Introduction
Glioma is the most common intracranial tumor of the
central nervous system (CNS) in adults, with >300,000 new cases
diagnosed worldwide each year, causing ~2.5% of all cancer-related
deaths (1). Historically, glioma
was considered to originate from the differentiated astrocytic and
oligodendrocytic components of the CNS (2). Based on the World Health Organization
(WHO) classification of CNS tumors, glioma is divided into primary
and secondary glioma, graded as WHO 1–4, and includes astrocytoma,
oligodendroglioma, glioblastoma (GBM) and other subtypes (3). Although a series of molecular
parameters have been incorporated in the classification, including
IDH1/2 mutation status, CDKN2A/B homozygosity,
EGFR amplification and +7/-10 chromosome alteration
(4–6), the grading and histological subtyping
of glioma is frequently challenging for pathologists. In addition,
due to its aggressive behavior and high recurrence rate, the 5-year
survival rate of patients with glioma is currently still low, at
<5% (7). Therefore, effective
biomarkers for accurate diagnosis and prognostic evaluation, and
therapeutic targets are urgently required. Serine and arginine rich
splicing factor 1 (SRSF1; also called SF2/ASF) was the first
defined alternative splicing protein, which mainly functions in RNA
splicing, and contains two N-terminal RNA recognition motifs and a
short C-terminal RS domain (8–11).
Previous studies have reported that SRSF1 is dysregulated in human
cancer. SRSF1 has been shown to promote colon cancer development by
activating DBF4B exon 6 splicing (12). Furthermore, SRSF1 can promote
mammary epithelial cell transformation by specifically regulating
the splicing of key targets downstream of mTOR and/or is
functionally linked to MYC (13).
SRSF1 can also inhibit autophagy through regulating Bcl-x splicing
and interacting with PIK3C3 in lung cancer (14). In addition to regulating target
genes, Das et al (15)
demonstrated that the deubiquitinating enzymes ubiquitin-specific
peptidase (USP)15 and USP4 regulate alternative splicing of SRSF1,
resulting in isoform-specific functions in lung cancer cells. The
METTL3 stabilized long non-coding RNA SNHG7 has also been reported
to accelerate tumor progression via the SRSF1/c-Myc axis in
prostate cancer (16). However, the
possible functions of SRSF1 in glioma, which may contribute to
enhanced malignancy, remain largely unknown.
It has been reported that SRSF1 is highly expressed
in GBM and oligodendroglioma (17).
However, to the best of our knowledge, whether SRSF1 expression is
associated with the histopathological subtype of glioma, WHO grade
and glioma-associated molecular characteristics remains unknown. To
address these issues, the present study aimed to investigate the
expression of SRSF1 in pilocytic astrocytoma, astrocytoma,
oligodendroglioma and GBM. The present study also sought to examine
SRSF1 expression in different grades of glioma to explore whether
immunohistochemistry (IHC) of this marker could be valuable as a
surrogate for the distinction of WHO grade. Moreover, SRSF1
immunoreactivity was assessed in IDH-mutant and
1p/19q co-deletion glioma. If the results confirmed the
relevance of SRSF1 with molecular characteristics, SRSF1 IHC has
the potential to enable pathologists to take advantage of the
accessibility and relatively low cost of IHC without the need for
molecular testing.
Materials and methods
Data acquisition and specimen
collection
The RNA-sequencing data and corresponding clinical
data of patients with glioma (n=224) were downloaded from the
Chinese Glioma Genome Atlas (CGGA) (http://www.cgga.org.cn). In addition,
paraffin-embedded glioma tissues (WHO grade 1–4; n=70) were
collected from The Third Affiliated Hospital of Kunming Medical
University (Yunnan Cancer Hospital; Kunming, China). The collected
clinical data included age, sex, tumor sites, grade, IDH
mutation status, 1p/19q codeletion status, histological type
and overall survival time from CGGA and clinical patients. All 70
patients with glioma were recruited between March 2011 and December
2021, and were followed up from the date of surgery until the date
of analysis, with the follow-up time ranging between 3 and 120
months. The study was approved by the Ethics Committee of The Third
Affiliated Hospital of Kunming Medical University (Yunnan Cancer
Hospital; approval no. KYLX2022090) and experiments were undertaken
with the understanding and written informed consent of all the
patients. The study conformed with The Declaration of
Helsinki).
Hematoxylin and eosin (H&E) and
IHC
H&E staining was used to observe morphology.
After incubation at 60°C for 3 h, 4-µm paraffin-embedded slices
were dewaxed in xylene and then placed successively in high to low
concentrations of alcohol to hydrate. Subsequently, the tissues
were stained with hematoxylin for 7 min at room temperature and
rinsed with water. After differentiation in hydrochloric alcohol,
until blue, the slices were stained with eosin for 1 min,
dehydrated in 70, 80, 95 and 100% ethanol for 1 min each, followed
by 100% ethanol I and 100% ethanol II for 2 min, and then the
tissues were cleared with xylene I and II for 8 min each. After air
drying, moderate neutral balsam was added quickly. Finally, slices
were covered with coverslips and were observed under a light
microscope (Leica Microsystems, Inc.).
IHC was performed to detect SRSF1 expression, Ki-67
index and IDH1 R132 expression in paraffin-embedded sections of
human WHO grade 1–4 glioma and normal tissues from other benign
lesions. After baking at 60°C for 3 h, the slices were dewaxed in
xylene, then hydrated with gradient alcohol and placed in a
solution of sodium citrate at pH 6.0, followed by high-pressure
heat to repair the antigens. After sealing the non-specific binding
site with 10% goat serum (Fuzhou MaixinBiotech Co., Ltd.) for 30
min at room temperature, the slices were incubated with the primary
rabbit polyclonal anti-SRSF1 (1:300; Santa Cruz Biotechnology,
Inc.), anti-Ki-67 (ready-to-use; Fuzhou Maixin Biotech Co., Ltd.)
and anti-IDH1 R132 (ready-to-use; Fuzhou Maixin Biotech Co., Ltd.)
antibodies for 60 min and secondary antibody for 30 min. DAB
(Fuzhou Maixin Biotech Co., Ltd.) was then added for 5 min.
Finally, the samples were restained with hematoxylin for 1 min.
Staining was examined under a DM1000 microscope (Leica
Microsystems, Inc.) to obtain IHC scores. The brown staining of the
cell nuclei was interpreted as positive SRSF1 and Ki-67 staining,
and staining of the cell cytoplasm was considered positive IDH1
R132 staining. The immunoreactive score (IRS) was calculated by
multiplying the staining intensity by the percentage of positive
cells, as previously reported (18). The staining intensity was scored as
follows: 0 (negative staining), 1 (weak staining), 2 (moderate
staining) and 3 (strong staining). The percentage of cells that
were positive was scored as follows: 0 (<5%), 1 (5–30%), 2
(31–50%), 3 (51–75%) and 4 (>75%). Low and high expression of
SRSF1 were defined as IRS <6 and IRS ≥6, respectively.
Cell culture and stable
transduction
The human U87MG cell line was purchased from Jinyuan
Biotechnology Co., Ltd., which was glioblastoma of unknown origin
and has been authenticated using STR profiling. The U251 cell line
was kindly provided by Mr. Qian Yao (Yunnan Cancer Hospital). All
cells were cultured in DMEM supplemented with 10% FBS (both from
Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified
incubator containing 5% CO2. The negative control
overexpression (LV-vector), SRSF1 overexpression (LV-SRSF1), short
hairpin (sh)RNA negative control (sh-NC) and shRNA targeting SRSF1
(sh1-SRSF1, sh2-SRSF1 and sh3-SRSF1) lentiviruses were synthesized
by Beijing Qingke Biotechnology Co., Ltd. The overexpression and
knockdown lentiviruses were constructed using the
pCDH-CMV-MCS-EF1-copGFP-T2A-Puro and pLVX-ShRNA2-Puro lentiviral
vectors, respectively, and were packaged in 293T cells (puromycin
resistance) by Qingke Biotechnology Co., Ltd. The 2nd generation
system was used and transfected with 2.5 µg lentiviral plasmid.
Lentiviral particles were collected and U87MG and U251 cells were
subsequently transduced with the LV-vector, LV-SRSF1, sh-NC,
sh1-SRSF1, sh2-SRSF1 and sh3 SRSF1 lentiviruses for 48 h at a
multiplicity of infection of 20. After transfection for 48 h, the
stably transduced cells were selected with 2.5 µg/ml puromycin for
5 days. A total of 0.5 µg/ml puromycin was used for maintenance.
The cells were then collected and used for the subsequent
experiments.
MTT assay
U87 and U251 cells were cultured in 96-well plates
at a density of 2×103 cells/well in 150 µl complete
medium and incubated for 0, 1, 3 and 5 days. Subsequently, 20 µl
MTT reagent (5 mg/ml; BestBio) was added to each well for 4 h at
37°C, then dissolved in 150 µl DMSO with agitation for 10 min at
room temperature. Absorbance was measured at 490 nm.
Colony formation assay
SRSF1 overexpression and knockdown cells were seeded
in 6-well plates at 0.5×103 cells/well and cultured for
2 weeks. Colonies were fixed with 4% paraformaldehyde (Fuzhou
Maixin Biotech Co., Ltd.) at room temperature and stained with 2 ml
0.1% crystal violet (MilliporeSigma) at room temperature for ≥2 h
and images were captured. A colony was defined as a group of >50
cells. The number of colonies was first counted manually under a
light microscope and then reconfirmed using ImageJ analysis
software (National Institutes of Health).
Wound healing assay
Transduced cells (8×104/well) were seeded
in a 6-well plate and scratched using a sterile pipette tip once
confluence reached >90%. The cell culture was replaced in
serum-free medium at room temperature. Cells were observed under a
light microscope (Olympus Corporation) at 0, 24 and 48 h, with
images captured under a light microscope (Olympus Corporation) at 0
and 48 h after scratching and quantified using ImageJ analysis
software (National Institutes of Health).
Transwell assay
Transduced U87 and U251 cells were seeded in at
5×104 cells/well in 200 µl serum-free medium into the
upper chamber (Corning Inc.) of a Transwell plate coated with
Matrigel (MilliporeSigma) on ice at 37°C for 2 h, whereas 800 µl
medium containing 20% FBS was added to the bottom chamber. After 48
h of incubation at 37°C, the invasive cells were stained with 1 ml
0.1% crystal violet for 4 h at room temperature. The images were
captured under a light microscope (Olympus Corporation) for data
analysis.
Western blot analysis
U87 and U251 cells were collected by centrifugation
at 800 × g for 10 min at 4°C, and lysed in RIPA buffer (Beijing
Solarbio Science & Technology Co., Ltd.) at 4°C for 30 min.
Samples were centrifugation at 12,000 × g for 5 min at 4°C and the
supernatants were collected. Total protein was extracted from
transduced U87 and U251 cells and detected using the BCA protein
assay (BestBio). Proteins (30 µg) were separated by SDS-PAGE on 10%
gels and were transferred onto PVDF membranes. After blocking with
5% non-fat milk for 1 h at room temperature, the membranes were
incubated with rabbit anti-SRSF1 (1:1,000; cat. no. SC33652; Santa
Cruz Biotechnology, Inc.) overnight at 4°C followed by incubation
with anti-Rabbit IgG-HRP (1:10,000; cat. no. SA00001-2;
ProteinTech) for 1 h at room temperature. Rabbit anti-β-tubulin
(1:1,000; cat. no. SC5274; Santa Cruz Biotechnology, Inc.) was used
as the loading control. The protein bands were visualized using ECL
(cat. no. WBKLS0100; MilliporeSigma). The bands in western blot
images were semi-quantified using ImageJ analysis software (version
1.8.0; National Institutes of Health).
Statistical analysis
All experiments were repeated three times and the
data are presented as the mean ± SD. Data were analyzed using SPSS
26.0 software (IBM Corp.). Unpaired Student's t-test was used to
compare the means of two groups of data. The associations between
SRSF1 expression and pathological characteristics were analyzed by
the χ2 test and Fisher's exact test. IHC scores were
compared between two groups using the Mann-Whitney U test, and
among multiple groups using the Kruskal-Wallis and Dunn's test. The
statistical significance among three or more groups was analyzed by
one-way analysis of variance, and the LSD (equal variances) or
Tamhane's T2 (unequal variances) test were used for post-hoc
pairwise comparisons after the homogeneity of variance test.
Spearman correlation analysis was used to detect the correlation
between two variables. Receiver operating characteristic (ROC)
curves were used to estimate the sensitivity and specificity of
SRSF1 for glioma grading, and the area under the curve (AUC) was
calculated. Survival analysis was analyzed by the Kaplan-Meier
method, with the log-rank test applied for comparisons. P<0.05
was considered to indicate a statistically significant
difference.
Results
Association between SRSF1 and
clinicopathological characteristics in the CGGA database
To preliminarily probe the relationship between the
expression of SRSF1 and primary glioma, data (mRNAseq_325) from the
CGGA database were assessed. The clinicopathological and molecular
features are summarized in Table I.
A total of 224 primary glioma cases were analyzed, including 83
astrocytoma, 56 oligodendroglioma and 85 GBM cases. Significant
differences between SRSF1 expression (by PCR) and age were
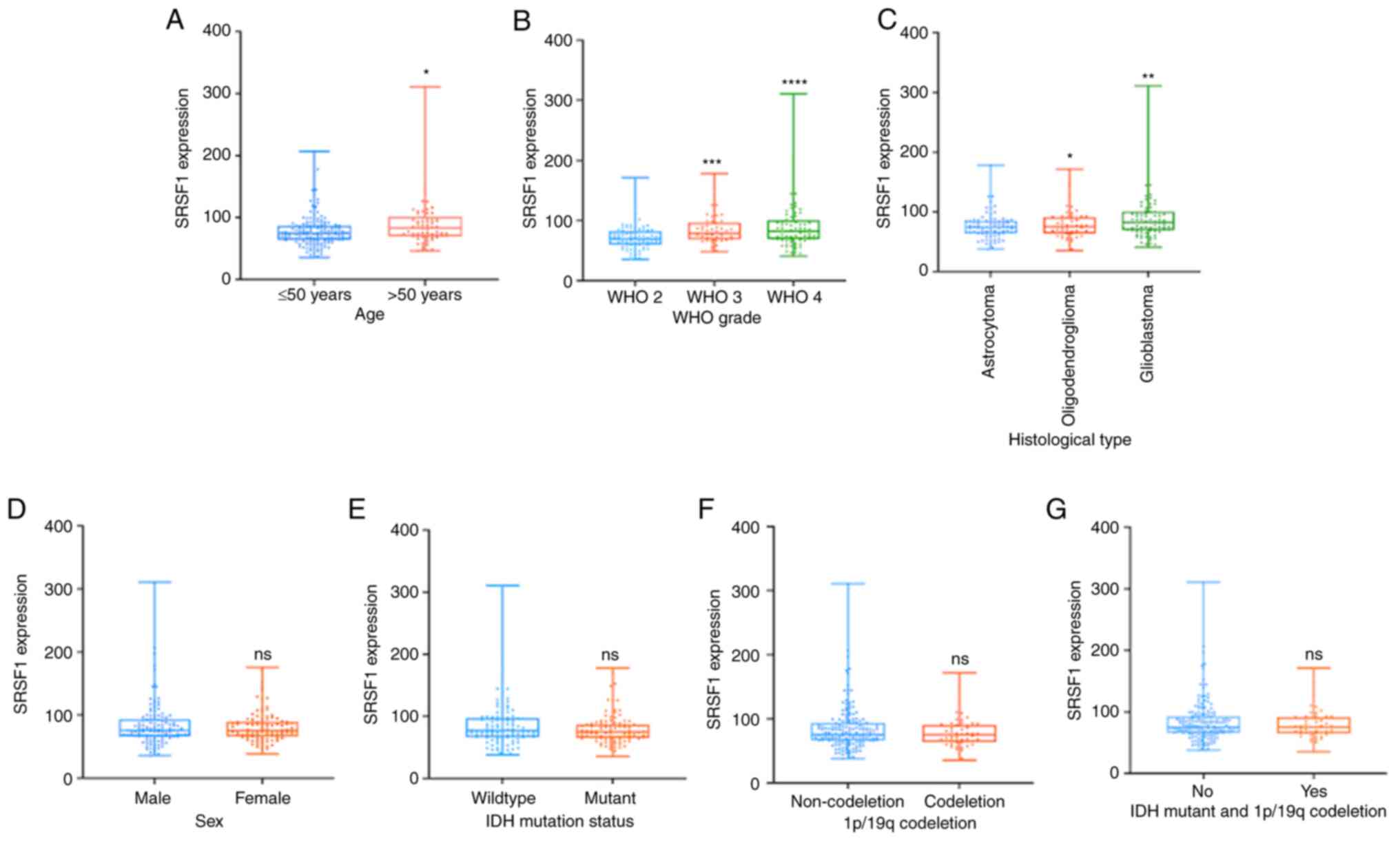
determined (Fig. 1A); however, no
statistically significant association between SRSF1 expression and
sex was observed (Fig. 1B).
Notably, the expression levels of SRSF1 were highly associated with
WHO grade. A total of 70/90 WHO 2 glioma cases (77.8%) showed low
expression of SRSF1, and the remaining 20/90 cases (22.2%) showed
high expression of SRSF1, which indicated that decreased expression
of SRSF1 was more frequent in WHO 2 glioma. The percentage of
patients with high SRSF1 level was higher in GBM (49.4%) than that
in WHO grade 3 (27.1%) and 2 (23.5%) glioma (Fig. 1C), as verified in clinical specimens
in Table II. In terms of
histological subtype, SRSF1 was diffusely strongly expressed in GBM
cases (49.4%), astrocytoma cases (28.2%) and oligodendroglioma
cases (22.4%), which indicated that the immunopositivity of SRSF1
was associated with GBM (Fig. 1D).
Additionally, the present study further investigated the
association between SRSF1 expression and molecular features in the
CGGA database, which showed that neither 1p/19q
co-deletion or IDH mutations were associated with SRSF1
expression (Fig. 1E and F). The
clinicopathological and molecular features are summarized in
Table I.
 | Table I.Association between SRSF1 expression
and the clinicopathological characteristics of patients with glioma
in the Chinese Glioma Genome Atlas cohort. |
Table I.
Association between SRSF1 expression
and the clinicopathological characteristics of patients with glioma
in the Chinese Glioma Genome Atlas cohort.
|
| SRSF1 expression, n
(%) |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Low (≤82.7) | High (>82.7) | Total | χ2 | P-value |
|---|
| Age, years |
|
|
| 5.190 | 0.022a |
|
≤50 | 105 (75.5) | 52 (61.2) | 157 |
|
|
|
>50 | 34 (24.5) | 33 (38.8) | 67 |
|
|
| Sex |
|
|
| 0.606 | 0.436 |
|
Male | 50 (36.0) | 35 (41.2) | 85 |
|
|
|
Female | 89 (64.0) | 50 (58.8) | 139 |
|
|
| Histological
type |
|
|
| 8.004 | 0.018a |
|
Astrocytoma | 59 (42.4) | 24 (28.2) | 83 |
|
|
|
Oligodendroglioma | 37 (26.6) | 19 (22.4) | 56 |
|
|
|
Glioblastoma | 43 (31.0) | 42 (49.4) | 85 |
|
|
| WHO grade |
|
|
| 15.88 |
<0.001b |
| WHO
2 | 70 (50.4) | 20 (23.5) | 90 |
|
|
| WHO
3 | 26 (18.7) | 23 (27.1) | 49 |
|
|
| WHO
4 | 43 (30.9) | 42 (49.4) | 85 |
|
|
| IDH |
|
|
| 1.989 | 0.158 |
|
Mutant | 74 (53.2) | 37 (43.5) | 111 |
|
|
|
Wildtype | 65 (46.8) | 48 (56.5) | 113 |
|
|
| 1p/19q |
|
|
| 0.512 | 0.474 |
|
Codeletion | 37 (26.6) | 19 (22.4) | 56 |
|
|
|
Non-codeletion | 102 (73.4) | 66 (77.6) | 168 |
|
|
| IDH mutant and
1p/19q codeletion |
|
|
| 2.382 | 0.123 |
|
Yes | 37 (26.6) | 15 (17.6) | 52 |
|
|
| No | 102 (73.4) | 70 (82.4) | 172 |
|
|
 | Table II.Association between SRSF1 expression
and clinicopathological characteristics of the 70 patients with
glioma. |
Table II.
Association between SRSF1 expression
and clinicopathological characteristics of the 70 patients with
glioma.
|
| SRSF1 IRS, n
(%) |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Low | High | Total | χ2 | P-value |
|---|
| Age, years |
|
|
| 8.750 | 0.003a |
|
≤50 | 12 (40.0) | 30 (75.0) | 42 |
|
|
|
>50 | 18 (60.0) | 10 (25.0) | 28 |
|
|
| Sex |
|
|
| 0.491 | 0.484 |
|
Male | 19 (63.3) | 22 (55.0) | 41 |
|
|
|
Female | 11 (36.7) | 18 (45.0) | 29 |
|
|
| Predominant
side |
|
|
| 2.619 | 0.269 |
|
Left | 12 (40.0) | 23 (57.5) | 35 |
|
|
|
Right | 13 (43.3) | 14 (35.0) | 27 |
|
|
|
Middle | 5 (16.7) | 3 (7.5) | 8 |
|
|
| Predominant
site |
|
|
| 5.417 | 0.168 |
| Frontal
lobe | 16 (53.3) | 23 (57.5) | 39 |
|
|
|
Temporal lobe | 4 (13.3) | 9 (22.5) | 13 |
|
|
|
Parietal lobe | 2 (6.7) | 4 (10.0) | 6 |
|
|
|
Others | 8 (26.7) | 4 (10.0) | 12 |
|
|
| WHO grade |
|
|
| 41.864 |
<0.001b |
| WHO
1 | 9 (30.0) | 1 (2.5) | 10 |
|
|
| WHO
2 | 18 (60.0) | 2 (5.0) | 20 |
|
|
| WHO
3 | 3 (10.0) | 17 (42.5) | 20 |
|
|
| WHO
4 | 0 (0.0) | 20 (50.0) | 20 |
|
|
| Ki-67 index |
|
|
| 24.083 |
<0.001b |
|
≤10% | 26 (86.7) | 11 (27.5) | 37 |
|
|
|
>10% | 4 (13.3) | 29 (72.5) | 33 |
|
|
| IDH1 R132 |
|
|
| 0.049 | 0.824 |
|
Mutant | 21 (70.0) | 27 (67.5) | 48 |
|
|
|
Wildtype | 9 (30.0) | 13 (32.5) | 22 |
|
|
Association between SRSF1 and
clinicopathological characteristics in clinical cases
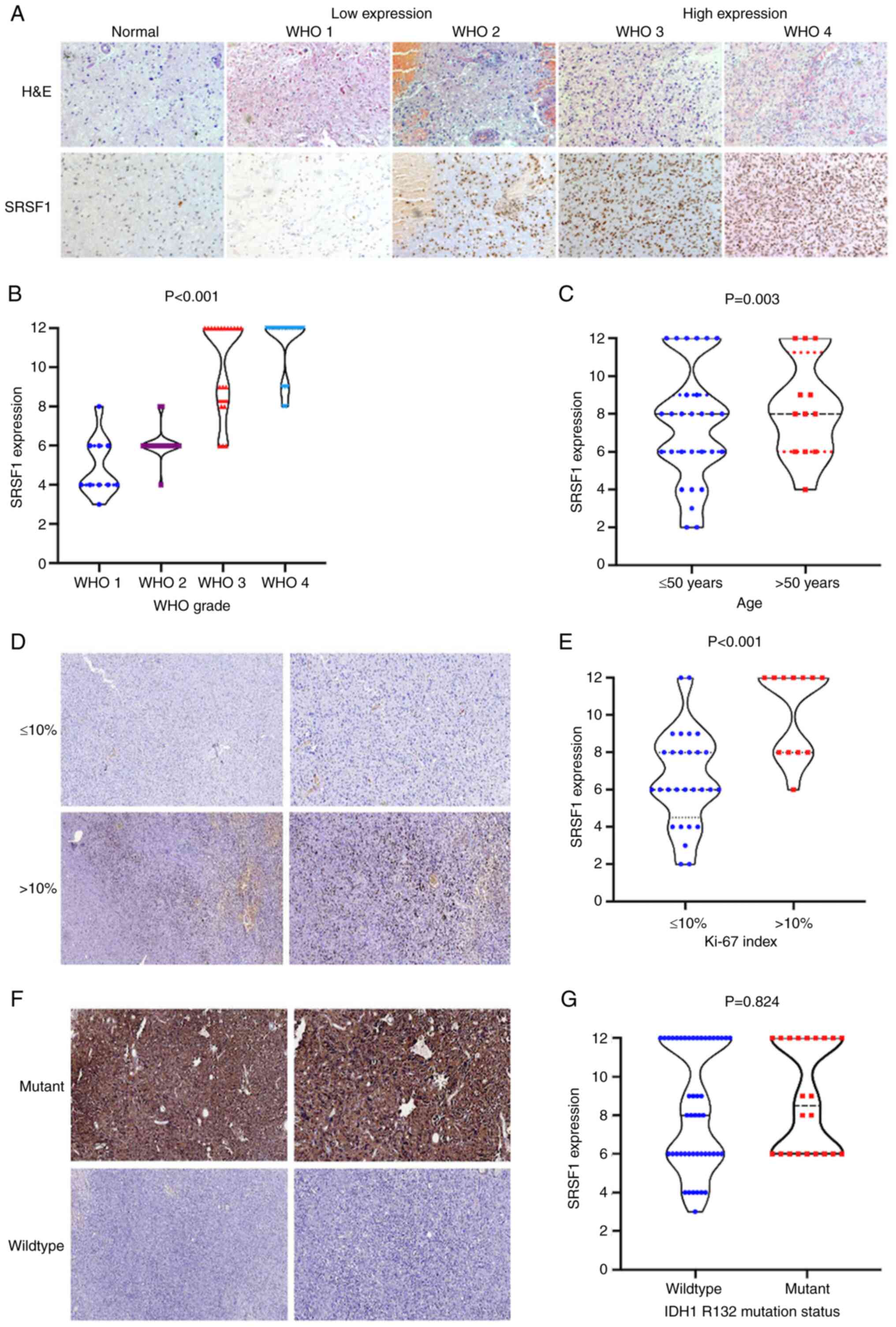
Consistent with the before mentioned findings, the
present study further analyzed the relationship between the
immunohistochemical expression of SRSF1 and the clinical features
of 70 primary glioma specimens (Table
II). The cohort included 41 men and 29 women with a median age
of 45 years (range, 2–81 years). High SRSF1 expression was more
frequently observed in younger patients (75%), and low expression
was more frequent in older patients (60%) (Fig. 2C). Regarding the predominant side,
50% of the cases were on the left, 39% were on the right, and the
remaining cases were in the middle. The tumors were mainly located
in the frontal lobe in 39/70 cases (56%), in the temporal lobe in
13/70 cases (19%) and the fewest cases were in the parietal lobe.
Notably, there were no significant associations between SRSF1
expression and the following clinicopathological variables: Sex,
predominant side and site. Furthermore, the present study revealed
that SRSF1 exhibited diffusely strong immunoreactivity in WHO grade
3 and 4 glioma (HGG) samples compared with that in WHO grade 1 and
2 glioma (LGG) and normal tissue samples (Fig. 2A). Among the 30 WHO 1 and WHO 2
(LGG) cases, 27 (90%) exhibited low IHC levels of SRSF1, and only
three (10%) exhibited high levels, whereas no detectable
immunostaining was observed in all pilocytic astrocytoma cases.
Consistently, all GBM samples presented strong and diffuse
immunostaining, and 85% of WHO grade 3 astrocytoma cases were
moderately immunostained, showing a weak immunopositivity for SRSF1
in 15% of HGG cases (Fig. 2B).
These data are consistent with those of the CGGA cohort. Notably,
Spearman analysis showed a positive correlation between SRSF1
immunoexpression and WHO grade (data not shown), indicating that
its expression gradually increased as WHO grade progressed. In
addition, 29 (72.5%) cases with an increased Ki-67 index exhibited
significantly higher SRSF1 expression levels than cases with a low
Ki-67 index, whereas 26 (86.7%) cases with a low Ki-67 index showed
low SRSF1 expression (Fig. 2D). As
revealed in Fig. 2E, high
expression of SRSF1 was closely asociated with Ki-67 index. Among
the 10 pilocytic astrocytoma cases, immunoexpression of IDH1 R132
was completely negative. With regard to IDH mutations, IDH1
R132 showed strong staining, but negative immunoexpression for the
IDH wild-type (Fig. 2F).
Furthermore, no statistically significant association was detected
between SRSF1 expression and IDH1 R132 immunoreactivity (Fig. 2G). The clinicopathological and
molecular features of the clinical cohort are summarized in
Table II
Potential use of SRSF1 for glioma
grading
From the ROC curve, the specificity of SRSF1 for GBM
was revealed to be 40%, the sensitivity was 100%, and the mean AUC
value was 0.8 (95% CI 0.7-0.9). The specificity of SRSF1 for WHO
grade 3 astrocytoma was 48%, the sensitivity was 85%, and the mean
AUC value was 0.701 (95% CI 0.57-0.831) (data not shown). These
findings indicated that SRSF1 performed well in distinguishing GBM
and WHO grade 3 astrocytoma from WHO grade 2 astrocytoma.
Association between SRSF1 expression
and survival
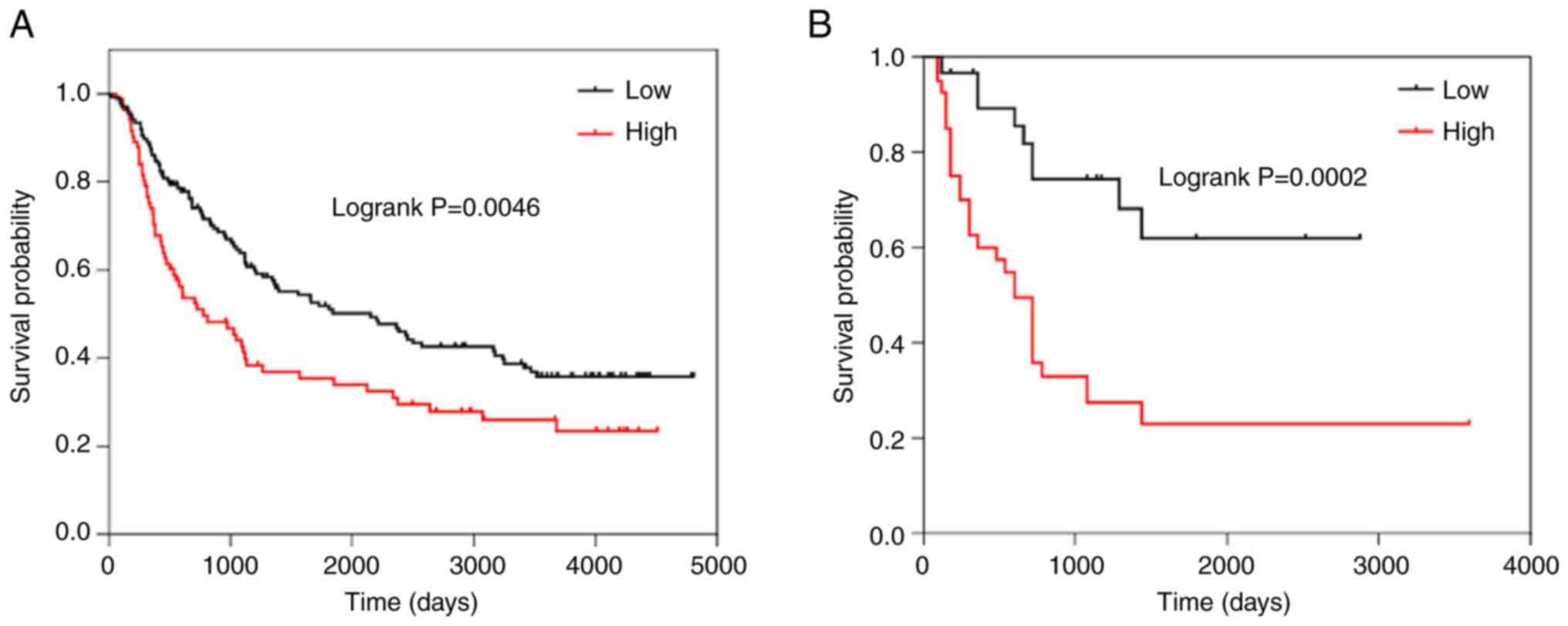
The Kaplan-Meier survival analysis revealed that
patients with HGG and high expression of SRSF1 had shorter OS times
than those with low expression of SRSF1 in the CGGA dataset
(Fig. 3A), which was further
confirmed in the 70 clinical glioma specimens (Fig. 3B). These findings indicated that
SRSF1 may be a detrimental biomarker for the survival of
gliomas.
Biological role of SRSF1 in vitro
To investigate the role of SRSF1 in the biological
processes of glioma, the present study knocked down SRSF1 using
individual-specific shRNAs (sh1-SRSF1, sh2-SRSF1 or sh3-SRSF1),
with sh-NC as the negative control (Fig. 4A and B), and overexpressed SRSF1 in
U87MG and U251 cells using LV-SRSF1 lentivirus, with LV-vector as
the negative control (Fig. 4C).
Western blot analysis confirmed efficient knockdown of SRSF1; SRSF1
expression was decreased by >70% in sh3-SRSF1-transduced cells
compared with that in the sh-NC group, which indicated that
sh3-SRSF1 had a higher interference efficiency compared with
sh1-SRSF1 and sh2-SRSF1. Consequently, only sh3-SRSF1 was used in
the subsequent experiments. Compared with LV-vector, overexpression
of SRSF1 significantly promoted cell proliferation, as determined
by MTT assay (Fig. 4D), whereas the
knockdown of SRSF1 severely inhibited the proliferation of cells,
with the largest difference detected after 3 days of culture
(Fig. 4E). Moreover, overexpression
of SRSF1 enhanced colony formation in comparison with the LV-vector
group (Fig. 4F), whereas knockdown
of SRSF1 reduced colony formation (Fig.
4G). In the U87MG and U251 cell lines, the wound healing assay
revealed that overexpression of SRSF1 significantly enhanced cell
migration, which was markedly suppressed by the knockdown of SRSF1
(Fig. 4H-J). Furthermore,
overexpression of SRSF1 significantly enhanced cell invasion, as
determined by Transwell assay (Fig.
4K). Conversely, knockdown of SRSF1 significantly suppressed
the invasion of U87MG and U251 cells (Fig. 4L). These results indicated that
SRSF1 promoted glioma progression and may act as an inducer of
glioma.
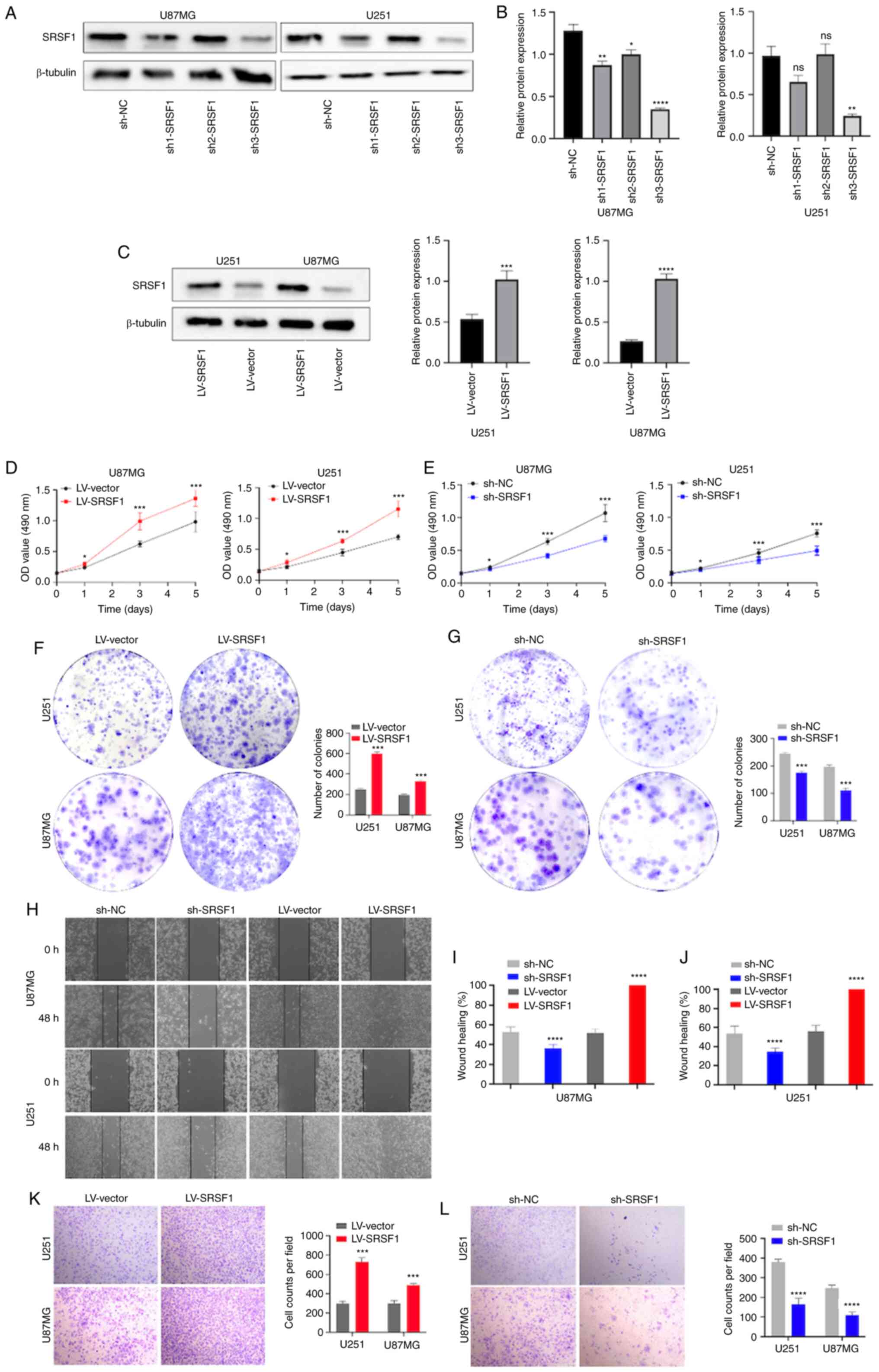 | Figure 4.SRSF1 increases the proliferation,
invasion and migration of U87MG and U251 cells. (A) Knockdown and
overexpression efficiency post-transduction with sh-NC, sh1-SRSF1,
sh2-SRSF1 and sh3-SRSF1, or LV-SRSF1 and LV-vector, as determined
by western blot analysis. Semi-quantification of SRSF1 protein
expression in (B) U87MG and (C) U251 cells. OD value at 490 nm of
the MTT assay in U251 and U87MG cells with stable (D)
overexpression or (E) knockdown of SRSF1. Results of the colony
formation assay in U251 and U87MG cells with stable (F)
overexpression or (G) knockdown of SRSF1. (H) Images of the wound
healing assay in U251 and U87MG cells with stable overexpression or
knockdown of SRSF1 (magnification, ×100). Semi-quantification of
wound healing assay in (I) U87MG and (J) U251 cells. Results of the
Transwell invasion assay in U251 and U87MG cells with stable (K)
overexpression (magnification, ×200) or (L) knockdown of SRSF1
(magnification, ×200). *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. LV, lentivirus; NC, negative control; ns, not
significant; sh, short hairpin; SRSF1, serine and arginine rich
splicing factor 1. |
Discussion
Accurate grading and histopathological subtyping are
critical for the treatment and prognosis of primary glioma;
however, no specific hallmarks are available to assist in the
grading and prognostic evaluation of glioma. The present study
hypothesized that it may be helpful to explore IHC biomarkers for
the solution of these issues.
Given prior evidence of SRSF1 expression, which is
expected to be highly expressed in GBM but at low levels in normal
tissues, it was hypothesized that IHC for SRSF1 may be useful for
distinguishing GBM and WHO grade 3 astrocytoma (HGG) from LGG.
Based on the CGGA database and immunohistochemical analysis of
clinical tissues, it was revealed that SRSF1 was upregulated in
glioma specimens, particularly in most GBM and WHO grade 3
astrocytoma cases. Notably, the present study further confirmed
that SRSF1 was immunostained with an increasing gradient in WHO 2–4
astrocytic tumors, showing homogeneously strong immunopositivity
for HGG and moderate immunopositivity for WHO grade 2 astrocytoma.
In GBM and WHO grade 3 astrocytoma, the present study confirmed
that immunohistochemical testing for SRSF1 provided the best
diagnostic values for distinguishing them from WHO grade 2
astrocytoma (40 and 48% specificity; 100 and 85% sensitivity,
respectively). In addition, SRSF1 expression was revealed to be
associated with the Ki-67 index, which is a significant parameter
for the increased grade of glioma (19). The present study revealed that the
increased ratio of the Ki-67 index paralleled the high levels of
SRSF1 expression. These results suggested a promising diagnostic
ability for the SRSF1 protein in distinguishing HGG from LGG, the
diagnosis of which may be challenging when tumors present no
typical histological features or, in particular, molecular
parameters are unavailable (20).
Immunohistochemical analysis of SRSF1 may serve as an objective
hallmark of glioma grading.
The present study also demonstrated that SRSF1
levels were associated with histopathological subtypes in the CGGA
cohort, consistent with the findings of Broggi et al
(21), which reported that SRSF1
was lower in astrocytoma and more frequently expressed in
anaplastic oligodendroglioma. Similarly, the present study
identified a high frequency of SRSF1 expression in GBM and WHO
grade 3 astrocytoma; however, the frequency of expression was low
in WHO grade 2 astrocytoma and oligodendroglioma. In this regard,
it is reasonable to propose that the potential application of SRSF1
in the distinction of GBM from low-grade astrocytoma and
oligodendroglioma. However, SRSF1 expression is not expected to be
useful in the differential diagnosis of astrocytoma and
oligodendroglioma. The association between SRSF1 and
oligodendroglioma remains to be determined. In contrast to
immunostains for SRSF1 in astrocytoma and oligodendroglioma,
pilocytic astrocytoma lacked SRSF1-positive cells; therefore, for
practical diagnostic purposes, in cases where the microscopic
features, clinical setting and imaging data are unclear, pure SRSF1
negativity may be considered supportive evidence for pilocytic
astrocytoma. The diagnostic use of the SRSF1 protein in
distinguishing adult diffuse astrocytoma from ependymoma and
pilocytic astrocytoma has also been reported (21). However, SRSF1 is not a reliable
marker for distinguishing sub-ependymal giant cell astrocytoma from
pleomorphic xanthoastrocytoma (21). On the basis of preliminary
observations comparing SRSF1 expression in limited
histopathological subtypes, larger-scale subtypes of CNS tumors are
required to elucidate the clinical utility of SRSF1
immunoreactivity for diagnosis.
Given the high frequency of SRSF1 expression in GBM
and WHO grade 3 astrocytoma, it was hypothesized that SRSF1 IHC may
be associated with glioma-related molecular markers, such as
IDH mutations and 1p/19q co-deletion status.
However, SRSF1 IHC expression was not revealed to be associated
with 1p/19q co-deletion and IDH mutations in
the present study. Since SRSF1 regulates pivotal alternative
splicing events of some tumor-related genes, further prospective
studies incorporating all clinically relevant molecular markers are
needed to evaluate for these molecular subgroups.
Beyond the significance for auxiliary diagnosis,
Kaplan-Meier survival analysis demonstrated that patients with HGG
and high SRSF1 expression had shorter OS times than those with low
SRSF1 expression in the CGGA datasets and among the clinical cases.
A preliminary study previously reported that SRSF1 is a predictive
factor for basal cell carcinoma recurrence (18). It has also been demonstrated that
SRSF1 is associated with various factors affecting the prognosis of
ovarian cancer (22),
hepatocellular carcinoma (23) and
hematological malignancies (24).
Moreover, the Ki-67 index, which was revealed to be positively
associated with SRSF1, predicted a poorer prognosis in HGG cases,
as confirmed in the present study (25). Thus, it was hypothesized that SRSF1
may serve as a prognostic factor for survival in glioma. Close
follow-up is essential for tumors with high levels of this
protein.
Based on the present clinical findings, SRSF1
appears to serve a crucial tumor-promoting role in primary glioma.
It has been reported that SRSF1 is downregulated by the
circ-PABPN1/microRNA-638 axis to suppress colorectal cancer
development in vitro and in vivo (26). Barbagallo et al (27) also reported that CircSMARCA5
regulates VEGFA mRNA splicing through SRSF1 in GBM. However, the
role of SRSF1 in CNS tumors has not been sufficiently elucidated.
Therefore, the present study investigated the biological functions
of SRSF1 in the U87MG and U251 cell lines. MTT and colony formation
assays showed that overexpression of SRSF1 promoted glioma cell
proliferation, which was suppressed after knockdown of SRSF1,
indicating that SRSF1 may be associated with the malignant
proliferation of glioma. Moreover, wound healing data confirmed
that overexpression of SRSF1 significantly enhanced cell migration,
suggesting that SRSF1 may promote glioma progression. The results
from a Transwell assay showed that cells stably overexpressing
SRSF1 exhibited markedly increased invasion, whereas invasion was
suppressed in cells with SRSF1 knockdown. Consistent with the
present study, Zhou et al (17) verified that the knockdown of SRSF1
impaired cell survival and invasion in GBM cell lines (17). These results demonstrated that SRSF1
could have a role in glioma development, further confirming the
pro-tumor activity of SRSF1.
In conclusion, the present study revealed that SRSF1
is diffusely expressed in GBM and WHO grade 3 astrocytoma. As
strong and diffuse SRSF1 expression is rare in WHO grade 2
astrocytoma, immunohistochemical testing for high SRSF1 has
potential clinical value as an auxiliary approach for the
distinction of HGG from WHO grade 2 astrocytoma. Since pilocytic
astrocytoma exhibited absent immunoreactivity for SRSF1 compared
with astrocytoma and oligodendroglioma, the detection of negative
SRSF1 expression may be used as an auxiliary biomarker for
pilocytic astrocytoma. Furthermore, SRSF1 could be considered a
prognostic indicator and the present study indicated that SRSF1
serves an active role in promoting glioma progression. Some
limitations in the present study need to be fully considered.
First, upstream and downstream mechanisms have not been
sufficiently investigated. Second, the utility of a single protein
can be limited; therefore, it must be incorporated into a series of
biomarkers, and more clinical evidence is needed to confirm the
conclusions of this study in neuro-oncology practice.
Acknowledgements
The authors would like to thank Mr. Qian Yao [The
Third Affiliated Hospital of Kunming Medical University (Yunnan
Cancer Hospital)] for providing cell line and technical
support.
Funding
The present study was supported by the Yunnan Province Science
and Technology Research Fund (grant no. 202101AT070005), the Joint
Special Funds for the Department of Science and Technology of
Yunnan Province-Kunming Medical University (grant no.
202001AY070001-244) and the China Postdoctoral Science Foundation
(grant no. 2021M693842).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LY conceived the study. LY, GB, FR JY and KX
interpreted and analyzed the data. KX and JY prepared the
manuscript. KX and GB revised the manuscript for important
intellectual content. LY and JY supervised the study. LY, GB, FR,
JY and KX confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in line with the
principles of The Declaration of Helsinki. Approval was granted by
the Ethics Committee of The Third Affiliated Hospital of Kunming
Medical University (Yunnan Cancer Hospital) (Date 2022; approval
no. KYLX2022090). Written informed consent was obtained from all
individual participants or their parents/guardians included in the
study.
Patient consent for publication
The authors affirm that human research participants
provided informed oral consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanai N, Alvarez-Buylla A and Berger MS:
Neural stem cells and the origin of gliomas. N Engl J Med.
353:811–822. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Wesseling P, Brat DJ,
Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM,
Reifenberger G, et al: The 2021 WHO Classification of tumors of the
central nervous system: A summary. Neuro Oncol. 23:1231–1251. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sejda A, Grajkowska W, Trubicka J,
Szutowicz E, Wojdacz T, Kloc W and Iżycka-Świeszewska E: WHO CNS5
2021 classification of gliomas: A practical review and road signs
for diagnosing pathologists and proper patho-clinical and
neuro-oncological cooperation. Folia Neuropathol. 60:137–152. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eckel-Passow JE, Lachance DH, Molinaro AM,
Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML,
Smirnov IV, et al: Glioma Groups Based on 1p/19q, IDH, and TERT
promoter mutations in tumors. N Engl J Med. 372:2499–2508. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Louis DN, Wesseling P, Aldape K, Brat DJ,
Capper D, Cree IA, Eberhart C, Figarella-Branger D, Fouladi M,
Fuller GN, et al: cIMPACT-NOW update 6: New entity and diagnostic
principle recommendations of the cIMPACT-Utrecht meeting on future
CNS tumor classification and grading. Brain Pathol. 30:844–856.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davis ME: Epidemiology and overview of
gliomas. Semin Oncol Nurs. 34:420–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cáceres JF and Krainer AR: Functional
analysis of pre-mRNA splicing factor SF2/ASF structural domains.
EMBO J. 12:4715–4726. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zuo P and Manley JL: Functional domains of
the human splicing factor ASF/SF2. EMBO J. 12:4727–4737. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Das S and Krainer AR: Emerging functions
of SRSF1, splicing factor and oncoprotein, in RNA metabolism and
cancer. Mol Cancer Res. 12:1195–1204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paz S, Ritchie A, Mauer C and Caputi M:
The RNA binding protein SRSF1 is a master switch of gene expression
and regulation in the immune system. Cytokine Growth Factor Rev.
57:19–26. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Luo C, Shen L, Liu Y, Wang Q,
Zhang C, Guo R, Zhang Y, Xie Z, Wei N, et al: SRSF1 Prevents DNA
damage and promotes tumorigenesis through regulation of DBF4B
Pre-mRNA splicing. Cell Rep. 21:3406–3413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Anczuków O, Rosenberg AZ, Akerman M, Das
S, Zhan L, Karni R, Muthuswamy SK and Krainer AR: The splicing
factor SRSF1 regulates apoptosis and proliferation to promote
mammary epithelial cell transformation. Nat Struct Mol Biol.
19:220–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lv Y, Zhang W, Zhao J, Sun B, Qi Y, Ji H,
Chen C, Zhang J, Sheng J, Wang T, et al: SRSF1 inhibits autophagy
through regulating Bcl-x splicing and interacting with PIK3C3 in
lung cancer. Signal Transduct Target Ther. 6:1082021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Das T, Lee EY, You HJ, Kim EE and Song EJ:
USP15 and USP4 facilitate lung cancer cell proliferation by
regulating the alternative splicing of SRSF1. Cell Death Discov.
8:242022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu J, Yuan JF and Wang YZ:
METTL3-stabilized lncRNA SNHG7 accelerates glycolysis in prostate
cancer via SRSF1/c-Myc axis. Exp Cell Res. 416:1131492022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou X, Wang R, Li X, Yu L, Hua D, Sun C,
Shi C, Luo W, Rao C, Jiang Z, et al: Splicing factor SRSF1 promotes
gliomagenesis via oncogenic splice-switching of MYO1B. J Clin
Invest. 129:676–693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Broggi G, Barbagallo D, Lacarrubba F,
Verzì AE, Micali G, Purrello M and Caltabiano R: The
immunohistochemical expression of the serine and arginine-rich
splicing factor 1 (SRSF1) is a predictive factor of the recurrence
of basal cell carcinoma: A preliminary study on a series of 52
cases. Medicina (Kaunas). 58:1392022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brat DJ, Prayson RA, Ryken TC and Olson
JJ: Diagnosis of malignant glioma: Role of neuropathology. J
Neurooncol. 89:287–311. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weller M, van den Bent M, Preusser M, Le
Rhun E, Tonn JC, Minniti G, Bendszus M, Balana C, Chinot O, Dirven
L, et al: EANO guidelines on the diagnosis and treatment of diffuse
gliomas of adulthood. Nat Rev Clin Oncol. 18:170–186. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Broggi G, Salvatorelli L, Barbagallo D,
Certo F, Altieri R, Tirrò E, Massimino M, Vigneri P, Guadagno E,
Maugeri G, et al: Diagnostic utility of the immunohistochemical
expression of serine and arginine rich splicing factor 1 (SRSF1) in
the differential diagnosis of adult gliomas. Cancers (Basel).
13:20862021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Ni X, Yu Z, Wu S and Liu Z: CRNDE
inducing cisplatin resistance through SRSF1/TIA1 signaling pathway
in ovarian cancer. Pathol Res Pract. 235:1539572022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Han T, Li Z, Han H, Yin Y, Zhang B,
Zhang H and Li L: A Novel circRNA hsa_circRNA_002178 as a
diagnostic marker in hepatocellular carcinoma enhances cell
proliferation, invasion, and tumor growth by stabilizing SRSF1
expression. J Oncol. 2022:41840342022.PubMed/NCBI
|
|
24
|
Salifu SP and Doughan A: New clues to
prognostic biomarkers of four hematological malignancies. J Cancer.
13:2490–2503. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tejada S, Becerra-Castro MV, Nuñez-Cordoba
J and Díez-Valle R: Ki-67 proliferative activity in the tumor
margins as a robust prognosis factor in glioblastoma patients. J
Neurol Surg A Cent Eur Neurosurg. 82:53–58. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao A and Liu Y: Propofol suppresses
colorectal cancer development by the circ-PABPN1/miR-638/SRSF1
axis. Anal Biochem. 631:1143542021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barbagallo D, Caponnetto A, Brex D,
Mirabella F, Barbagallo C, Lauretta G, Morrone A, Certo F, Broggi
G, Caltabiano R, et al: CircSMARCA5 Regulates VEGFA mRNA splicing
and angiogenesis in glioblastoma multiforme through the binding of
SRSF1. Cancers (Basel). 11:1942019. View Article : Google Scholar : PubMed/NCBI
|