Introduction
Renal cell carcinoma (RCC) is one of the most common
malignancies of the urogenital system. In 2019, ~73,820 new cases
and 14,770 deaths in the United States were due to RCC (1–3). The
occurrence of RCC has been increasing at a rate of 2–4% per year
(4). Based on the histological
phenotype, RCC is categorized into four subcategories: Chromophobe,
clear cell, collecting duct and papillary, with clear cell RCC
(ccRCC) accounting for 80–90% of all RCC tumors (5). Attributable to the paucity of apparent
early symptoms, 30% of patients are diagnosed with advanced-stage
RCC in the first instance, leading to distant metastasis of tumor
cells to the bones, the lungs, the brain and other important organs
(6–9). Unfortunately, patients with metastatic
ccRCC are insensitive to chemoradiotherapy (10), and the 5-year survival rate is only
8% (11). Although targeted therapy
has been developed and approved for the treatment of ccRCC, patient
outcomes have not been notably improved due to extensive
intertumoral heterogeneity (12).
Thus, given the high incidence and mortality of ccRCC, it is
necessary to identify novel molecular biomarkers for early
detection and for improving prognosis.
Vacuolar H+-ATPases (V-ATPases)
participate in the regulation of intracellular and extracellular pH
since they are a family of ATP-dependent proton pumps that acidify
various intracellular organelles and extracellular environments;
they play a vital role in several biological processes, such as
membrane transport, protein processing and degradation, small
molecule coupling transport, and other physiological processes,
including urinary acidification and bone reabsorption (13). V-ATPases are composed of different
subunits, which are divided into two main regions: A cytoplasmic
domain, V1, for ATP hydrolysis, and a membrane domain, V0, which
facilitates proton transport (14).
The V0 domain subunit has four distinct subtypes a1-a4, which
regulate the targeting of V-ATPases to different cellular membranes
(15–17). ATP6V0A4 gene encoding a4 subunit,
primarily expressed in kidneys and the epididymis, allows
localization of V-ATPases to the plasma membrane of renal
α-intercalated cells where it secretes H+ into the urine
(18).
Previous research showed high expression of ATP6V0A4
in breast cancer tissues, and knockdown of ATP6V0A4 using siRNA in
the highly invasive MDA-MB-231 human breast cancer cell, suppressed
invasiveness in vitro (18).
Additionally, treatment of MDA-MB-231 cells with specific V-ATPase
inhibitors, such as bafilomycin or concanamycin, exhibited a
promising inhibitory effect on invasion (19). Moreover, knockdown of V-ATPases
expression using siRNA in HCCLM3 cells (a human hepatocellular
carcinoma cell line with high metastatic potential) effectively
inhibited tumor growth and metastasis by reducing proton secretion
and downregulating matrix metalloproteinase-2 and gelatinase
activities (20). However, there
are few studies on the differential expression, development and
molecular mechanism of ATP6V0A4 in malignant tumors (14,21).
In the present study, given that ATP6V0A4 was
specifically expressed in the kidney and epididymis, the
correlation between ATP6V0A4 expression and clinicopathological
characteristics of patients with ccRCC, as well as prognosis, was
examined to ascertain the potential role of ATP6V0A4 as a candidate
biomarker in the occurrence and progression of ccRCC.
Materials and methods
Dataset analysis
In the present study, eight ccRCC-related gene
microarrays (GSE76351, GES6344, GSE15641, GSE16449, GSE47032,
GSE66270, GSE53000 and GSE53757) (22–29)
were acquired from the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/).
The inclusion criteria were: i) ccRCC datasets; ii) specimens
consisting of tumor and adjacent normal kidney tissues; iii)
expression profiling assessed by microarray analysis; and iv)
profiling data collected from human samples. In total, 255 ccRCC
specimens and 165 adjacent normal kidney specimens were
analyzed.
Identification of differentially
expressed genes (DEGs)
The limma package (30) in R version 4.0 (31) was used to compare gene expression
profiles between ccRCC tissues and the adjacent normal kidney
tissues in the GEO datasets. The cut-off criterion for identifying
DEGs was: |log2 fold-change (FC)|>2. P<0.05 was
considered to indicate a statistically significant difference.
Robust Rank Aggregation (RRA)
method
Robust DEGs were identified using the core algorithm
of the RRA R package (32), which
was ranked consistently better than anticipated. This algorithm was
parameter-free and was less affected by outliers, noise and errors.
The screening criterion of |log2 FC|>2 and an
adjusted P-value of <0.05 was considered to indicate a
statistically robust DEG.
Functional enrichment analyses
DEG analysis using Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were conducted using DAVID (version 6.8) (33,34),
and their potential functional relevance was explored. Biological
processes (BPs), molecular functions (MFs) and cell components
(CCs) were assessed by GO enrichment analysis, while pathway
enrichment analysis was performed using KEGG. An adjusted P-value
of <0.05 was considered to indicate a statistically significant
difference.
Protein-protein interaction (PPI)
network analysis
The PPI network of DEGs was established using STRING
(http://string-db.org) (35). The parameter of interactive
relationships among DEGs was set as highest confidence >0.9.
Cytoscape (version 3.7.1) software (36) was used to visualize and analyze the
network.
Hub gene selection and analysis
Identification of hub genes based on these DEGs was
conducted using the cytoHubba (37)
plug-in in Cytoscape, which provides 12 computed methods to analyze
results. Hub genes were obtained by the intersection of the top 50
genes evaluated using the 12 methods. The receiver operating
characteristic (ROC) curves were comprehensive indexes, which were
utilized to reflect the sensitivity and specificity of hub genes
and evaluate the potential value of hub genes as biomarkers in
distinguishing the ccRCC tissues from adjacent normal kidney
tissues.
The Cancer Genome Atlas (TCGA)
database validation
TCGA (http://ualcan.path.uab.edu) was used to obtain
information relating to the differential expression of hub genes in
ccRCC and normal kidney tissues and determine the associations of
the expression levels of these genes with prognosis. We chose the
hub gene that was most significantly associated with survival for
further analysis. The gene expression profiles were downloaded from
TCGA (xenabrowser.net) to analyze the association between the hub
gene and clinicopathological characteristics of patients with
ccRCC.
Cell culture and ccRCC tissue
specimens
In total, four ccRCC cell lines (ACHN, Caki-1, 786-O
and 769-P), a papillary RCC cell line (Caki-2) and a human proximal
tubular epithelial cell line (HK-2) were acquired from the American
Type Culture Collection. All cell lines were cultured in DMEM, 1640
or McCoy's 5A (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% FBS and 100 U/ml penicillin/streptomycin in a humidified
incubator supplied with 5% CO2 air at 37°C. In addition,
a total of 23 pairs of ccRCC tissues and adjacent normal kidney
tissues were collected from ccRCC patients [including 12 males and
11 females, and the median age was 58 (43–78) years] who had not received
chemoradiotherapy. Patients were diagnosed at the Department of
Urology, Peking University Shenzhen Hospital (Shenzhen, China)
between March 2020 and March 2021. All experiments strictly
followed the Declaration of Helsinki and ethical approval was
obtained from Peking University Shenzhen Hospital Ethics Committee
(approval no. 20090017). All patients were informed of the intent
to collect their specimen, the potential risks and the purposes of
the study, and they provided written informed consent.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA from cell lines and tissue was extracted
using TRIzol® reagent (Takara Bio, Inc.), and 1 µg total
RNA was reverse transcribed using a PrimeScript™ RT reagent Kit
with gDNA Eraser according to the manufacturer's protocol (Takara
Bio, Inc.). Next, qPCR was performed on a LightCycler 480 (Roche
Diagnostics) using a SYBR Premix Ex Taq™ II Kit (Takara Bio, Inc.).
Thermocycling conditions were as follows: Initial denaturation at
95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec, 60°C for
30 sec and 72°C for 30 sec, then final extension at 95°C for 1 sec,
65°C for 1 min and 95°C for 15 sec. The primers used in this study
were synthesized by Sangon Biotech, Co., Ltd., with the following
sequences: GAPDH forward, 5′-CCACTCCTCCACCTTTGACG-3′ and reverse,
5′-CTGGTGGTCCAGGGGTCTTA-3′; ATP6V0A4 forward,
5′-CTGCCGAGGAAACGTGTACTT-3′ and reverse,
5′-GGCTCGAAACCCATCACAGA-3′; SUCNR1 forward,
5′-TGCTGGCAGAGTTCCTGTCAAG-3′ and reverse,
5′-AGTTGCATTCCATGCCATGATCC-3′; and SERPINE1 forward,
5′-AGAGCGCTGTCAAGAAGACC-3′ and reverse, 5′-AGTTCTCAGAGGTGCCTTGC-3′.
The expression of hub genes was normalized to the respective GAPDH
expression and calculated by the 2−∆∆Cq method (38).
Western blotting
Protein samples from cells were collected using RIPA
lysis buffer and protease inhibitor (Beyotime Institute of
Biotechnology). Protein concentrations were determined using a BCA
protein assay kit (Takara Bio, Inc.). A total of 20 µg protein/lane
was loaded on 10% SDS-gels, resolved using SDS-PAGE and transferred
to PVDF membranes, and then the membranes were blocked using 5%
skimmed milk for 2 h at room temperature. Subsequently, the
membranes were incubated with the primary antibodies overnight at
4°C, followed by subsequent incubation with the relevant secondary
antibody for 1 h at room temperature. The primary antibodies used
were an anti-ATP6V0A4 antibody (1:3,000; cat. no. ab97440; Abcam)
and an anti-GAPDH antibody (1:10,000; cat. no. AC002; ABclonal
Biotech Co., Ltd.). The secondary antibodies used were an
anti-rabbit (1:2,000; cat. no. 7074; Cell Signaling Technology,
Inc.) and an anti-mouse IgG HRP-linked antibody (1:2,000; cat. no.
7076; Cell Signaling Technology, Inc.). Signals were visualized
using a chemiluminescence imaging system (Tanon-5200Multi; Tanon
Science and Technology Co., Ltd.) and BeyoECL Star reagent
(Beyotime Institute of Biotechnology). ImageJ v1.53 (National
Institutes of Health) was used for densitometry.
Immunohistochemistry staining
Single spot tissue microarray (TMA) slides (30
points of ccRCC, 30 points of adjacent normal kidney tissues; cat.
no. HKid-CRC060CS-01, Shanghai Outdo Biotech Company) were prepared
and incubated with the ATP6V0A4 rabbit antibody (1:500; cat. no.
21570-1-AP; ProteinTech Group, Inc.) at 4°C overnight. And the
rabbit streptavidin-biotin detection system (OriGene Technologies,
Inc.) was used for staining. Two experienced pathologists evaluated
the results according to the staining intensity [0 (negative),
0.5+, 1+, 2+ and 3+] and the positive staining rate (0–100%). Next,
the total score (0–300%) was calculated as the product of the
staining intensity score and positive staining rate score. Low
expression was defined as expression less than or equal to the
median score (0%), whereas a score greater than the median was
classed as high expression.
Statistical analysis
The expression of ATP6V0A4 between ccRCC tissues and
adjacent normal kidney tissues was analyzed using Fisher's exact
test. The association between ATP6V0A4 and each clinicopathological
variable was analyzed using a χ2 test. Data are
presented as the mean ± SD of at least three repeats. Unpaired
student's t-test was used for comparing expression levels of hub
genes between ccRCC tissues and adjacent normal kidney tissues.
Survival curves were analyzed using the Kaplan-Meier method for
patients with high or low expression levels of hub genes and were
evaluated for statistical significance using the log-rank test.
One-way ANOVA followed by Dunnett's post-hoc test was utilized for
comparing the expression of ATP6V0A4 among ccRCC cell lines.
GraphPad Prism version 7 (GraphPad Software, Inc.) was used for all
statistical analyses. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEGs in each GEO
dataset
Integrated bioinformatics analyses were used to
screen for the significant DEGs in the ccRCC datasets (Fig. 1). First, the core phrase ‘clear cell
renal cell carcinoma’ was searched in the GEO database, and the
GSE76351, GES6344, GSE15641, GSE16449, GSE47032, GSE66270, GSE53000
and GSE53757 microarray datasets were downloaded. In total, 255
cases of ccRCC specimens and 165 cases of adjacent normal kidney
specimens were analyzed. DEGs in each GEO dataset were identified
based on the cut-off criteria. Amongst all the DEGs in each
dataset, there were 247 upregulated and 284 downregulated genes
(GES6344), 88 upregulated and 179 downregulated genes (GSE15641),
183 upregulated and 428 downregulated genes (GSE16449), 185
upregulated and 337 downregulated genes (GSE47032), 58 upregulated
and 201 downregulated genes (GSE53000), 365 upregulated and 521
downregulated genes (GSE53757), 718 upregulated and 591
downregulated genes (GSE66270), and 252 upregulated and 471
downregulated genes (GSE76351). Each of these was plotted as
volcano plots (Fig. 2A-H).
Selection of robust DEGs using the RRA
method
The core algorithm of RRA was utilized to identify
robust DEGs in the different datasets. A total of 344 significantly
robust DEGs, consisting of 115 upregulated DEGs and 229
downregulated DEGs, were identified in the eight GEO datasets. The
top 20 DEGs are presented using a heatmap (Fig. 2I).
Functional and pathway enrichment
analyses
GO and KEGG pathway enrichment analyses were used to
evaluate the biological functions of the significant DEGs. The GO
results suggested a significant enrichment of DEGs in ‘organic
anion transport’ and ‘small molecule catabolic process’ in BPs, in
‘apical plasma membrane’ and ‘apical part of the cell’ in CCs, and
in ‘anion transmembrane transporter activity’ and ‘active
transmembrane transporter activity’ in MFs (Fig. 3A). The KEGG pathway enrichment
analysis indicated that DEGs were significantly enriched in the
‘phagosome’, the ‘PPAR signaling pathway’, ‘complement and
coagulation cascades’, the ‘HIF-1 signaling pathway’ and ‘carbon
metabolism’ (Fig. 3B).
Construction of the PPI network and
identification of hub genes
After introducing the significant DEGs into the
STRING database, the interaction among DEGs was plotted as a PPI
network. Based on the aforementioned results, 7 hub genes (SUCNR1,
CXCR4, VCAN, CASR, ATP6V0A4, VEGFA and SERPINE1) were identified
based on the intersection of the top 50 genes (Fig. 4A). Cytoscape was utilized for
further visualizing the network of hub genes (Fig. 4B). ROC curves based on data obtained
from TCGA were plotted to evaluate the diagnostic values of the hub
genes. The area under the curve values of SERPINE1, CXCR4, VEGFA,
VCAN, ATP6V0A4, SUCNR1 and CASR were 0.7907, 0.9745, 0.9624,
0.8137, 0.9545, 0.8514, and 0.9829, respectively (P<0.0001;
Fig. 4C-I).
 | Figure 4.Construction of the PPI network and
identification of hub genes. (A) Intersectional plot of the top 50
genes evaluated using 12 methods in the 8 datasets. (B) PPI network
of the hub genes. (C-I) Receiver operating characteristic curves of
the hub genes, (C) SERPINE1, (D) CXCR4, (E) VEGFA, (F) VCAN, (G)
ATP6V0A4, (H) SUCNR1 and (I) CASR. AUC, area under the curve; MNC,
Maximum Neighborhood Component; DMNC, Density of Maximum
Neighborhood Component; MCC, Maximal Clique Centrality; EPC, Edge
Percolated Component. |
Validation and survival analysis based
on TCGA
To verify the differential expression of hub genes,
the corresponding gene expression profiles of kidney renal clear
cell carcinoma (KIRC) were downloaded from TCGA. As shown in
Fig. 5A, all hub genes were
significantly differentially expressed. Combined with the survival
analysis, it was found that ATP6V0A4, SUCNR1, CASR, CXCR4, and
SERPINE1 were significantly associated with prognosis (Fig. 5B). Since CASR and CXCR4 have been
studied in ccRCC (39,40), the remaining three genes were
selected for further validation.
Validation of hub genes by
RT-qPCR
The expression levels of three hub genes (ATP6V0A4,
SUCNR1 and SERPINE1) were examined by RT-qPCR in 23 pairs of ccRCC
tissues and adjacent normal tissues. The results indicated that the
expression levels of ATP6V0A4 (P=0.0007) and SUCNR1 (P<0.0001)
in ccRCC tissues were significantly lower than those in the
adjacent normal kidney tissues (Fig.
6A-D), while the expression of SERPINE1 did not differ
significantly between the cancerous and paracancerous tissues
(Fig. 6E and F). Since the
difference in expression of ATP6V0A4 between the cancerous and
paracancerous tissues was greater than that of SUCNR1 and ATP6V0A4
was specifically expressed in the kidney, a focus was placed on the
role of ATP6V0A4 in ccRCC progression.
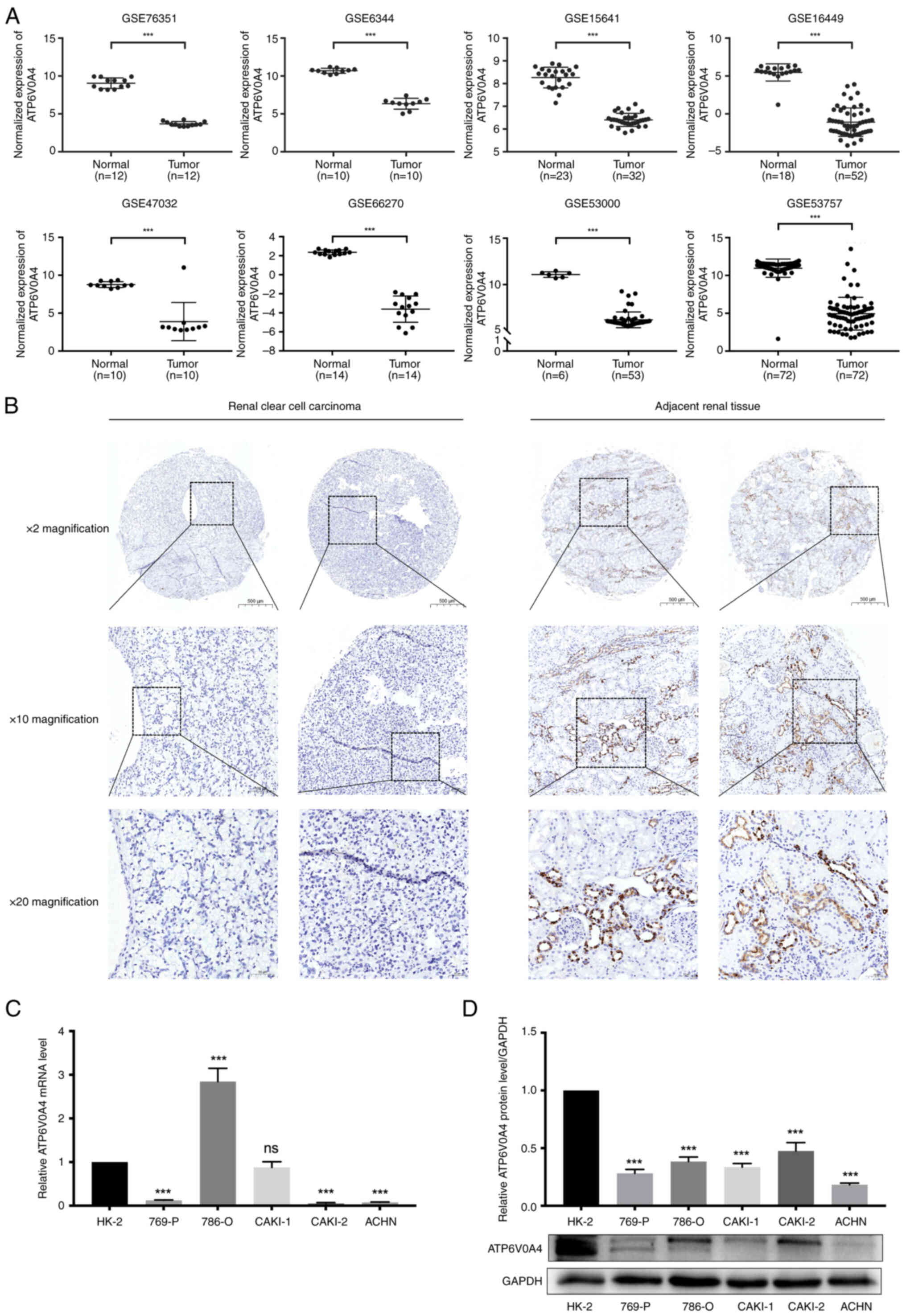
ATP6V0A4 expression is downregulated
in ccRCC
The difference in the expression profiles of
ATP6V0A4 between ccRCC tissues and the adjacent normal kidney
tissues in eight GEO datasets (GSE76351, GES6344, GSE15641,
GSE16449, GSE47032, GSE66270, GSE53000 and GSE53757) was analyzed.
The results showed significantly lower expression of ATP6V0A4 in
ccRCC tissues compared with that in the adjacent normal kidney
tissues (all P<0.0001) (Fig.
7A). These results were also confirmed by immunohistochemistry.
There was a significant downregulation of the ATP6V0A4 protein
expression levels in the ccRCC tissues compared with that in the
adjacent normal kidney tissues (P<0.001; Fig. 7B; Table
I). In addition, how ATP6V0A4 expression was associated with
the clinicopathological characteristics was determined based on
data from 534 patients with ccRCC obtained from TCGA. It was shown
that dysregulated expression of ATP6V0A4 in ccRCC was associated
with the 5-year survival rate, but not with age, sex, primary tumor
(T stage), lymph node involvement (N stage), distant metastasis (M
stage) and AJCC stage of patients with ccRCC (Table II). Combined with previous
Kaplan-Meier survival analysis, it was shown that patients with
ccRCC and high ATP6V0A4 expression had a better prognosis.
 | Table I.Expression of ATP6V0A4 in the
cancerous and paracancerous tissues. |
Table I.
Expression of ATP6V0A4 in the
cancerous and paracancerous tissues.
|
|
| ATP6V0A4
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Tissue sample | No. of cases | High | Low | P-value |
|---|
| ccRCC tissues | 30 | 1 | 29 | <0.001 |
| Adjacent
tissues | 29 | 29 | 0 |
|
 | Table II.Association between ATP6V0A4
expression and clinicopathological characteristics of patients with
clear cell renal cell carcinoma based on data obtained from The
Cancer Genome Atlas.8800240497. |
Table II.
Association between ATP6V0A4
expression and clinicopathological characteristics of patients with
clear cell renal cell carcinoma based on data obtained from The
Cancer Genome Atlas.8800240497.
|
| ATP6V0A4
expression |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variable | Low, n (%) | High, n (%) | χ2
value | P-value |
|---|
| No. of cases | 267 | 267 | - | - |
| Sex |
|
| 0.825 | 0.3637 |
|
Male | 179 (67.0%) | 169 (63.3%) |
|
|
|
Female | 88 (33.0%) | 98 (36.7%) |
|
|
| Age, years |
|
| 2.719 | 0.0991 |
|
<60 | 114 (42.7%) | 133 (49.8%) |
|
|
|
≥60 | 153 (57.3%) | 134 (50.2%) |
|
|
| Pathological T
stage |
|
| 4.415 | 0.2200 |
| T1 | 130 (48.7%) | 144 (53.9%) |
|
|
| T2 | 40 (15.0%) | 30 (11.2%) |
|
|
| T3 | 89 (33.3%) | 88 (33.0%) |
|
|
| T4 | 8 (3.0%) | 3 (1.1%) |
|
|
| Pathological N
stage |
|
| 4.124 | 0.1272 |
| N0 | 122 (45.7%) | 118 (44.2%) |
|
|
| N1 | 4 (1.5%) | 12 (4.5%) |
|
|
| NX | 141 (52.8%) | 137 (51.3%) |
|
|
| Pathological M
stage |
|
| 0.3309 | 0.8475 |
| M0 | 212 (79.4%) | 212 (79.4%) |
|
|
| M1 | 37 (13.9%) | 41 (15.4%) |
|
|
| MX | 16 (6.0%) | 14 (5.2%) |
|
|
| AJCC stage |
|
| 2.333 | 0.5063 |
| I | 128 (47.9%) | 140 (52.4%) |
|
|
| II | 34 (12.7%) | 24 (9.0%) |
|
|
|
III | 63 (23.6%) | 60 (22.5%) |
|
|
| IV | 41 (15.4%) | 41 (15.4%) |
|
|
| Survival time,
years |
|
| 6.101 | 0.0135a |
|
<5 | 204 (76.4%) | 177 (66.3%) |
|
|
| ≥5 | 62 (23.2%) | 87 (32.6%) |
|
|
For further study, both RT-qPCR and western blotting
experiments were conducted to explore the expression level of
ATP6V0A4 in RCC cell lines. The results showed that ATP6V0A4 was
significantly downregulated in 769-P, ACHN, and Caki-2 cell lines
compared with that in HK-2 cells at the transcription level
(Fig. 7C). While ATP6V0A4 level was
remarkably decreased in all 5 RCC cell lines compared with that in
HK-2 cells at translational level (Fig.
7D).
Discussion
As the most common subtype of RCC, ccRCC has an
increasing prevalence, and is associated with a poor prognosis and
considerable metastatic potential (41,42).
Despite the several advances in establishing prediction models
based on the clinical characteristics of ccRCC, exploration of the
molecular mechanisms of tumorigenesis, increased understanding of
the development and metastasis of ccRCC, and the countless targeted
and immunosuppressive therapeutics, the strategies for curing ccRCC
remain limited (43–45). Additionally, the generation of drug
resistance remains a thorny issue during treatment. Consequently,
there is an urgent need to identify highly specific and sensitive
candidate biomarkers, elucidate the molecular mechanisms underlying
tumorigenesis and metastasis, and develop novel and effective
treatment regimens for patients with ccRCC.
Bioinformatics is a transdisciplinary research
approach, specifically used to screen out DEGs to understand the
molecular and genetic basis of diseases. In the present study,
eight GEO datasets were analyzed using bioinformatics analysis. A
total of 344 significantly robust DEGs, including 115 upregulated
and 229 downregulated, were identified using the RRA method. Among
them, 7 hub genes (SUCNR1, CXCR4, VCAN, CASR, ATP6V0A4, VEGFA and
SERPINE1) were identified based on the intersection of the top 50
genes evaluated using cytoHubba. Furthermore, data obtained from
TCGA was combined with the clinical specimens to evaluate the
expression of ATP6V0A4 in ccRCC and its association with
clinicopathological factors and clinical outcomes. The data
suggested that the expression of ATP6V0A4 was significantly
downregulated in ccRCC, and significantly associated with age and
survival. However, due to the deficiency of clinicopathological
characteristics and follow-up information for the tissue samples,
clinical correlation and survival analysis could not be
performed.
Previous studies have reported that ATP6V0A4 is
highly expressed in highly invasive breast cancer and glioma
(14,18). However, in the present study, it was
found that ATP6V0A4 expression was significantly downregulated in
ccRCC tissues and that this downregulation was indicative of a poor
prognosis. Similar results were also observed in vitro in
ccRCC cell lines. Since proton pumps at the plasma membrane are
highly enriched in renal α-intercalated cells, ATP6V0A4 is
tissue-restricted and replaces generic subunits of V-ATPases
(46), which indicated that
ATP6V0A4 was abundant in normal kidney tissues. Normal renal cells
underwent a neoplastic transformation and lost the ability to
differentiate and mature to varying degrees, which led to the loss
of normal structure in the kidney and replacement by tumor cells.
It was speculated that given the high enrichment of ATP6V0A4 in
normal renal tissues, the decreased expression of ATP6V0A4 resulted
in the loss of normal tissue structure in renal cancer.
As previously reported, the dysregulation of
ATP6V0A4 plays a vital role in tumor progression. The knockdown of
ATP6V0A4 has been shown to significantly inhibit cell invasion in
breast cancer by decreasing the targeting of V-ATPases to the
plasma membrane of MDA-MB-231 cells (18). The V-ATPases have since been found
to be present at the plasma membranes of several invasive tumor
cells, including melanoma, Ewing sarcoma, and liver, lung, ovarian,
esophageal, prostate and pancreatic cancer (47–54).
The V-ATPases are heavily involved in tumor cell hallmarks such as
migration and invasion by activating secreted cathepsin through
local acidification of the extracellular environment to degrade the
basement membrane and extracellular matrix proteins or by
activating other secreted proteases such as matrix
metalloproteinases (55,56). Several studies have demonstrated
that pharmacological inhibition of V-ATPases results in potent
antitumor and anti-metastatic effects in vitro and in
vivo (13,57–61).
Treatment of highly invasive MDA-MB-231 and MCF10-CA1a human breast
cancer cells with specific V-ATPase inhibitors such as concanamycin
A and bafilomycin appreciably suppressed the invasive ability in
vitro (19,62). Additionally, Kulshrestha et
al (51) found that targeted
inhibition of V-ATPase restrained ovarian tumor invasion via
regulation of matrix metalloproteinase activity. Further research
needs to evaluate the expression patterns of multiple subunits and
determine whether the activity of V-ATPases can be regulated by
changes in the expression of ATP6V0A4 in ccRCC.
In conclusion, the bioinformatics analyses performed
in the present study showed that ATP6V0A4 gene expression was
significantly different between renal cancer tissues and
paracancerous tissues. The upregulated expression of ATP6V0A4 was
associated with the improved survival of patients with ccRCC, which
may provide a basis for targeted drug therapy. Exploring the
mechanisms underlying the effects of ATP6V0A4 in RCC may assist in
identifying novel druggable targets to improve the management of
patients with ccRCC.
Acknowledgements
Not applicable.
Funding
This study was funded by the Sanming Project of Medicine in
Shenzhen (approval no. SZSM202111007), the Shenzhen Key Medical
Discipline Construction Fund (approval no. SZXK020), the Science,
Technology and Innovation Commission of Shenzhen Municipality
(approval no. JCYJ20220530150812027), and the Hospital project of
Shenzhen Second People's Hospital (approval no. 20223357025).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX, JJ and CY performed the experiments and analyzed
the data. JX, JJ and YW wrote the manuscript. JX, YW and BS
designed the study. JX, JJ and BS confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Patients were diagnosed at the Department of
Urology, Peking University Shenzhen Hospital, China. All
experiments strictly adhered to the guidelines described in the
Declaration of Helsinki, and ethical approval was obtained from the
Ethics Committee of Peking University Shenzhen Hospital (approval
no. 20090017). All patients were informed of the intent to collect
their specimen, the potential risks and the purposes of the study,
and they provided written informed consent.
Patient consent for publication
All patients provided consent for their information
to be published.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ccRCC
|
clear cell renal cell carcinoma
|
|
GEO
|
Gene Expression Omnibus
|
|
DEG
|
differentially expressed gene
|
|
PPI
|
protein-protein interaction
|
|
TCGA
|
The Cancer Genome Atlas
|
|
RRA
|
Robust Rank Aggregation
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
ROC
|
receiver operating characteristic
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
V-ATPases
|
vacuolar H+-ATPases
|
References
|
1
|
Xu WH, Xu Y, Wang J, Wan FN, Wang HK, Cao
DL, Shi GH, Qu YY, Zhang HL and Ye DW: Prognostic value and immune
infiltration of novel signatures in clear cell renal cell carcinoma
microenvironment. Aging (Albany NY). 11:6999–7020. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Butz H, Szabó PM, Nofech-Mozes R, Rotondo
F, Kovacs K, Mirham L, Girgis H, Boles D, Patocs A and Yousef GM:
Integrative bioinformatics analysis reveals new prognostic
biomarkers of clear cell renal cell carcinoma. Clin Chem.
60:1314–1326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Q, Tang H, Luo X, Chen J, Zhang X, Li
X, Li Y, Chen Y, Xu Y and Han S: Immune-associated gene signatures
serve as a promising biomarker of immunotherapeutic prognosis for
renal clear cell carcinoma. Front Immunol. 13:8901502022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sacco E, Pinto F, Sasso F, Racioppi M,
Gulino G, Volpe A and Bassi P: Paraneoplastic syndromes in patients
with urological malignancies. Urol Int. 83:1–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Flanigan RC, Campbell SC, Clark JI and
Picken MM: Metastatic renal cell carcinoma. Curr Treat Options
Oncol. 4:385–390. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jonasch E, Walker CL and Rathmell WK:
Clear cell renal cell carcinoma ontogeny and mechanisms of
lethality. Nat Rev Nephrol. 17:245–261. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ke J, Chen J and Liu X: Analyzing and
validating the prognostic value and immune microenvironment of
clear cell renal cell carcinoma. Anim Cells Syst (Seoul). 26:52–61.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lai Y, Tang F, Huang Y, He C, Chen C, Zhao
J, Wu W and He Z: The tumour microenvironment and metabolism in
renal cell carcinoma targeted or immune therapy. J Cell Physiol.
236:1616–1627. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Choueiri TK and Motzer RJ: Systemic
therapy for metastatic renal-cell carcinoma. N Engl J Med.
376:354–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Z, Xu H, Yu L, Wang J, Meng Q, Mei H,
Cai Z, Chen W and Huang W: Patient-derived renal cell carcinoma
organoids for personalized cancer therapy. Clin Transl Med.
12:e9702022. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stransky L, Cotter K and Forgac M: The
function of V-ATPases in cancer. Physiol Rev. 96:1071–1091. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gleize V, Boisselier B, Marie Y,
Poëa-Guyon S, Sanson M and Morel N: The renal v-ATPase a4 subunit
is expressed in specific subtypes of human gliomas. Glia.
60:1004–1012. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cotter K, Stransky L, McGuire C and Forgac
M: Recent insights into the structure, regulation, and function of
the V-ATPases. Trends Biochem Sci. 40:611–622. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Breton S and Brown D: Regulation of
luminal acidification by the V-ATPase. Physiology (Bethesda).
28:318–329. 2013.PubMed/NCBI
|
|
17
|
Sun-Wada GH and Wada Y: Vacuolar-type
proton pump ATPases: Acidification and pathological relationships.
Histol Histopathol. 28:805–815. 2013.PubMed/NCBI
|
|
18
|
Hinton A, Sennoune SR, Bond S, Fang M,
Reuveni M, Sahagian GG, Jay D, Martinez-Zaguilan R and Forgac M:
Function of a subunit isoforms of the V-ATPase in pH homeostasis
and in vitro invasion of MDA-MB231 human breast cancer cells. J
Biol Chem. 284:16400–16408. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sennoune SR, Bakunts K, Martínez GM,
Chua-Tuan JL, Kebir Y, Attaya MN and Martínez-Zaguilán R: Vacuolar
H+-ATPase in human breast cancer cells with distinct
metastatic potential: Distribution and functional activity. Am J
Physiol Cell Physiol. 286:C1443–C1452. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu X, Qin W, Li J, Tan N, Pan D, Zhang H,
Xie L, Yao G, Shu H, Yao M, et al: The growth and metastasis of
human hepatocellular carcinoma xenografts are inhibited by small
interfering RNA targeting to the subunit ATP6L of proton pump.
Cancer Res. 65:6843–6849. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su K, Collins MP, McGuire CM, Alshagawi
MA, Alamoudi MK, Li Z and Forgac M: Isoform a4 of the vacuolar
ATPase a subunit promotes 4T1-12B breast cancer cell-dependent
tumor growth and metastasis in vivo. J Biol Chem. 298:1023952022.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gumz ML, Zou H, Kreinest PA, Childs AC,
Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et
al: Secreted frizzled-related protein 1 loss contributes to tumor
phenotype of clear cell renal cell carcinoma. Clin Cancer Res.
13:4740–4749. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tun HW, Marlow LA, von Roemeling CA,
Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ and
Copland JA: Pathway signature and cellular differentiation in clear
cell renal cell carcinoma. PLoS One. 5:e106962010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jones J, Otu H, Spentzos D, Kolia S, Inan
M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brannon AR, Reddy A, Seiler M, Arreola A,
Moore DT, Pruthi RS, Wallen EM, Nielsen ME, Liu H, Nathanson KL, et
al: Molecular stratification of clear cell renal cell carcinoma by
consensus clustering reveals distinct subtypes and survival
patterns. Genes Cancer. 1:152–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valletti A, Gigante M, Palumbo O, Carella
M, Divella C, Sbisà E, Tullo A, Picardi E, D'Erchia AM, Battaglia
M, et al: Genome-wide analysis of differentially expressed genes
and splicing isoforms in clear cell renal cell carcinoma. PLoS One.
8:e784522013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wotschofsky Z, Gummlich L, Liep J, Stephan
C, Kilic E, Jung K, Billaud JN and Meyer HA: Integrated microRNA
and mRNA signature associated with the transition from the locally
confined to the metastasized clear cell renal cell carcinoma
exemplified by miR-146-5p. PLoS One. 11:e01487462016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gerlinger M, Horswell S, Larkin J, Rowan
AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos
CR, et al: Genomic architecture and evolution of clear cell renal
cell carcinomas defined by multiregion sequencing. Nat Genet.
46:225–233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
von Roemeling CA, Radisky DC, Marlow LA,
Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW and Copland JA:
Neuronal pentraxin 2 supports clear cell renal cell carcinoma by
activating the AMPA-selective glutamate receptor-4. Cancer Res.
74:4796–4810. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing Vienna, Austria: ISBN 3-900051-07-0. 2012, http://www.R-project.org/
|
|
32
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Joeckel E, Haber T, Prawitt D, Junker K,
Hampel C, Thüroff JW, Roos FC and Brenner W: High calcium
concentration in bones promotes bone metastasis in renal cell
carcinomas expressing calcium-sensing receptor. Mol Cancer.
13:422014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rasti A, Abolhasani M, Zanjani LS, Asgari
M, Mehrazma M and Madjd Z: Reduced expression of CXCR4, a novel
renal cancer stem cell marker, is associated with high-grade renal
cell carcinoma. J Cancer Res Clin Oncol. 143:95–104. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Zhu X, Qiao X, Sun L, Xia T and
Liu C: Clinical implications of HSC70 expression in clear cell
renal cell carcinoma. Int J Med Sci. 18:239–244. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Luo Y, Shen D, Chen L, Wang G, Liu X, Qian
K, Xiao Y, Wang X and Ju L: Identification of 9 key genes and small
molecule drugs in clear cell renal cell carcinoma. Aging (Albany
NY). 11:6029–6052. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Du GW, Yan X, Chen Z, Zhang RJ, Tuoheti K,
Bai XJ, Wu HH and Liu TZ: Identification of transforming growth
factor beta induced (TGFBI) as an immune-related prognostic factor
in clear cell renal cell carcinoma (ccRCC). Aging (Albany NY).
12:8484–8505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cui T, Guo J and Sun Z: A computational
prognostic model of lncRNA signature for clear cell renal cell
carcinoma with genome instability. Expert Rev Mol Diagn.
22:213–222. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yap NY, Ong TA, Pailoor J, Gobe G and
Rajandram R: The significance of CD14 in clear cell renal cell
carcinoma progression and survival prognosis. Biomarkers. 28:24–31.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Smith AN, Borthwick KJ and Karet FE:
Molecular cloning and characterization of novel tissue-specific
isoforms of the human vacuolar H(+)-ATPase C, G and d subunits, and
their evaluation in autosomal recessive distal renal tubular
acidosis. Gene. 297:169–177. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nishisho T, Hata K, Nakanishi M, Morita Y,
Sun-Wada GH, Wada Y, Yasui N and Yoneda T: The a3 isoform vacuolar
type H+-ATPase promotes distant metastasis in the mouse
B16 melanoma cells. Mol Cancer Res. 9:845–855. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Avnet S, Di Pompo G, Lemma S, Salerno M,
Perut F, Bonuccelli G, Granchi D, Zini N and Baldini N: V-ATPase is
a candidate therapeutic target for Ewing sarcoma. Biochim Biophys
Acta. 1832:1105–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu J, Xie R, Liu X, Wen G, Jin H, Yu Z,
Jiang Y, Zhao Z, Yang Y, Ji B, et al: Expression and functional
role of vacuolar H(+)-ATPase in human hepatocellular carcinoma.
Carcinogenesis. 33:2432–2440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lu Q, Lu S, Huang L, Wang T, Wan Y, Zhou
CX, Zhang C, Zhang Z and Li X: The expression of V-ATPase is
associated with drug resistance and pathology of non-small-cell
lung cancer. Diagn Pathol. 8:1452013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kulshrestha A, Katara GK, Ibrahim S,
Pamarthy S, Jaiswal MK, Gilman Sachs A and Beaman KD: Vacuolar
ATPase ‘a2’ isoform exhibits distinct cell surface accumulation and
modulates matrix metalloproteinase activity in ovarian cancer.
Oncotarget. 6:3797–3810. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang L, Lu Q, Han Y, Li Z, Zhang Z and Li
X: ABCG2/V-ATPase was associated with the drug resistance and tumor
metastasis of esophageal squamous cancer cells. Diagn Pathol.
7:1802012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Michel V, Licon-Munoz Y, Trujillo K,
Bisoffi M and Parra KJ: Inhibitors of vacuolar ATPase proton pumps
inhibit human prostate cancer cell invasion and prostate-specific
antigen expression and secretion. Int J Cancer. 132:E1–E10. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chung C, Mader CC, Schmitz JC, Atladottir
J, Fitchev P, Cornwell ML, Koleske AJ, Crawford SE and Gorelick F:
The vacuolar-ATPase modulates matrix metalloproteinase isoforms in
human pancreatic cancer. Lab Invest. 91:732–743. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cotter K, Capecci J, Sennoune S, Huss M,
Maier M, Martinez-Zaguilan R and Forgac M: Activity of plasma
membrane V-ATPases is critical for the invasion of MDA-MB231 breast
cancer cells. J Biol Chem. 290:3680–3692. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Forgac M: Vacuolar ATPases: Rotary proton
pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol.
8:917–929. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
McGuire C, Cotter K, Stransky L and Forgac
M: Regulation of V-ATPase assembly and function of V-ATPases in
tumor cell invasiveness. Biochim Biophys Acta. 1857:1213–1218.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Schneider LS, von Schwarzenberg K, Lehr T,
Ulrich M, Kubisch-Dohmen R, Liebl J, Trauner D, Menche D and
Vollmar AM: Vacuolar-ATPase inhibition blocks iron metabolism to
mediate therapeutic effects in breast cancer. Cancer Res.
75:2863–2874. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Schempp CM, von Schwarzenberg K, Schreiner
L, Kubisch R, Müller R, Wagner E and Vollmar AM: V-ATPase
inhibition regulates anoikis resistance and metastasis of cancer
cells. Mol Cancer Ther. 13:926–937. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wiedmann RM, von Schwarzenberg K,
Palamidessi A, Schreiner L, Kubisch R, Liebl J, Schempp C, Trauner
D, Vereb G, Zahler S, et al: The V-ATPase-inhibitor archazolid
abrogates tumor metastasis via inhibition of endocytic activation
of the Rho-GTPase Rac1. Cancer Res. 72:5976–5987. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kubisch R, Fröhlich T, Arnold GJ,
Schreiner L, von Schwarzenberg K, Roidl A, Vollmar AM and Wagner E:
V-ATPase inhibition by archazolid leads to lysosomal dysfunction
resulting in impaired cathepsin B activation in vivo. Int J Cancer.
134:2478–2488. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Capecci J and Forgac M: The function of
vacuolar ATPase (V-ATPase) a subunit isoforms in invasiveness of
MCF10a and MCF10CA1a human breast cancer cells. J Biol Chem.
288:32731–32741. 2013. View Article : Google Scholar : PubMed/NCBI
|