Introduction
The 5 year survival rate for liver cancer was 20%
between 2010 and 2016 (1). Recent
trends in liver cancer are promising because of the long-term rise
in mortality slowed in female patients and stabilized among men in
the United States (1). Treatment
strategies for hepatocellular carcinoma (HCC), a prominent
histological type of liver cancer include surgical resection, local
radiofrequency ablation, liver transplantation and systemic therapy
with different drugs (2,3). Currently, there are effective
therapeutic options available for patients with HCC that
significantly improve survival rates, but a significant number of
patients are unresponsive to locoreginal or systemic therapy and
ultimately succumb to their disease (2,3).
Therefore, it is very significant to develop effective therapeutic
regiments for patients with HCC patients.
Curcumin is a diphenolic compound derived from
curcuma longa. Curcumin alone or in combination with other
drugs (including metformin, N-n-butyl haloperidol iodide,
resveratrol, and quercetin) has been reported to demonstrate
anticancer activity in preclinical and clinical trials (4–7). As a
potential chemo-preventive and therapeutic drug for liver cancer,
curcumin has been reported to demonstrate good prospects in
preclinical studies of liver cancer (5,8). The
anticancer effects of curcumin are mainly manifested via activation
of the apoptosis pathway and inhibition of cell proliferation for
HCC cells and affects the tumor microenvironment, such as
inhibiting inflammatory processes angiogenesis and metastasis
(4,9). Previous studies have reported that
curcumin can inhibit the proliferation of liver cancer cells by
regulating certain genes and signaling pathways, such as
hypoxia-inducible factor-1α, nuclear factor E2-related factor 2
(Nrf2), IL-6/STAT3 pathway, and so on (10,11).
The antitumor mechanism of curcumin is unclear,
particularly in HCC cells. Therefore, the present study utilized
the pharmacology database and analysis platform of the traditional
Chinese medicine (TCM) system in combination with bioinformatics
technology to screen potential target genes of curcumin, and
subsequently verified these targets through an animal xenograft
tumor model of human HCC treated by curcumin.
Materials and methods
Screening of candidate genes
Possible target genes of curcumin were searched
using the term ‘curcumin’ through the TCM Systems Pharmacology
Database and Analysis Platform (TCMSP; tcmspw.com/tcmsp.php).
RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA;
http://portal.gdc.cancer.gov) for liver
hepatocellular carcinoma (LIHC) and other types of cancer
(including bladder, thyroid, kidney, colon, lung, prostate, cervix
uteri, bile duct, and breast) were used to analyze the expression
levels and correlation among the candidate target genes (12). Kaplan-Meier plots were generated
using the survminer (version 0.4.4) package in R (version 4.0.3,
http://www.R-project.org/) according
TCGA HCC gene expression data and clinical data (TCGA-LIHC). The
enrichment of Gene Ontology (GO) terms and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathways was performed using the R
packages. After ID conversion of the candidate target genes using
the org.Hs.eg.db package (version 3.11.4), enrichment analysis was
performed using clusterProfiler package (version 3.12.0) and bubble
plots of GO terms and KEGG pathways were generated using the
ggplot2 package (version 2.0.0) (13). Homologous sequences of candidate
genes were summarized using the GeneCards database
(genecards.org/). Specifically, ‘PTPN1’ was used as the search
term, and then paralogs for the protein tyrosine phosphatase
non-receptor type 1 (PTPN1) gene were examined. The protein
interaction network was generated using the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) database
(cn.string-db.org), which is a protein-protein interaction
prediction website, and the confidence interval for the minimum
required interaction score was set to 0.7. The igrap package
(version 1.2.6) and ggrap package (version 2.0.5) in R were used to
analyze and visualize protein-protein interactions. Cytoscape
(version 3.8.0; Cytoscape Consortium; www.cytoscape.org) was used for visualization of the
protein network. The CytoHubba plugin (version 0.1, http://apps.cytoscape.org/apps/cytohubba) were used to
screen the top 5 genes as hub genes by the maximal clique
centrality (MCC) algorithm.
Establishment of human HCC xenograft
model in mice
The human HCC cell line HuH7 (Procell Life Science
& Technology Co., Ltd.) was cultured in Roswell Park Memorial
Institute 1640 containing 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) in a 37°C incubator with 5%
CO2. A total of 12 6-week-old male BALB/c-nu/nu mice
(18±2 g) were purchased from Ji'nan Penyue Laboratory Animal
Breeding Co., Ltd. Mice were examined every day for their health
status and behavior. The food and water supply is inspected
regularly to ensure that it is free from contamination and meets
the nutritional requirements of mice before they were sacrificed.
There were no dead mice during the experiment. Mice were maintained
in standard conditions, with an ambient temperature of 21±2°C, 50%
humidity and a 12-h light/dark cycle. All animal experiments were
approved by the Institutional Animal Care and Use Committee of the
Binzhou Medical University (approval no. 2022-607).
Treatment of xenograft model with
curcumin
After 1 week of adaptive feeding, each nude mouse
was implanted with a subcutaneous injection of 1×106
HuH7 cells in 100 µl phosphate buffered saline. When the tumor
volume was 60–100 mm3 (the 9th day after injection), the
12 nude mice were randomly divided into two groups (the control and
curcumin groups). Animals in the curcumin group were
intraperitoneally injected with curcumin (100 mg/kg/day dissolved
in corn oil at 10 mg/ml) (14), and
the animals in the control group were treated with corn oil (10
ml/kg/day). Curcumin was purchased from Dalian Meilun Biology
Technology Co., Ltd. Corn oil was purchased from COFCO Corporation.
The length and width of the tumors were measured with a vernier
caliper every two days. The tumor volume was calculated using the
following formula: Volume=0.5 × length × width2. The day
on which the treatment began was recorded as day 0. The relative
tumor volume was calculated as follows: Relative tumor
volume=(tumor volume on the day of measurement)/(tumor volume on
day 0). Mice were sacrificed using cervical dislocation after 12
days of treatment. Following euthanasia by cervical dislocation,
mice mortality was verified by the absence of a heartbeat,
respiratory arrest and the lack of reaction to a painful stimulus.
Tumors were then removed, and Image-Pro Plus software (IPP; version
6.0; Media Cybernetics, Inc.) was used to measure the length and
width of each tumor to calculate the tumor volume.
Immunohistochemistry
Tumor tissues from each group were fixed using 4%
paraformaldehyde for 24 h at room temperature and embedded in
paraffin. Sections (4 µm thick) were obtained from the paraffin
blocks, which were then dewaxed in xylene for 15 min and rehydrated
in water. Antigen retrieval was performed using citrate buffer (pH
6.0) for 15 min at 95–100°C. After blocking in 3% hydrogen peroxide
for 10 min at 37°C and 10% goat serum (SL038, Solarbio, China) for
10 min at 37°C successively, sections were incubated with the
following primary antibodies (all purchased from Proteintech Group,
Inc.); proliferating cell nuclear antigen (PCNA; 1:400; cat. no.
24036-1AP), BAX (1:500; cat. no. 50599-2Ig), PTPN1 (1:200; cat. no.
11334-1AP) and PTPN11 (1:400; 24570-1AP) overnight at 4°C. Sections
were then incubated with the enzyme labeled sheep anti mouse/rabbit
IgG polymer in the GTVision Detection System/Mo&Rb (GK600505,
Gene Tech Biotechnology Co., Ltd.) at room temperature for 30 min
and visualized using the DAB solution (GK600505, Gene Tech
Biotechnology Co., Ltd.). Sections were counterstained with Harris
hematoxylin for 2 min at room temperature. The sections were
dehydrated in graded alcohol, cleared in xylene and mounted. Images
were acquired using an AJ-VERT station (Chongqing Optec Instrument
Co., Ltd.) with a light microscope. IPP (version 6.0; Media
Cybernetics, Inc.) software was used for semi-quantitative analysis
by randomly selecting three high-power fields from each section.
The integrated optical density (IOD) of BAX, PTPN1 and PTPN11
expression levels and the positive cell numbers of PCNA expression
levels in tumors were quantified.
Statistical analysis
All data are expressed as mean ± standard error. For
the statistical comparisons of the protein expression levels and
tumor volume in control and curcumin groups, the normal
distribution and homogeneity of variances were tested using the
Shapiro-Wilk test and F test. To evaluate differences between study
groups, unpaired and paired Student's t-tests were used for the
data with normal distribution, and homogeneity of variance and
Mann-Whitney U tests were used for the data with non-normal
distribution and inhomogeneity of variance from unpaired samples
and Wilcoxon signed rank tests were used for the data from paired
samples. Statistical analyses were performed using GraphPad Prism 8
(GraphPad Software; Dotmatics). Survival curves of candidate target
genes to HCC were plotted using the survminer package in R, and
data were analyzed using a log-rank test (Mantel-Cox). Patients
were grouped according to the target genes expression levels
relative to the median of the group (high > median and low ≤
median). The two-stage test using the TSHRC package (version 0.1.6)
was used when there was an intersection in the survival curve.
Correlations between PTPN1 and its paralogous genes were assessed
using Spearman's rank correlation test in R. P<0.05 was
considered to indicate a statistically significant difference.
Results
PTPN1, heat shock protein 90 α-family
class a member 1 (HSP90AA1) and potassium voltage-gated channel
subfamily h member 2 (KCNH2) are overexpressed in HCC
The following target genes of curcumin were
identified using TCMSP: PTPN1, HSP90AA1, coagulation factor X
(F10), prostaglandin-endoperoxide synthase 2 (PTGS2) and KCNH2. To
understand the relationships among the target proteins,
protein-protein interaction analysis was performed using the STRING
search tool. With a confidence cut-off of 0.4, a network was
generated. The results showed interactions among these target
proteins (Fig. S1A). Enrichment
analysis of target genes indicated that the most enriched GO terms
and pathways, included the ‘IL-17 signaling pathway’, ‘protein
tyrosine kinase binding’, ‘ubiquitin protein ligase binding’,
‘scaffold protein binding’, ‘endoplasmic reticulum lumen’,
‘cytoplasmic side of endoplasmic reticulum membrane’, and positive
regulation of reactive oxygen species processes (Fig. S1B).
To assess the expression levels of these target
genes in HCC, the TCGA database was used to compare and analyze the
expression levels of the aforementioned target genes in HCC. mRNA
expression levels of PTPN1, HSP90AA1 and KCNH2 were significantly
higher in tumor tissues compared with in normal liver tissues
(Fig. S1C). The expression levels
of PTGS2 in tumor tissues was significantly lower compared with
that in normal tissues and the expression levels of F10 were
markedly increased compared with those in normal tissue (Fig. S1C). Additionally, the correlation
of the five target genes was analyzed, and the results demonstrated
that the correlation coefficients between these genes were between
−0.5 and 0.5 which suggested that any correlations between these
genes was weak (Fig. S1D).
Patients with HCC and high PTPN1
expression levels have a poor prognosis
RNA-seq data from the TCGA LIHC dataset were divided
into high and low expression groups according to the median
expression level of each target gene. Survival data were analyzed
using Cox regression analysis. The results demonstrated that HCC
patients with high PTPN1 and HSP90AA1 expression levels had shorter
overall survival compared with those with low expression levels
(Fig. S2A and B). Differences in
the median overall survival between the high expression level group
of PTGS2, F10 and KCNH2 and the low expression level group were not
statistically significant (Fig.
S2C-E). The median disease specific survival rates in the high
expression level group for PTPN1, HSP90AA1 and KCNH2 were shorter
compared with those in the low expression group (Fig. S2F, G and I). However, the
expression levels of PTGS2, F10 and were not associated with median
disease specific survival (Fig. S2H
and J). The progress free interval in patients with high PTPN1
expression levels was significantly shorter compared with that in
patients with low PTPN1 expression levels (Fig. S2K). The expression levels of
HSP90AA1, PTGS2, F10 and KCNH2 were not associated with the
progress free interval (Fig.
S2L-O). These results suggested that patients with HCC and high
PTPN1 expression levels had a worse prognosis. Furthermore, the
expression level of PTPN1 in different types of cancer was analyzed
using RNA-seq data from the TCGA project. mRNA expression levels of
PTPN1 in tumor tissues were significantly higher compared with
those in normal tissues from certain types of cancer, including
breast carcinoma, cholangiocarcinoma, colon adenocarcinoma,
esophageal adenocarcinoma, head and neck squamous cell carcinoma,
kidney renal clear cell carcinoma, kidney renal papillary cell
carcinoma, HCC, rectum adenocarcinoma and stomach adenocarcinoma
(Fig. S3). However, expression
levels of PTPN1 were significantly lower in tumor tissues compared
with normal tissues from lung adenocarcinoma, lung squamous cell
carcinoma and thyroid carcinoma (Fig.
S3).
Curcumin inhibits the expression of
PTPN1 in HCC xenograft tumor
To verify whether curcumin could inhibit expression
of PTPN1, a human HCC xenograft model was established by
subcutaneously injecting HuH7 cells into nude mice. Intraperitoneal
injection of curcumin was then administered for 12 days. The
results demonstrated that the tumor growth rate of the curcumin
group was significantly reduced compared with that of the control
group on day 6, 8, 10 and 12 after treatment (Fig. 1A), and the tumor volume of the
curcumin group was significantly smaller compared with that of the
control group (Fig. 1B).
Immunohistochemistry indicated that the protein expression levels
of PCNA in the tumor tissue of the curcumin group were
significantly reduced compared with those of the control group
(Fig. 1C). The protein expression
levels of BAX in the tumor tissue of the curcumin group were
significantly increased compared with those of the control group.
These results suggested that curcumin inhibited cell proliferation
and promoted apoptosis in HCC. Next, the expression levels of PTPN1
in tumor tissues were assessed. The results demonstrated that the
IOD values for PTPN1 protein expression levels in tumor tissues of
the curcumin group were significantly reduced compared with those
in the control group (Fig. 1C),
which suggested that curcumin could inhibit PTPN1 protein
expression.
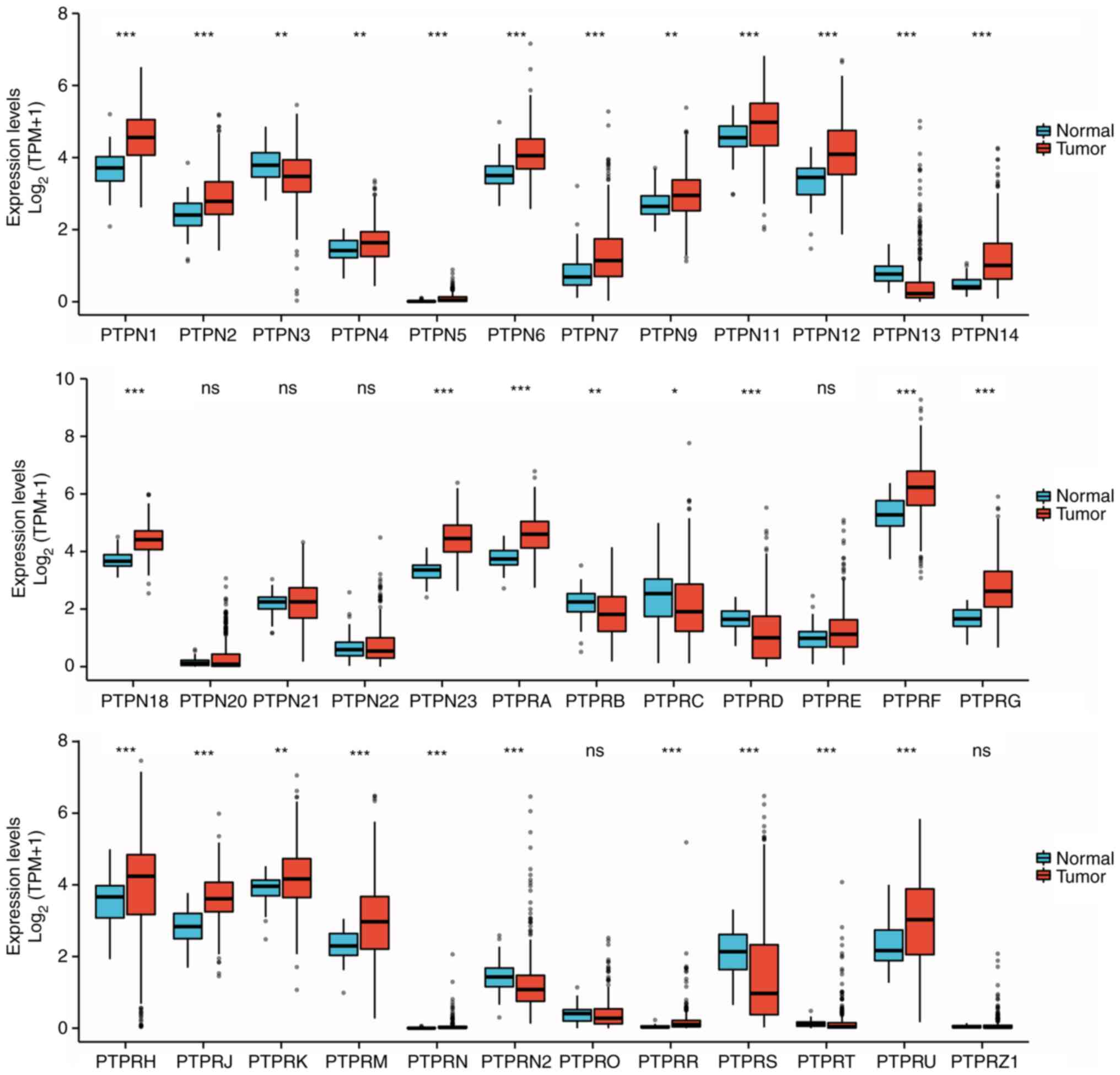
Differential expression of classical
PTP family genes in HCC
PTPN1 belongs to the classical PTP family. Using the
data from TCGA LIHC dataset, unpaired sample statistical analysis
was performed on the mRNA expression levels of classical PTPs, the
results demonstrated that certain PTP family genes were highly
expressed in HCC tumor tissues, including PTPN1, PTPN2, PTPN4-7,
PTPN9, PTPN11, PTPN12, PTPN14, PTPN18, PTPN23, PTPRA, PTPRF, PTPRG,
PTPRH, PTPRJ, PTPRK, PTPRM, PTPRN, PTPRR and PTPRU. PTPs with
reduced expression levels in HCC tumor tissue included PTPN3,
PTPN13, PTPRB, PTPRC, PTPRD, PTPRN2, PTPRS and PTPRT (Fig. 2). Statistical analysis of paired
samples demonstrated that PTPs with significantly increased
expression levels in HCC tumor tissues included PTPN1, PTPN2,
PTPN4-7, PTPN9, PTPN11, PTPN12, PTPN14, PTPN18, PTPN23, PTPRA,
PTPRF, PTPRG, PTPRJ, PTPRK, PTPRM, PTPRN, PTPRR and PTPRU. PTPs
with significantly reduced expression levels in HCC tumor tissues
included PTPN3, PTPN13, PTPRB, PTPRC, PTPRD and PTPRS (Fig. 3). Using the STRING online tool, a
protein interaction network diagram for classical PTP family
proteins was constructed (Fig. 4A).
The GO and KEGG pathways enrichment of high- and low-expression
genes indicated that these genes were mainly enriched in
‘peptidyl-tyrosine dephosphorylation’, ‘protein dephosphorylation’,
‘dephosphorylation’, ‘cytoplasmic side of membrane’, ‘cytoplasmic
side of plasma membrane’, ‘secretory granule membrane’, protein
tyrosine entries such as ‘protein tyrosine phosphatase activity’,
‘phosphoprotein phosphatase activity’, ‘phosphatase activity’,
‘adherens junction’, ‘cell adhesion molecules’ and ‘MAPK signaling’
(Fig. 4B).
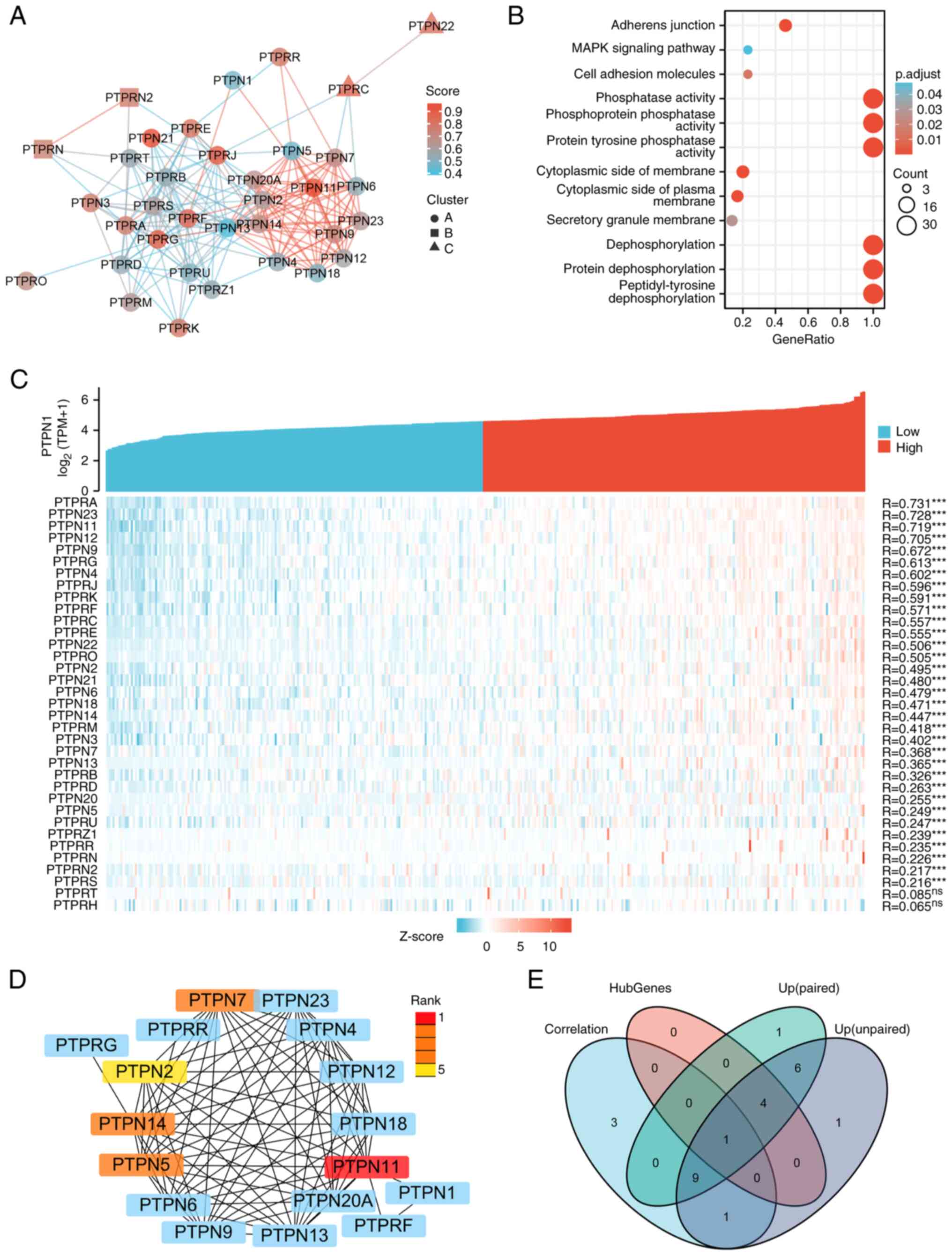 | Figure 4.Screening the key genes in the PTP
gene family in HCC. (A) Interactive relationship of the PTP gene
family. The Search Tool for the Retrieval of Interacting
Genes/Proteins online tool (https://cn.string-db.org/cgi/input.pl) was utilized
to analyze the relationship of the target genes. (B) Gene Ontology
(https://bioconductor.org/packages/release/data/annotation/html/GO.db.html)
and Kyoto Encyclopedia of Genes and Genomes
(bioconductor.org/packages//2.7/data/annotation/html/KEGG.db.html)
enrichment of the PTP gene family. ID conversion of the candidate
target genes using the org.Hs.eg.db package (version 3.11.4),
enrichment analysis was performed using clusterProfiler package
(version 3.12.0) and bubble plots were generated using the ggplot2
package (version 2.0.0). (C) The correlation between PTPN1 and its
paralogous genes in HCC. Correlation coefficients were calculated
with a Spearman's rank correlation coefficient test. (D) Hub genes
of PTP family genes were screened from the protein-protein
interaction network using the cytoHubba plugin (version 0.1,
http://apps.cytoscape.org/apps/cytohubba) in
Cytoscape (version 3.8.0; Cytoscape Consortium; www.cytoscape.org). The maximal clique centrality
algorithm was used to calculate the score of each protein node and
to identify the top 5 hub genes. (E) Venn diagrams indicating the
overlap between Correlation (the correlation coefficient with PTPN1
is >0.5), HubGenes (Hub genes of PTP family genes), Up (paired)
(Up regulated genes in paired samples of HCC), and Up (unpaired)
(Up regulated genes in unpaired samples of HCC). Venn diagrams were
generated using the venn Diagram package (version 1.7.3 in R. HCC,
hepatocellular carcinoma; PTP, protein tyrosine phosphatase; PTPN,
PTP non-receptor type; PTPR, PTP receptor-type; TPM, transcripts
per kilobase million. |
Expression levels of PTPN11 are
correlated with PTPN1 in HCC
RNA-seq data from the TCGA LICH dataset was used to
analyze the correlation between the expression levels of the
classical PTP family and PTPN1 in HCC. The results demonstrated
that there were 14 genes whose correlation coefficients with PTPN1
were >0.5 and which were statistically significant (Fig. 4C). The cytohubba plug-in for
Cytoscape, was used to screen five core genes (PTPN11, PTPN7,
PTPN14, PTPN5 and PTPN2) according to the score of the MCC and
Degree algorithms (Fig. 4D).
Finally, PTPN11 was screened by intersecting the upregulated
differentially expressed genes in HCC, genes strongly correlated
with PTPN1 and Hub genes (Fig. 4E).
Together, these results suggested that both PTPN1 and PTPN11 may be
the targets of curcumin.
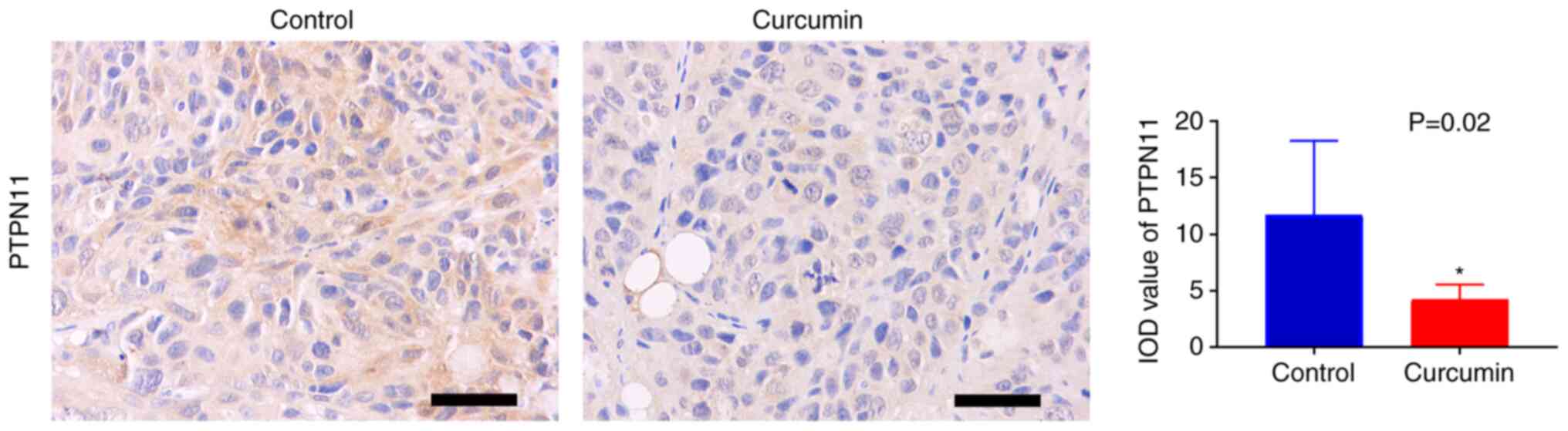
Curcumin inhibits the expression of
PTPN11 in HCC xenograft tumor tissue
The effect of curcumin on expression levels of
PTPN11 was assessed using immunohistochemistry. The results
demonstrated that the integrated optical density of PTPN11 protein
expression in the tumor tissue of the curcumin group was
significantly lower compared with that of the control group
(Fig. 5), which indicated that
curcumin may inhibit the growth of HCC tumors via downregulation of
PTPN11 expression.
Discussion
A previous clinical trial reported that curcumin has
considerable potential for treating patients with cancer (15). In the present study, curcumin was
demonstrated to promote apoptosis, inhibit cell proliferation and
inhibit HCC tumor growth, which was consistent with the
experimental results reported in the literature (16). Curcumin is a phenolic compound
extracted from turmeric, which has antioxidant, anti-angiogenic and
anti-inflammatory effects (17,18).
The effectiveness of curcumin has been previously reported by
detailed in vitro, in vivo and clinical trials (15,19).
Curcumin can modulate certain signaling pathways, including growth
factors, cytokines, transcription factors, and genes which modulate
cellular proliferation and apoptosis in cancer cells, which can
lead to mortality or inhibition of cancer cell proliferation
(11,16).
In the current study, TCMSP database combined with
the RNA-seq data from the TCGA LIHC dataset were used to screen the
target gene, PTPN1, of curcumin in HCC. PTPN1 is a member of the
classical PTP family (20,21). PTPs remove phosphate groups from
proteins involved in cell signal transduction and regulate cell
growth, differentiation, metabolism, gene transcription and immune
responses (21). Aberrant tyrosine
phosphorylation levels, which result from PTPN1 dysfunction are
associated with the progression of numerous diseases including
cancer (21,22), diabetes (23,24)
and obesity (24). Recent studies
have reported that PTPN1 inhibition may be an option for the
treatment of such diseases (21,23).
In the present study, the expression levels of PTPN1 in 18 cancer
types, from TCGA data, were analyzed, and the results indicated
that 10 types of cancer had significantly higher expression levels
of PTPN1 and 3 types of cancer had significantly lower expression
levels of PTPN1 compared with normal tissues. Therefore, modulation
of PTPN1 phosphatase activity with inhibitors may contribute to
innovative targeted pharmacotherapy for certain cancer types.
Previous studies have reported that curcumin, and its derivatives
with a cytotoxic effect against breast cancer cells, exhibited an
inhibitory effect against PTPN1 phosphatase (25,26).
Another study reported that curcumin was an activator of PTPN1 and
could reduce cell motility in colon cancer through
dephosphorylation of cortactin (27). Curcumin inhibits PTPN1 phosphatase
activity in breast cancer and activates it in colon cancer cells,
so the effect of curcumin on PTPN1 in HCC cells needs to be
verified.
Furthermore, the differential expression of typical
tyrosine-specific PTPs in HCC tissues and their correlation with
PTPN1 expression levels was analyzed. The results demonstrated that
the mRNA expression levels of PTPN1 in HCC were significantly
correlated with that of PTPN11. The SH2 domain-containing
phosphatase 2 (SHP2) encoded by PTPN11 is involved in
transcriptional regulation, cytokine signaling, cell
differentiation, and tumor cell proliferation and migration
(28,29). Mutations in PTPN11 cause certain
types of cancer, such as lung, neuroblastoma, breast, skin and
cervical cancer.
Previous studies have reported that SHP2 is
overexpressed in the majority of HCC and is more highly expressed
during metastasis (30,31). SHP2 promotes the growth of liver
cancer cells through the Ras/Raf/MEK/ERK cascade and the growth of
liver cancer cells through the PI3K/Akt/mTOR signaling pathway
enhances the potential malignancy of liver cancer (30,32).
SHP2 can promote the self-renewal ability of liver cancer stem
cells by activating the β-catenin signaling pathway (33). Therefore, high SHP2 expression
levels often indicate a poor prognosis in patients with liver
cancer. The results of the present study suggest that curcumin
inhibits PTPN11 protein expression in HCC cells. Further
experiments are needed to elucidate the mechanism by which curcumin
inhibits the expression of PTPN11. It is also necessary to clarify
whether the reduction in protein expression levels of SHP2 is the
consequence or the cause of curcumin-mediated inhibition of HCC
cell proliferation.
The present study had certain limitations. First,
bioinformatic techniques were used to identify potential targets of
curcumin for HCC and in vivo experiments were conducted to
validate the findings; however, an in-depth investigation of its
specific mechanism was not performed. Second, it was difficult to
obtain sufficient tumor samples for western blotting analysis to
accurately measure the candidate protein. While western blotting is
considered the benchmark method for protein expression
quantification in tissues, it is imperative to also observe the
localization of candidate proteins within tumor cells. Therefore,
the present study prioritized the preparation of specimens for
immunohistochemical techniques.
In summary, the results of the present study suggest
that curcumin can inhibit the proliferation of HCC cells and
promote apoptosis. Furthermore, curcumin inhibits PTPN1 and PTPN11
expression. Curcumin may inhibit the proliferation of HCC cells by
inhibiting the expression of PTPN1 and PTPN11 genes. However, this
conclusion must be verified experimentally. Future experiments are
required to elucidate the mechanism of curcumin inhibition of PTPN1
and PTPN11 expression in HCC cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Provincial Medicine and
Health Science Technology Development Program Shandong (grant no.
2017WS822) and the Innovation and Entrepreneurship Training Program
for College Students (grant no. S202010440077).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ, YL, XW, ZW, EX, JL and DW contributed to the
study conception and design. Material preparation and data
collection were performed by XW and DW. Data collection and
analysis were performed by JZ, YL, XW, ZW and EX. The first draft
of the manuscript was written by JZ, JL and DW. All authors
commented on previous versions of the manuscript. All authors read
and approved the final version of the manuscript. JZ and DW confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luo XY, Wu KM and He XX: Advances in drug
development for hepatocellular carcinoma: Clinical trials and
potential therapeutic targets. J Exp Clin Cancer Res. 40:1722021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang HH, Zhang Y, Cheng YN, Gong FL, Cao
ZQ, Yu LG and Guo XL: Metformin incombination with curcumin
inhibits the growth, metastasis, and angiogenesis of hepatocellular
carcinoma in vitro and in vivo. Mol Carcinog. 57:44–56. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baby J, Devan AR, Kumar AR, Gorantla JN,
Nair B, Aishwarya TS and Nath LR: Cogent role of flavonoids as key
orchestrators of chemoprevention of hepatocellular carcinoma: A
review. J Food Biochem. 45:e137612021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghobadi N and Asoodeh A: Co-administration
of curcumin with other phytochemicals improves anticancer activity
by regulating multiple molecular targets. Phytother Res.
37:1688–1702. 2023. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khan H, Ni Z, Feng H, Xing Y, Wu X, Huang
D, Chen L, Niu Y and Shi G: Combination of curcumin with N-n-butyl
haloperidol iodide inhibits hepatocellular carcinoma malignant
proliferation by downregulating enhancer of zeste homolog 2
(EZH2)-lncRNA H19 to silence Wnt/β-catenin signaling.
Phytomedicine. 91:1537062021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bai C, Zhao J, Su J, Chen J, Cui X, Sun M
and Zhang X: Curcumin induces mitochondrial apoptosis in human
hepatoma cells through BCLAF1-mediated modulation of
PI3K/AKT/GSK-3β signaling. Life Sci. 306:1208042022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khan H, Ullah H and Nabavi SM: Mechanistic
insights of hepatoprotective effects of curcumin: Therapeutic
updates and future prospects. Food Chem Toxicol. 124:182–191. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu J, Lin H, Wu G, Zhu M and Li M:
IL-6/STAT3 is a promising therapeutic target for hepatocellular
carcinoma. Front Oncol. 11:7609712021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shao S, Duan W, Xu Q, Li X, Han L, Li W,
Zhang D, Wang Z and Lei J: Curcumin suppresses hepatic stellate
cell-induced hepatocarcinoma angiogenesis and invasion through
downregulating CTGF. Oxid Med Cell Longev. 2019:81485102019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sreepriya M and Bali G: Chemopreventive
effects of embelin and curcumin against
N-nitrosodiethylamine/phenobarbital-induced hepatocarcinogenesis in
Wistar rats. Fitoterapia. 76:549–555. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karaboga Arslan A, Uzunhisarcıklı E, Yerer
M and Bishayee A: The golden spice curcumin in cancer: A
perspective on finalized clinical trials during the last 10 years.
J Cancer Res Ther. 18:19–26. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Zhu Z, Han L, Zhao L, Weng J, Yang
H, Wu S, Chen K, Wu L and Chen T: A curcumin derivative, WZ35,
suppresses hepatocellular cancer cell growth via downregulating
YAP-mediated autophagy. Food Funct. 10:3748–3757. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Giordano A and Tommonaro G: Curcumin and
cancer. Nutrients. 11:23762019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Elkhamesy A, Refaat M, Gouida MSO, Alrdahe
SS and Youssef MM: Diminished CCl4-induced
hepatocellular carcinoma, oxidative stress, and apoptosis by
co-administration of curcumin or selenium in mice. J Food Biochem.
46:e138452022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salehi B, Stojanović-Radić Z, Matejić J,
Sharifi-Rad M, Anil Kumar NV, Martins N and Sharifi-Rad J: The
therapeutic potential of curcumin: A review of clinical trials. Eur
J Med Chem. 163:527–545. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tonks NK, Diltz CD and Fischer EH:
Purification of the major protein-tyrosine-phosphatases of human
placenta. J Biol Chem. 263:6722–6730. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen PJ and Zhang YT: Protein tyrosine
phosphatase 1B (PTP1B): Insights into its new implications in
tumorigenesis. Curr Cancer Drug Targets. 22:181–194. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sivaganesh V, Sivaganesh V, Scanlon C,
Iskander A, Maher S, Lê T and Peethambaran B: Protein tyrosine
phosphatases: Mechanisms in cancer. Int J Mol Sci. 22:128652021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Teimouri M, Hosseini H, ArabSadeghabadi Z,
Babaei-Khorzoughi R, Gorgani-Firuzjaee S and Meshkani R: The role
of protein tyrosine phosphatase 1B (PTP1B) in the pathogenesis of
type 2 diabetes mellitus and its complications. J Physiol Biochem.
78:307–322. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maheshwari N, Karthikeyan C, Trivedi P and
Moorthy NSHN: Recent advances in protein tyrosine phosphatase 1B
targeted drug discovery for type II diabetes and obesity. Curr Drug
Targets. 19:551–575. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kostrzewa T, Przychodzen P,
Gorska-Ponikowska M and Kuban-Jankowska A: Curcumin and
Cinnamaldehyde as PTP1B inhibitors with antidiabetic and anticancer
potential. Anticancer Res. 39:745–749. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kostrzewa T, Wołosewicz K, Jamrozik M,
Drzeżdżon J, Siemińska J, Jacewicz D, Górska-Ponikowska M,
Kołaczkowski M, Łaźny R and Kuban-Jankowska A: Curcumin and its new
derivatives: Correlation between cytotoxicity against breast cancer
cell lines, degradation of PTP1B phosphatase and ROS generation.
Int J Mol Sci. 22:103682021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Radhakrishnan VM, Kojs P, Young G,
Ramalingam R, Jagadish B, Mash EA, Martinez JD, Ghishan FK and
Kiela PR: pTyr421 cortactin is overexpressed in colon cancer and is
dephosphorylated by curcumin: Involvement of non-receptor type 1
protein tyrosine phosphatase (PTPN1). PLoS One. 9:e857962014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen C, Cao M, Zhu S, Wang C, Liang F, Yan
L and Luo D: Discovery of a novel inhibitor of the protein tyrosine
phosphatase Shp2. Sci Rep. 5:176262015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Asmamaw MD, Shi XJ, Zhang LR and Liu HM: A
comprehensive review of SHP2 and its role in cancer. Cell Oncol
(Dordr). 45:729–753. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu JJ, Li Y, Chen WS, Liang Y, Wang G,
Zong M, Kaneko K, Xu R, Karin M and Feng GS: Shp2 deletion in
hepatocytes suppresses hepatocarcinogenesis driven by oncogenic
β-catenin, PIK3CA and MET. J Hepatol. 69:79–88. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han T, Xiang DM, Sun W, Liu N, Sun HL, Wen
W, Shen WF, Wang RY, Chen C, Wang X, et al: PTPN11/Shp2
overexpression enhances liver cancer progression and predicts poor
prognosis of patients. J Hepatol. 63:651–660. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yue X, Han T, Hao W, Wang M and Fu Y: SHP2
knockdown ameliorates liver insulin resistance by activating IRS-2
phosphorylation through the AKT and ERK1/2 signaling pathways. FEBS
Open Bio. 10:2578–2587. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiang D, Cheng Z, Liu H, Wang X, Han T,
Sun W, Li X, Yang W, Chen C, Xia M, et al: Shp2 promotes liver
cancer stem cell expansion by augmenting β-catenin signaling and
predicts chemotherapeutic response of patients. Hepatology.
65:1566–1580. 2017. View Article : Google Scholar : PubMed/NCBI
|