorectal cancer via
regulating MMP-9. Cell Adh Migr. 13:273–284. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W, Chen Y, Deng J, Zhou J, Gu X, Tang
Y, Zhang G, Tan Y, Ge Z, Huang Y, et al: Cullin1 is a novel
prognostic marker and regulates the cell proliferation and
metastasis in colorectal cancer. J Cancer Res Clin Oncol.
141:1603–1612. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang S, Wu X, Chen Y, Zhang J, Ding J,
Zhou Y, He S, Tan Y, Qiang F, Bai J, et al: Prognostic and
predictive role of JWA and XRCC1 expressions in gastric cancer.
Clin Cancer Res. 18:2987–2996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang T, Gao X, Chen S, Li D, Chen S, Xie
M, Xu Z and Yang G: Genome-wide identification and expression
analysis of ethylene responsive factor family transcription factors
in Juglans regia. PeerJ. 9:e124292021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Austin H, Henley SJ, King J, Richardson LC
and Eheman C: Changes in colorectal cancer incidence rates in young
and older adults in the United States: What does it tell us about
screening. Cancer Causes Control. 25:191–201. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bhandari A, Woodhouse M and Gupta S:
Colorectal cancer is a leading cause of cancer incidence and
mortality among adults younger than 50 years in the USA: A
SEER-based analysis with comparison to other young-onset cancers. J
Investig Med. 65:311–315. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Howlader N, Noone AM, Krapcho M, Garshell
J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z,
et al: SEER cancer statistics review, 1975–2012. National Cancer
Institute; Bethesda, MD: 2014
|
|
16
|
Singh KE, Taylor TH, Pan CG, Stamos MJ and
Zell JA: Colorectal cancer incidence among young adults in
California. J Adolesc Young Adult Oncol. 3:176–184. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Young JP, Win AK, Rosty C, Flight I, Roder
D, Young GP, Frank O, Suthers GK, Hewett PJ, Ruszkiewicz A, et al:
Rising incidence of early-onset colorectal cancer in Australia over
two decades: Report and review. J Gastroenterol Hepatol. 30:6–13.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hay ED: An overview of
epithelio-mesenchymal transformation. Acta Anat (Basel). 154:8–20.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalluri R and Weinberg RA: The basics of
epithelial- mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nieto MA: The snail superfamily of
zinc-finger transcription factors. Nat Rev Mol Cell Biol.
3:155–166. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen WJ, Wang H, Tang Y, Liu CL, Li HL and
Li WT: Multidrug resistance in breast cancer cells during
epithelial-mesenchymal transition is modulated by breast cancer
resistant protein. Chin J Cancer. 29:151–157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moody SE, Perez D, Pan TC, Sarkisian CJ,
Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD and
Chodosh LA: The transcriptional repressor snail promotes mammary
tumor recurrence. Cancer Cell. 8:197–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX
and Barsky SH: ERalpha suppresses slug expression directly by
transcriptional repression. Biochem J. 416:179–187. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX,
Shetuni B and Barsky SH: ERalpha signaling through slug regulates
E-cadherin and EMT. Oncogene. 29:1451–1462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kurrey NK, Jalgaonkar SP, Joglekar AV,
Ghanate AD, Chaskar PD, Doiphode RY and Bapat SA: Snail and slug
mediate radioresistance and chemoresistance by antagonizing
p53-mediated apoptosis and acquiring a stem-like phenotype in
ovarian cancer cells. Stem Cells. 27:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|