Introduction
Osteosarcoma (OS) is one of the most commonly
encountered malignancies in the world, with an occurrence rate of
one to four in a million (1,2). It is
more commonly noted in the epiphysis of young bones. OS is a highly
invasive disease. It easily spreads and metastasizes through the
bloodstream during the initial stages of its development and the
resulting metastases to the lungs are the primary reasons of
patient mortality (3). Despite
significant progress in the diagnosis and treatment of OS, the
survival rate is low and the prognosis is poor. Currently, it is
considered that the majority of cases with OS are developed by
interactions between the environment and the cytogenetic material,
with oxidative stress possibly being one of the main attributing
factors (4,5).
Oxidative stress is a physiological disorder induced
by an unbalance between the production of free radicals during
oxidation and the ability to scavenge them; it is characterized by
increased levels of reactive oxygen species (ROS). Increased ROS
production has been found in a variety of tumors to date (6,7).
Elevated levels of ROS have long been considered to be a
carcinogenic factor as increased ROS can damage cellular genetic
material (8,9). However, recent studies have reported
that ROS can act as a signal in tumors, blocking cancer
differentiation (10,11). The importance of ROS in the
regulation of cellular function cannot be overstated.
It has been shown that the generation of ROS is
critical in regulating osteocyte function; in addition, the
pathophysiology of mineralized tissues is affected by oxidative
stress (12). The role of oxidative
stress in OS has rarely been reported in the literature. It has
been shown that OS cell proliferation and migration are regulated
in part by redox-activated sodium-hydrogen antiporter 1 (13). Oxidative stress induced by low-dose
doxorubicin promotes the invasiveness of OS (14). Therefore, the role of oxidative
stress in OS remains to be further explored. It is important to
build a prognostic signature for OS management by screening for
oxidative stress-related genes (OXSRGs) that are associated with OS
prognosis.
In the present study, an OXSRG signature was
identified to be associated with OS prognosis following the
analysis of data from The Cancer Genome Atlas (TCGA). The
interaction between this signature and the tumor microenvironment
(TME) was subsequently investigated. In addition, molecular
analysis was performed to further explore the role of OXSRGs in
OS.
Materials and methods
Acquisition of hub OXSRGs associated
with OS prognosis
The RNA sequencing (RNA-seq) data and the clinical
characteristics of OS were obtained from the TARGET program of the
TCGA database (https://portal.gdc.cancer.gov/). The keywords used for
the searching data were ‘transcriptome profiling’, ‘gene expression
quantification’, ‘RNA-seq’ and ‘bones, joints, and articular
cartilage of limbs’. Following exclusion of the patients with
unknown survival status, data from 84 cases were included. This
dataset was used as the training cohort. GSE21257, which was used
as an external validation set, was a dataset of 53 OS samples with
prognostic information obtained from the Gene Expression Omnibus
database.
The GeneCards website was searched with the keyword
‘oxidative stress’ and the pertinent genes were obtained. The genes
with a score ≥7 were selected for the next step of the univariate
COX regression analysis. The least absolute shrinkage and selection
operator regression (LASSO) with default parameters was used for
subsequent screening of the hub OXSRGs.
Functional enrichment analysis of hub
OXSRGs
The Spearman test was used to calculate the
correlation between the expression levels of the hub OXSRGs.
Furthermore, a network of hub OXSRGs and their 20 co-expression
genes was analyzed via GeneMANIA (15). In addition, enrichment analyses of
the hub OXSRGs in Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) were performed using the clusterProfiler
package (16).
Construction and validation of the
prognostic signature
The risk score was calculated as follows: Risk
score=(Exp=expression level; Coe=regression coefficient). To assess
the probability of survival in OS, a nomogram was constructed,
combining clinical characteristics and the prognostic signature.
The calibration curves were used for assessing the nomogram.
The patients were separately classified into a
low-risk and a high-risk group according to the median value of the
risk score. Kaplan-Meier (KM) survival analysis was used to
estimate the overall survival of the two groups. Receiver operating
characteristic (ROC) curves were performed to calculate the
sensitivity and specificity. These analyses were repeated in the
validation dataset. Finally, COX regression was used to determine
whether the signature was an independent prognostic factor.
In addition, KM survival analysis and ROC curves
were used for each of the hub genes to assess the significance of
each single gene.
Analysis of TME
Estimate package was applied to calculate the TME
scores and tumor purity (17). The
Wilcoxon rank sum test was used to analyze the differences between
two groups. In addition, the correlation between the risk score and
the tumor purity was assessed by the Spearman test.
Single sample gene set enrichment analysis was
applied to explore the immune infiltration levels in OS. Pearson
correlation was used to assess the correlation between risk score
and immune infiltration.
Identification and functional
enrichment analysis of differentially expressed genes (DEGs)
The comparison of low- and high-risk groups with the
limma package resulted in the identification of DEGs in the
high-risk group (18). When the
DEGs were obtained, false discovery rate (FDR)<0.05 and
|log2FC|>2 was considered to indicate a statistically
significant difference. The Protein-Protein Interaction (PPI)
network, which was based on DEGs, was constructed by the STRING
database. The confidence level was set as a combined score >0.9.
Finally, a functional enrichment analysis of DEGs was performed in
KEGG and GO.
Cell culture and transfection
Three OS cell lines (MG63, U2OS, and HOS) and one
osteoblast cell line (hFOB1.19) were used in this study. The OS
cell lines were purchased from Zhong Qiao Xin Zhou Biotechnology
Co. The osteoblast cell line (hFOB1.19) was purchased from Procell
Life Science & Technology Co., Ltd. Three human OS cell lines
(HOS, MG63, and U2OS) were maintained in a humidified incubator at
37°C in the presence of 5% CO2, while the human osteoblast cell
line (hFOB1.19) was maintained at 34°C. MG63, U2OS, and HOS cells
were cultured in MEM complete medium and hFOB1.19 cells were
cultured in DMEM/F-12 complete medium. All complete media contained
10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% antibiotics
(100 units/ml penicillin and 100 units/ml streptomycin).
A mitogen-activated protein kinase kinase kinase 5
(MAP3K5) overexpression plasmid was purchased from Shanghai
GeneChem Co., Ltd. and used to infect HOS cells according to the
manufacturer's protocol. The name of the plasmid backbone for the
overexpression vector was CMV enhancer-MCS-SV40-puromycin. The
control was empty plasmid, which was constructed using the same
method but without the target gene inserted.
Total RNA was extracted from cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the instructions of the manufacturer. PCR was
subsequently performed to determine the mRNA expression levels of
the genes according to previous studies (19). β-actin was set as an internal
control and the relative gene expression was calculated by the
2−ΔΔCq method. The sequences of primers were included in
Table SI.
Detection of the effect of MAP3K5 on
OS cells
The cells were seeded at a density of
1×103 cells/well in a 96-well plate. A total of 10 µl
Cell Counting Kit-8 (CCK-8) reagent was added at specific
timepoints. Following incubation at 37°C for 1 h, the optical
density was measured at 450 nm.
The EdU Detection Kit (Guangzhou RiboBio Co., Ltd.)
was used to assess the fraction of DNA-replicating cells, which
represented the cell proliferative status.
The wound healing assays were performed by using a
sterile pipette tip to scratch and form a gap when cells were
completely confluent in the 6-well plate. Subsequently, serum-free
medium was added. Following incubation for 24 h, the gap was
imaged.
Transwell assays were performed according to a
previous study (20). Initially,
1×104 cells per group were inoculated in the upper
chamber. Following 24 h of cell incubation, the cells in the bottom
chamber were fixed, stained, and photographed.
The cell suspensions were made in a
phosphate-buffered saline solution. Annexin V-FITC Apoptosis
Detection Kit (Vazyme Biotech Co., Ltd.) and flow cytometry were
used to detect apoptosis.
According to the manufacturer's protocol, ROS
generation was detected via a Reactive Oxygen Species Assay Kit
(Beyotime Institute of Biotechnology). The incremental production
of ROS was expressed as a percentage of control.
Statistical analysis
The present study was based on the R software
(version 4.1.2) to analyze the data and generate graphs. All
experiments were performed at least three times independently.
Continuous variables were expressed using the mean ± standard
deviation. Flexible statistical methods were used in the
statistical analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of hub OXSRGs and
functional enrichment analysis
The data were collated to obtain a gene expression
matrix containing 84 patients, of whom 55 survived and 29 did not.
The pertinent information is presented in Table I. Following screening, 807 genes
associated with oxidative stress were obtained from the GeneCards
website. Subsequently, 17 genes associated with prognosis of OS
were obtained by univariate COX regression (Fig. 1A). The five most powerful prognostic
genes were identified by LASSO regression (Fig. 1B and C).
 | Table I.Demographic and clinical
characteristics of patients. |
Table I.
Demographic and clinical
characteristics of patients.
|
Characteristics | TARGET cohort
(n=84) | GSE21257
(n=53) |
|---|
| Sex
(male/female) | 47/37 | 34/19 |
| Age (years) | 14.98±4.78 | 18.71±12.20 |
| Survival status
(alive/dead) | 55/29 | 30/23 |
| Average survival
time (months) | 76.21±68.81 | 38.17±40.45 |
The data indicated a significant correlation in the
expression levels between the five hub OXSRGs (Fig. 2A). GeneMANIA revealed that mitogen
activated protein kinase 1 (MAPK1) and MAP3K5 were involved in
several biological functions related to oxidative stress, such as
response to oxidative stress and ROS and cellular response to
oxidative radicals (Fig. 2B). The
GO and KEGG enrichment results also indicated that hub OXSRGs were
involved in a variety of oxidative stress-related pathways and
functions including the forkhead box signaling pathway. Notably,
certain hub genes were also associated with inflammatory and
immune-related pathways, such as the IL-17 and the TNF signaling
pathways (Fig. 2C-F).
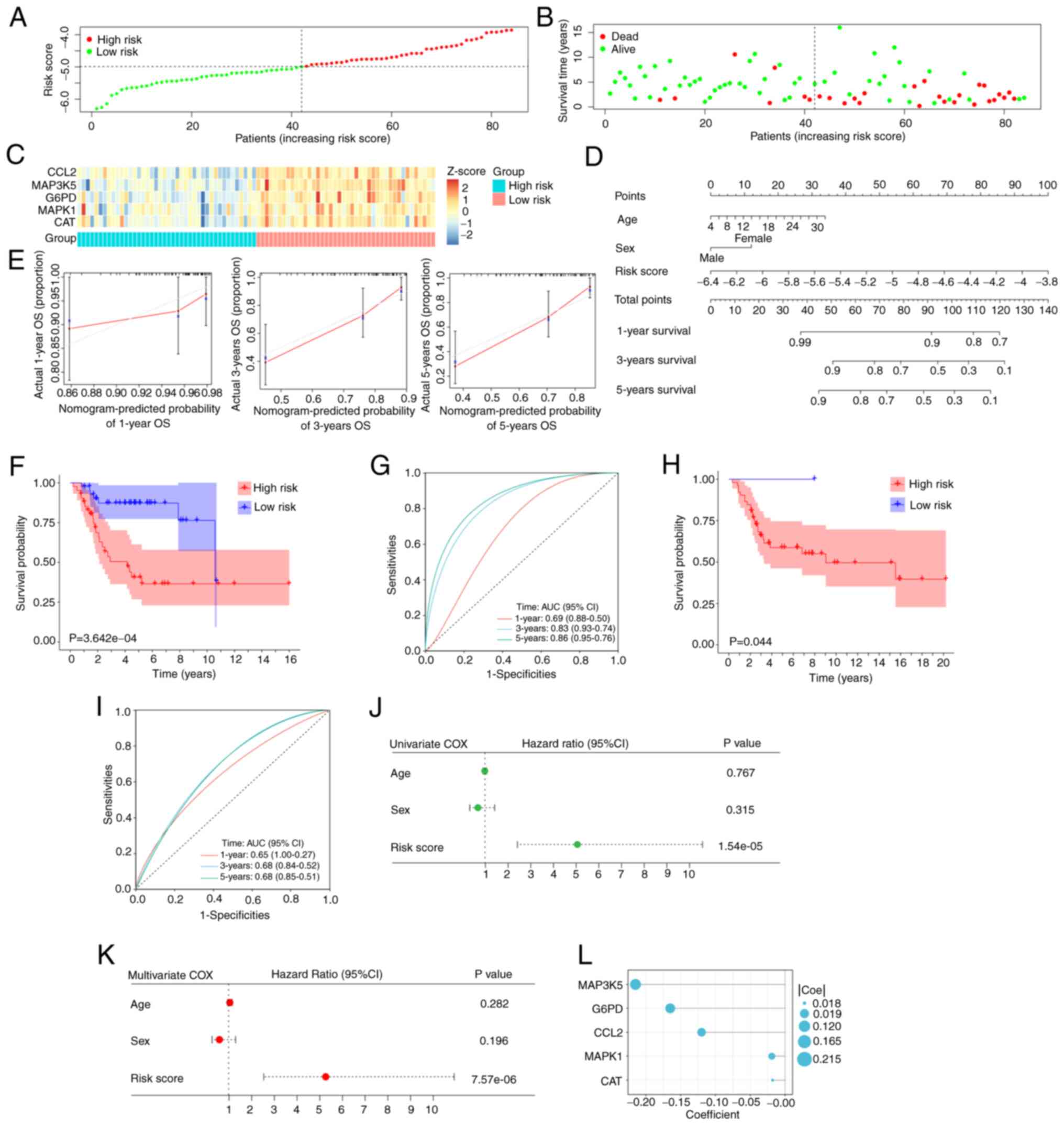
Establishment of the prognostic
signature based on five hub OXSRGs
Regression coefficients were assigned to the formula
to calculate a combined risk score for the five genes to assess the
prognosis of patients. Patients were grouped according to the
median risk score (Fig. 3A). The
risk score was higher in patients with poor survival as shown in
the scatter plot (Fig. 3B). The
heat map indicated the expression profiles of the five hub genes in
the training cohort (Fig. 3C).
Finally, a nomogram including age, sex, and the risk score based on
the signature was successfully constructed (Fig. 3D). The calibration plots
demonstrated that the predictive results of the nomogram were
highly consistent with the actual observations (Fig. 3E).
A significant difference was noted in the survival
probability between the two groups (P<0.001; Fig. 3F). The time-dependent ROC curves
suggested optimal specificity and sensitivity for the prognostic
evaluation of the signature in patients with OS (Fig. 3G). The data from the validation set
indicated that the prognostic signature remained highly stable
(Fig. 3H and I).
Finally, the univariate and multivariate COX
regression analyses were used to investigate the predictive power
of the signature. The results of both analyses indicated that the
risk score was an independent prognostic factor for patients with
OS (P<0.001; Fig. 3J and K). The
coefficients of five hub OXSRGs are depicted (Fig. 3L).
Correlation analysis between risk
score and TME
The results indicated that the high-risk group
exhibited lower stromal scores, immune scores, and higher tumor
purity, compared with those of the low-risk group (Fig. 4A and B). Scatter plot analysis
indicated a positive correlation between the risk score and the
tumor purity (R=0.558; Fig.
4C).
The butterfly diagram illustrated the correlation
between the risk score and the immune infiltration phenotype
(Fig. 4D). The risk score was
negatively correlated with the level of infiltration of almost all
immune cells and immune-related functions (P<0.05). The
aforementioned results indicated that the prognostic signature
could assess the immune status of patients with OS to a certain
extent.
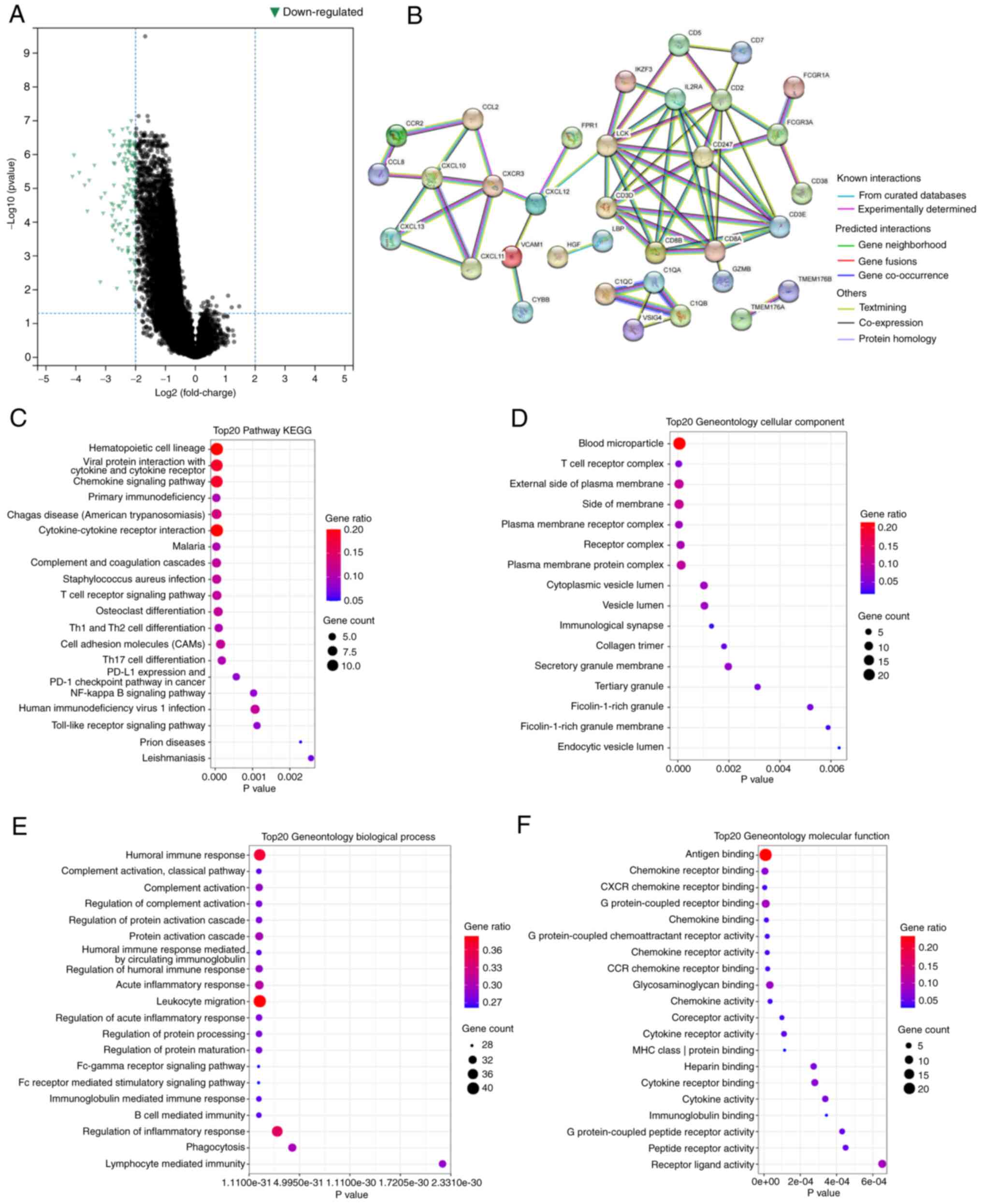
Functional enrichment analysis of
DEGs
The data indicated that in the high-risk group, the
majority of the genes were downregulated (Fig. 5A). The PPI network demonstrated the
interactions between these DEGs (Fig.
5B).
GO and KEGG enrichment analysis revealed that these
DEGs were significantly enriched in a large number of pathways. For
instance, in KEGG, DEGs were involved in the NF-κB signaling
pathway (Fig. 5C). This pathway has
been demonstrated to regulate the amount of intracellular ROS and
to be involved in a variety of immune processes (21,22).
GO enrichment analysis indicated regulation of T cell activation
and regulation of the inflammatory response, and other
immune-related pathways (Fig.
5D-F). Table II describes the
detailed information related to the five hub genes.
 | Table II.Detailed information of hub
OXSRGs. |
Table II.
Detailed information of hub
OXSRGs.
| Gene symbols | Full names |
Log2FC | FDR | Coefficient |
|---|
| CAT | Catalase | −1.03 | <0.001 | −0.018 |
| MAPK1 | Mitogen-Activated
Protein Kinase 1 | −0.84 | <0.001 | −0.019 |
| G6PD | Glucose-6-Phosphate
Dehydrogenase | −1.26 | <0.001 | −0.165 |
| MAP3K5 | Mitogen-Activated
Protein Kinase Kinase Kinase 5 | −1.68 | <0.001 | −0.215 |
| CCL2 | C-C Motif Chemokine
Ligand 2 | −2.42 | <0.001 | −0.120 |
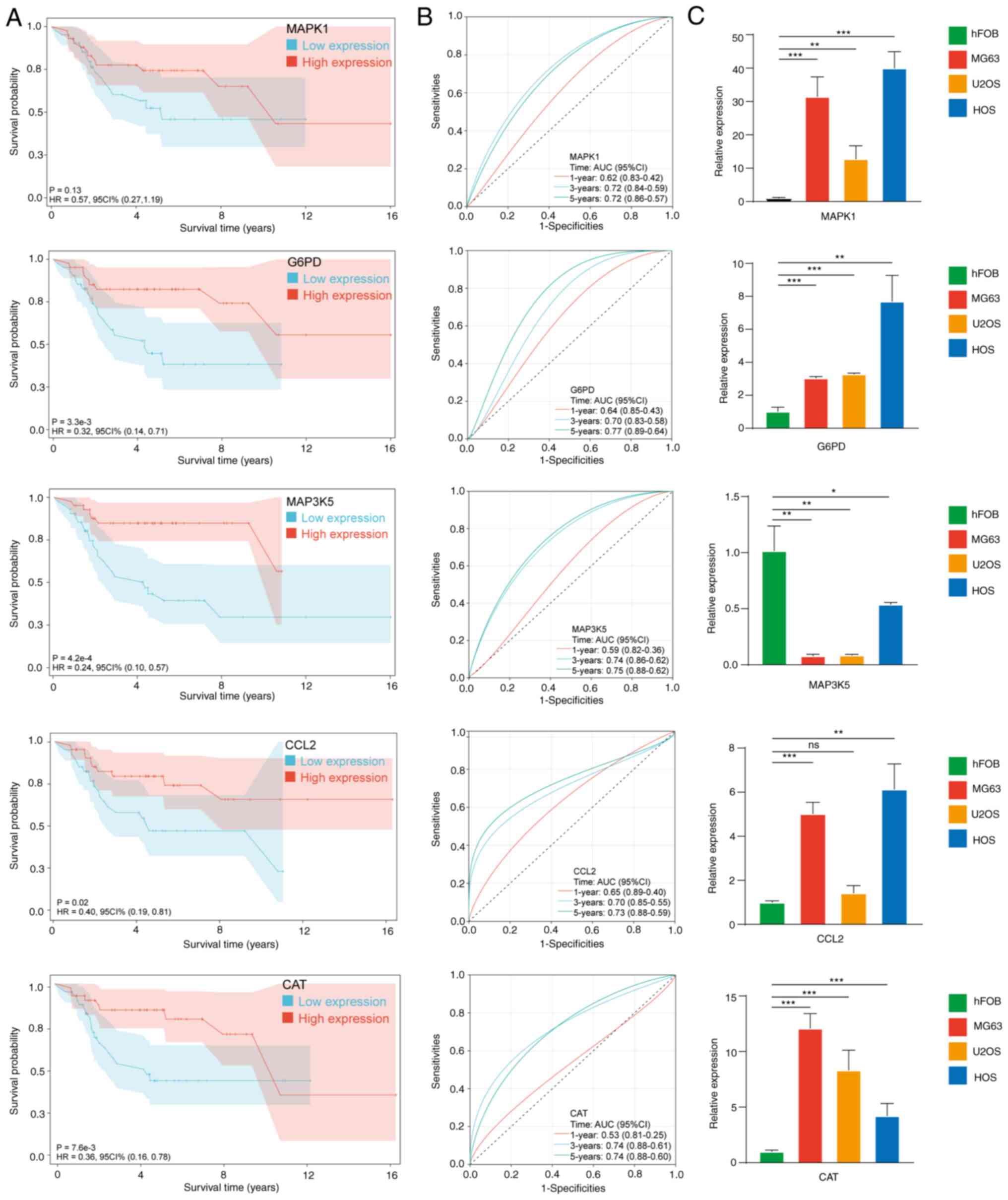
Analysis of single gene
In Fig. 6A and B,
single-gene analysis indicated that all hub OXSRGs with the
exception of MAPK1 (P=0.13) were significantly associated with
patient prognosis. ROC curves indicated the satisfactory predictive
value of hub OXSRGs.
In Fig. 6C, compared
with hFOB 1.19, the mRNA levels of chemokine ligand 2 (CCL2),
catalase (CAT), MAPK1 and glucose-6-phospate dehydrogenase (G6PD)
were significantly elevated in the OS cell line, whereas the
expression levels of MAP3K5 were significantly decreased
(P<0.05). In the five hub genes, the absolute values of the
regression coefficients of MAP3K5 were the highest. In addition,
MAP3K5 was the only gene that was downregulated in the OS cell
lines. Therefore, the role of MAP3K5 was assessed in OS.
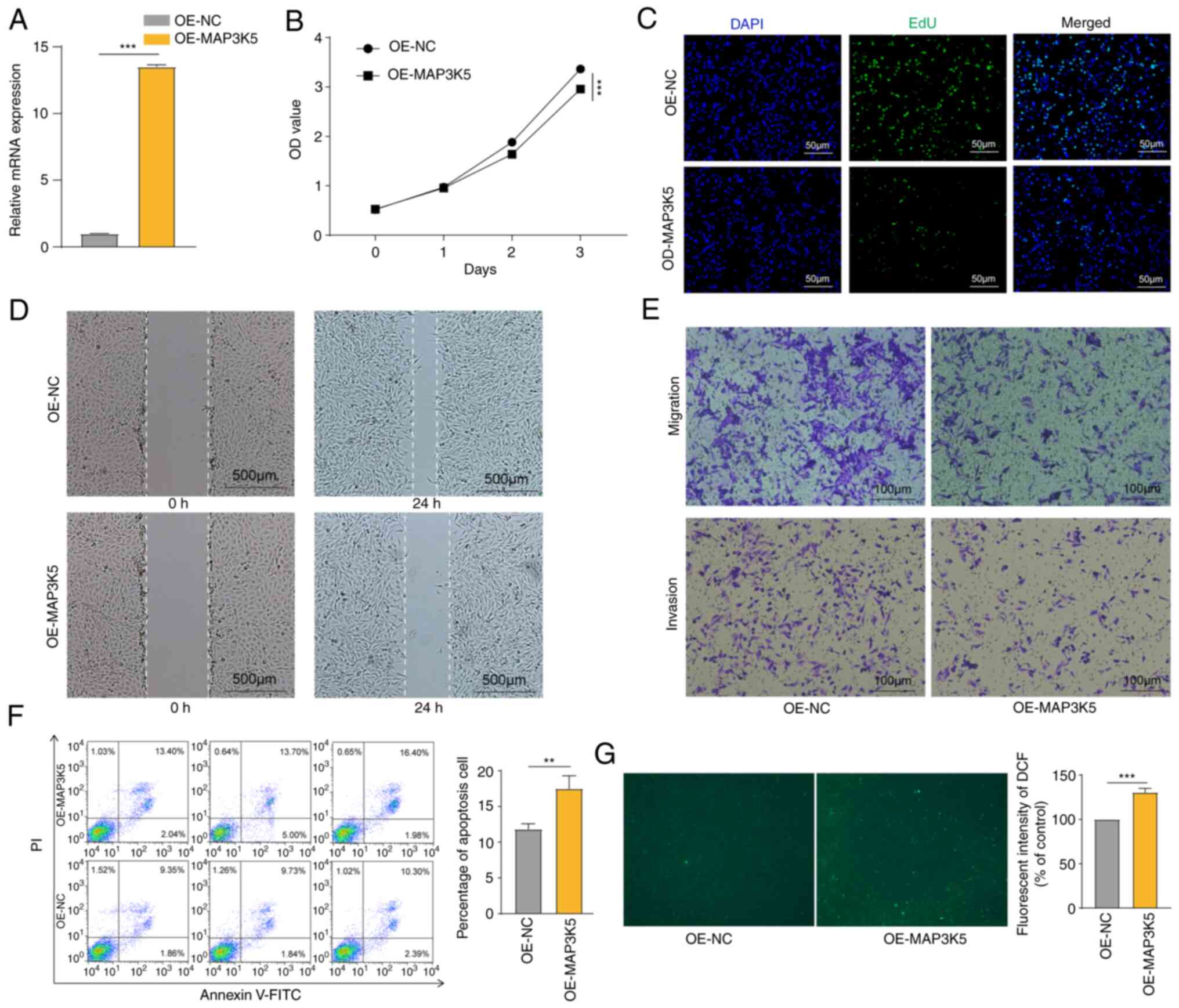
MAP3K5 inhibited the OS cell and
induced ROS generation
MAP3K5 expression was successfully upregulated in
HOS cells (Fig. 7A). CCK-8 assays
and EdU staining revealed that overexpression of MAP3K5 inhibited
cell proliferation (Fig. 7B and C).
By using wound healing assays, it was shown that MAP3K5 attenuated
cell motility (Fig. 7D). Moreover,
transwell assays further validated these findings (Fig. 7D). The data from flow cytometry
analysis revealed that MAP3K5 caused increased apoptosis in the
cells (Fig. 7F). Finally, higher
ROS generation was detected in HOS cells following MAP3K5
overexpression (Fig. 7G). The
aforementioned data indicated that MAP3K5 exhibited inhibitory
effects on OS cells, which may be mediated via ROS production.
Discussion
OS is a frequent type of primary orthopedic
malignancy. As the clinical significance of specific molecular
markers is continuously explored, accumulating studies have
concluded that molecular markers can predict tumor prognosis. A
high number of predictors and therapeutic targets are being
identified. The usage of risk models to predict tumor prognosis is
becoming increasingly accepted. However, no studies have been
conducted to construct an effective prognostic signature based on
OXSRG in OS. In the present study, OXSRGs were studied using a
bioinformatics approach and their prognostic power for OS was
demonstrated. Ultimately, a five-gene signature (CAT, MAPK1, G6PD,
MAP3K5, and CCL2) was constructed. The combination of clinical
characteristics and the signature created a nomogram with optimal
applicable value.
Regulation of oxidative stress is an important
factor in tumor development and in the response to antitumor
therapy (23). One of the
characteristics of tumors is the Warburg effect and the high levels
of oxidative stress (24,25). High ROS levels may inhibit
tumorigenesis. However, ROS can also promote tumor formation
through the induction of DNA mutations and pro-oncogenic signaling
pathways (26). This paradox has
important implications for developing therapies against cancer by
regulating ROS levels. Therefore, the advantage of the present
study was that it improved the understanding of oxidative stress in
OS. In addition, the present study also provided suggestions for
treating OS via regulation of oxidative stress.
A study by Babior et al reported that human
phagocytes (mainly neutrophils and macrophages) utilized membrane
NADPH oxidase to produce large amounts of O2, which was
subsequently decomposed by superoxide dismutase into H2O2 and
oxygen (27). Subsequently, the
study examining the balance between oxidative and antioxidant
systems in physiological and pathophysiological conditions has had
a significant influence on the scientific field of immunology. It
is now becoming more and more apparent that ROS has an important
role in the immune system and that it is closely associated with
various aspects of the immune response, such as immune cell
interactions and activation, immunosuppression, and host defense
(28). The interaction between
immune function and tumors has received increasing attention and a
large number of studies have been reported in the literature.
Mifamurtide is an immune adjuvant, which has been at the forefront
of OS treatment; however, there is a lack of large-scale clinical
trials to demonstrate its efficacy and safety. Therefore, Kansara
et al have suggested that the understanding of the
relationship between bone tumors and immune function is still in
its infancy (1). The data of the
present study indicated a significant difference in the immune
infiltration between the high-risk and low-risk groups. In the
highly risk scores, both immune scores and levels of immune
infiltration were significantly lower (P<0.05). This may explain
the poor outcomes noted in the high-risk group of patients with
OS.
Some of these five genes involved in the signature
have already been studied by a number of studies. MAPK1 encodes a
member of the MAP kinase family, which serves as an integration
point for various biochemical signals involved in several cellular
processes, such as proliferation, differentiation, transcriptional
regulation, and development. The study by Wu et al indicated
that microRNA (miR)-511 mimics inhibited OS cell proliferation and
invasion, and reduced tumor load in metastatic OS in nude mice by
targeting MAPK1 (29). Although
G6PD is a relatively well-known gene, its role in the development
of OS remains understudied. Wang et al demonstrated that
long non-coding RNA OR3A4 regulated the growth of OS cells by
regulating miR-1207-5p/G6PD signaling (30). Chemokines are a family of secreted
proteins involved in immune regulation and inflammatory processes.
It has been shown that CCL2 is highly expressed in OS and acts by
promoting the proliferation and invasion of OS cells via the
receptor activator of nuclear factor kappa-Β ligand signaling
pathway (31).
Several studies have been published reporting the
role of MAP3K5 in various human diseases (32,33).
Currently, MAP3K5 has been identified as one of the key components
in the regulation of ROS (34).
Activation of MAP3K5 can produce pro-inflammatory cytokines, which
are critical to the innate immune response (35). However, a limited number of studies
have focused on the association of MAP3K5 with OS. OS. Gao et
al demonstrated that miR-494 repressed OS development by
modulating the formation of MAP3K5-related apoptosis complexes
(36). In the present study,
molecular experiments were performed to further investigate the
role of MAP3K5 in OS. The data revealed that MAP3K5 was a negative
factor to OS. Patients with high expression of MAP35 had higher
survival rates. MAP3K5 exhibited inhibitory effects on OS cells. In
addition, MAP3K5 induced ROS generation in OS cells.
However, certain limitations are evident in the
present study. Firstly, the total number of patients with OS in the
present study was limited and future studies require a larger
dataset to further validate this prediction model. Secondly,
biological experiments are required to investigate the molecular
mechanisms by which OXSRGs affect OS prognosis. Importantly,
bioinformatic methods were applied in interpreting these results in
the data and presenting the new findings of the current study.
These results are, to some extent, enlightening for subsequent
mechanistic studies.
In the current study, an OXSRG signature was
developed and validated to predict the prognosis of patients with
OS, which provided a novel idea for the treatment of OS. It was
also discovered that MAP3K5 may be a therapeutic target of OS. The
detection of MAP3K5 expression in the osteosarcoma tissue of
patients is a limitation of our study and an aim of future
experiments.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated during and/or analyzed during the
current study are available from the corresponding author upon
reasonable request.
Authors' contributions
JL planned the study and supervised the analyses. BX
collected data, performed the experiments and analyses. ST and CL
drafted the manuscript and have made significant contributions to
the analysis and interpretation of the data. JL and BX confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang T, Wang L, Zhang L, Long Y, Zhang Y
and Hou Z: Single-cell RNA sequencing in orthopedic research. Bone
Res. 11:102023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu CC and Livingston JA: Genomics and the
immune landscape of osteosarcoma. Adv Exp Med Biol. 1258:21–36.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun W, Wang B, Qu XL, Zheng BQ, Huang WD,
Sun ZW, Wang CM and Chen Y: Metabolism of reactive oxygen species
in osteosarcoma and potential treatment applications. Cells.
9:872019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu W, Zhao Y, Wang G, Feng S, Ge X, Ye W,
Wang Z, Zhu Y, Cai W, Bai J and Zhou X: TRIM22 inhibits
osteosarcoma progression through destabilizing NRF2 and thus
activation of ROS/AMPK/mTOR/autophagy signaling. Redox Biol.
53:1023442022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sarmiento-Salinas FL, Perez-Gonzalez A,
Acosta-Casique A, Ix-Ballote A, Diaz A, Treviño S, Rosas-Murrieta
NH, Millán-Perez-Peña L and Maycotte P: Reactive oxygen species:
Role in carcinogenesis, cancer cell signaling and tumor
progression. Life Sci. 284:1199422021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheung EC and Vousden KH: The role of ROS
in tumour development and progression. Nat Rev Cancer. 22:280–297.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roy K, Wu Y, Meitzler JL, Juhasz A, Liu H,
Jiang G, Lu J, Antony S and Doroshow JH: NADPH oxidases and cancer.
Clin Sci (Lond). 128:863–875. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
García JG, Ansorena E, Izal I, Zalba G, de
Miguel C and Milagro FI: Structure, regulation, and physiological
functions of NADPH oxidase 5 (NOX5). J Physiol Biochem. Mar
11–2023.(Epub ahead of print). View Article : Google Scholar
|
|
10
|
Moloney JN, Stanicka J and Cotter TG:
Subcellular localization of the FLT3-ITD oncogene plays a
significant role in the production of NOX- and
p22phox-derived reactive oxygen species in acute myeloid
leukemia. Leuk Res. 52:34–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sabharwal SS and Schumacker PT:
Mitochondrial ROS in cancer: Initiators, amplifiers or an Achilles'
heel? Nat Rev Cancer. 14:709–721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marcucci G, Domazetovic V, Nediani C,
Ruzzolini J, Favre C and Brandi ML: Oxidative stress and natural
antioxidants in osteoporosis: Novel preventive and therapeutic
approaches. Antioxidants (Basel). 12:3732023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bai H, Chen G, Fang C, Yang X, Yu S and
Hai C: Osteosarcoma cell proliferation and migration are partly
regulated by redox-activated NHE-1. J Clin Transl Res. 1:168–179.
2015.PubMed/NCBI
|
|
14
|
Shin SH, Choi YJ, Lee H, Kim HS and Seo
SW: Oxidative stress induced by low-dose doxorubicin promotes the
invasiveness of osteosarcoma cell line U2OS in vitro. Tumour Biol.
37:1591–1598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
R-Forge: estimate: Estimate of stromal and
immune cells in malignant tumor tissues from expression data.
https://R-Forge.R-project.org/projects/estimate/
|
|
18
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Jiao G, Ren S, Zhang X, Li C, Wu
W, Wang H, Liu H, Zhou H and Chen Y: Exosomes from bone marrow
mesenchymal stem cells enhance fracture healing through the
promotion of osteogenesis and angiogenesis in a rat model of
nonunion. Stem Cell Res Ther. 11:382020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng D, Xia K, Yu L, Gong C, Shi Y, Li W,
Qiu Y, Yang J and Guo W: A novel six metastasis-related prognostic
gene signature for patients with osteosarcoma. Front Cell Dev Biol.
9:6992122021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morgan MJ and Liu ZG: Crosstalk of
reactive oxygen species and NF-κB signaling. Cell Res. 21:103–115.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun SC: The noncanonical NF-κB pathway.
Immunol Rev. 246:125–140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sahoo BM, Banik BK, Borah P and Jain A:
Reactive oxygen species (ROS): Key components in cancer therapies.
Anticancer Agents Med Chem. 22:215–222. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Parkinson EK, Haferkamp S and Mycielska
ME: Cancer cell metabolism. Int J Mol Sci. 23:72102022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
PavlovaN N, Zhu J and Thompson CB: The
hallmarks of cancer metabolism: Still emerging. Cell Metab.
34:355–377. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie
MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al: Association
of reactive oxygen species levels and radioresistance in cancer
stem cells. Nature. 458:780–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Babior BM, Kipnes RS and Curnutte JT:
Biological defense mechanisms. The production by leukocytes of
superoxide, a potential bactericidal agent. J Clin Invest.
52:741–744. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Y, Bazhin AV, Werner J and
Karakhanova S: Reactive oxygen species in the immune system. Int
Rev Immunol. 32:249–270. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu J, Zhang C and Chen L: MiR-511 mimic
transfection inhibits the proliferation, invasion of osteosarcoma
cells and reduces metastatic osteosarcoma tumor burden in nude mice
via targeting MAPK1. Cancer Biomark. 26:343–351. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Chen K and Zhao Z: LncRNA OR3A4
regulated the growth of osteosarcoma cells by modulating the
miR-1207-5p/G6PD signaling. Onco Targets Ther. 13:3117–3128. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen Q, Sun W, Liao Y, Zeng H, Shan L, Yin
F, Wang Z, Zhou Z, Hua Y and Cai Z: Monocyte chemotactic protein-1
promotes the proliferation and invasion of osteosarcoma cells and
upregulates the expression of AKT. Mol Med Rep. 12:219–225. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Challa TD, Wueest S, Lucchini FC, Dedual
M, Modica S, Borsigova M, Wolfrum C, Blüher M and Konrad D: Liver
ASK1 protects from non-alcoholic fatty liver disease and fibrosis.
EMBO Mol Med. 11:e101242019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao H, Chen X, Hu G, Li C, Guo L, Zhang
L, Sun F, Xia Y, Yan W, Cui Z, et al: Small extracellular vesicles
from brown adipose tissue mediate exercise cardioprotection. Circ
Res. 130:1490–1506. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ichijo H, Nishida E, Irie K, ten Dijke P,
Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K and Gotoh
Y: Induction of apoptosis by ASK1, a mammalian MAPKKK that
activates SAPK/JNK and p38 signaling pathways. Science. 275:90–94.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matsuzawa A, Saegusa K, Noguchi T,
Sadamitsu C, Nishitoh H, Nagai S, Koyasu S, Matsumoto K, Takeda K
and Ichijo H: ROS-dependent activation of the TRAF6-ASK1-p38
pathway is selectively required for TLR4-mediated innate immunity.
Nat Immunol. 6:587–592. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao G and Jian Y: MicroRNA-494 represses
osteosarcoma development by modulating ASK-1 related apoptosis
complexes. Transl Cancer Res. 9:4121–4130. 2020. View Article : Google Scholar : PubMed/NCBI
|