Introduction
Renal cell carcinoma (RCC) is one of the most common
forms of cancer in individuals and can be classified into three
types: Kidney renal clear cell carcinoma (KIRC), kidney renal
papillary cell carcinoma (KIRP) and malignancies of the
chromophobe. KIRC, which is one of the most common forms of urinary
cancer with a growing incidence (1), accounts for 70–85% of histologic
subtypes of RCC, which derives from the tubule epithelium of renal
parenchyma (2). Even though a
number of targeted pharmaceuticals and immunosuppressives have been
developed, surgical operation remained the most effective and
primary method for treating this condition (3). Early-stage KIRC does not usually
manifest any symptoms and 20–30% eventually progress to metastatic
RCC (mRCC) (4). In recent years,
there has been an increase in indolent cancers being discovered
incidentally and the clinical treatment of active surveillance,
robot-assisted nephron-saving surgery and minimally invasive
techniques, such as thermal ablation, have become more popular. The
surgery for kidney cancer at an early stage can potentially be
curative, but recurrences after surgery remain common and
inoperable kidney cancer at a late stage is usually fatal. It is
estimated that ~40% of patients are resistant to conventional
chemotherapy and radiation therapy and patients with mRCC who have
experienced treatment failure have a 5-year survival rate of
<20% (5). Somatic mutant genes
in KIRC have been identified by whole genome sequencing and their
involvement in pathogenesis and mechanisms has been explored
(6). To date, the molecular
pathology of renal cancer remains unclear. However, there is an
urgent need to discover more ways of identifying these biomarkers
in order to facilitate early detection and stop the devastating
progression of KIRC. At the same time, there is a very active
search for new biomarkers in the field of renal oncology that have
the potential to further improve diagnosis, treatment and prognosis
of RCC.
Fascin (FSCN) is an actin-binding protein of 55 kDa
that is responsible for the formation and stability of microspikes,
filopodia and invadopodia, which is critical for cell adhesion,
motility and migration (7–9). The FSCN family contains three
isoforms, namely FSCN1, FSCN2 and FSCN3, which are encoded by
FSCN1, FSCN2 and FSCN3 genes, respectively (10). The actin-binding protein FSCN1
exists in mammalian cells such as neurons, endothelial cells and
mesenchymal cells, but is significantly reduced or absent in normal
epithelial cells which acts as a migration factor associated with
epithelial-to-mesenchymal transition (10–12).
Migration and metastasis of colon cancer cells are significantly
accelerated by overexpression of FSCN1 (13), while tumor metastasis and cell
motility in prostate cancer are diminished when FSCN1 is knocked
down in cellular models (14).
FSCN2, which is expressed by retinal photoreceptor cells, serves a
critical role in stabilizing stereocilia after development, is
abundant in stereocilia and is developmentally regulated, appearing
in inner-hair-cell stereocilia during final stages of elongation
(8,15). A study has found that FSCN2 is
essential for maintaining ear and eye function, is an actin
cross-linking protein that is mainly localized in retinas and in
the stereocilia of hair cells (16). FSCN3, which is testis specific, may
function in terminal elongation of the spermatid head (17). Currently, however, little
information is available on the relationships between FSCN2/3 and
tumors and the role of the FSCN family in KIRC remains to be
elucidated.
The current study examined the expression and
functional role of FSCN1-3 in KIRC by using various public
databases. Additionally, the relationship between FSCN family
expression levels and clinicopathological features, prognosis,
tumor immune cell infiltration and drug sensitivity was studied in
patients with KIRC.
Therefore, the present study provided improved
knowledge about the molecular mechanisms of KIRC to facilitate
further studies.
Materials and methods
Ethics statement
The Ethics Committee of the First Affiliated
Hospital of Nanchang University approved the research protocol
(approval no. 12-110). All datasets were gathered from public
databases with written consent.
Patient and tumor samples
There were 20 KIRC tissues and adjacent normal
tissues collected from patients whose pathology was independently
confirmed by two pathologists. In total, 20 matched pairs of KIRC
tissues and adjacent normal kidney tissues were stored in liquid
nitrogen between 2021 and 2022. The tissue samples are the same as
those used in the previous article (18). Written informed consent was obtained
from the patients involved.
RNA and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted using TRIzol (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
RNA samples were stored at −80°C until use. The extracted RNA was
reverse-transcribed into cDNA using the First-Strand cDNA Synthesis
kit (Qiagen, Inc.) according to the manufacturer's protocols. Each
cDNA sample was added to a 20 µl reaction volume containing an
appropriate primer set and SYBR green supermix. Triplicates of all
samples were analyzed. The SYBR Real-Time PCR kit (Qiagen, Inc.)
was used under the following conditions according to the
manufacturer's protocols: 95°C for 2 min, followed by 40 cycles of
95°C for 5 sec and 60°C for 10 sec. Relative expression was
normalized to GAPDH and calculated according to the
2−∆∆Cq method (19). In
the present study, the following primers were used: GAPDH forward
primer GCCACATCGCTCAGACACCAT, GAPDH reverse primer:
CCCATACGACTGCAAAGACCC, Human FSCN1 forward: GACGAGCTCTTTGCTCTGGA,
Human FSCN1 reverse: TCGGTCTCCTCGTCCTGATT, Human FSCN2 forward:
TGGAGGAGAGTCACCCACAG, Human FSCN2 reverse: TCAGGAAGGTCTCGTGGTCT,
Human FSCN3 forward: GCTTCGTTCAGCCAATGGCTAC, Human FSCN3 reverse:
ATCCTGCCACAGTTCCAGTGCA. The QuantiTect SYBR Green PCR kit (Qiagen,
Inc.) was used to perform real-time quantitative PCR. GAPDH was
used as an internal control. Experiments were replicated three
times.
Gene expression profiling interactive
analysis (GEPIA) dataset
The GEPIA dataset (http://gepia.cancer-pku.cn/) was used to analyze The
Cancer Genome Atlas (TCGA; (https://tcga-data.nci.nih.gov/tcga/) tumors compared
with TCGA normal and the genotype-tissue expression (GTEx) normal
datasets and the box plots for the expression of FSCN1, FSCN2 and
FSCN3 between KIRC tissues and the adjacent tissues were obtained
(20). P<0.05 was considered to
indicate a statistically significant difference.
UALCAN
The UALCAN database (http://ualcan.path.uab.edu) contains 31 types of
patients with cancer with clinical and RNA-seq data. UALCAN is an
interactive portal, which can be used to study the relationship
between the expression of target genes in TCGA and the clinical
data of patients. In the present study, correlation of the FSCNs
expression with clinical pathological parameters, including
individual cancer, tumor grade and KIRC subtypes, were analyzed.
P<0.05 was considered to indicate a statistically significant
difference. *P<0.05, **P<0.01 and ***P<0.001.
cBioPortal analysis
The c-BioPortal (https://www.cbioportal.org) is an online database for
interactive exploration of multidimensional cancer genomic datasets
(21). The present study analyzed
the genetic alterations of FSCN1-3, which contained genomic
profiles counted on mutations and putative copy-number alterations
(CNA) from GISTIC 2.0 (22).
OncoPrint v.3.3.1 was constructed in cBioPortal (https://www.cbioportal.org/) to directly reflect all
types of changes including gene amplification, deep deletion, mRNA
upregulation and mRNA downregulation in patients with KIRC. In
addition, genetic alterations in FSCNs genes were correlated with
OS of patients with KIRC and the log-rank test was used to perform
the difference between altered group and unaltered group. Following
the c-BioPortal's online instruction, 50 frequent neighbor genes of
FSCNs family and the coexpression correlation of coefficient
between FSCN genes were achieved.
STRING analysis
The STRING database (http://string-db.org/) provided the significant
protein-protein interactions. The PPI network of FSCN1-3 and 50
frequent neighbor genes was generated using STRING (23).
Tumor immune estimation resource
database (TIMER)
TIMER includes >10,000 samples representing 32
types of cancer from the TCGA, which was an easy-to-operate online
tool established for systematically analyzing the abundance of
immune infiltration (24). The gene
module explored the relationship between members of the FSCN family
and immune cell infiltration, including B cells, CD4+ T cells, CD8+
T cells, neutrophils, macrophages and dendritic cells in KIRC.
Using the somatic copy number alterations (SCNA) module, the tumor
infiltration levels was compared with different somatic copy number
alterations in FSCNs.
Statistical analysis
The present study used R software (version 3.6.2;
http://www.R-project.org/) to conduct
the statistical analyses. Based on KIRC samples, the RNAseq data
was downloaded from TCGA, which primarily included the lncRNA
dataset (level 3) and clinical data for patients with RCC. Using R
and the Wilcox test, the different expressions of FSCNs in KIRC
were analyzed using the ggplot2 package. In order to estimate the
prognosis of FSCNs, Kaplan-Meier survival analysis and Cox
proportional hazards regression analysis were performed. Univariate
and multifactorial Cox regression analysis were used to analyze the
relationship between FSCN1-3 genes and clinicopathological
parameters. Univariate analysis and multivariate analysis were used
to evaluate the independent prognostic significance of FSCN1-3 mRNA
expression. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) enrichment were performed using the R package
clusterProfiler.
Results
Transcriptional levels of different
FSCN family members in patients with clear cell renal cell
carcinoma
The present study first examined the mRNA levels of
FSCN family members in KIRC based on RNA-seq data from TCGA KIRC
cohort. As shown in Fig. 1A-C, the
FSCN1 and FSCN3 expressions were significantly higher in KIRC
tissue compared with normal tissue samples, whereas the FSCN2
expression was lower in cancerous tissue than in normal tissue. The
mRNA transcription levels of the three FSCN members for patients
with KIRC were then examined according to the GEPIA database. As
shown in Fig. 1D-F, the expression
level of FSCN1 mRNA in KIRC tissues was significantly higher than
that in normal kidney tissues, whereas no significant difference in
the expression of FSCN2 and FSCN3 was found between KIRC and
non-cancerous kidney tissue. To validate this conclusion, we
analyzed the FSCN1/2/3 mRNA expression in 15 pairs of KIRC samples
and adjacent histologically normal tissues using real-time PCR
(RT-qPCR). As shown in Fig. 1G-I,
studies indicated that FSCN1/3 are highly expressed in kidney
cancer, while FSCN2 is expressed at low levels in adjacent cancer
tissues compared to normal tissues. On the basis of the above
results, it was inferred that FSCN1 and FSCN3 transcriptional
levels were significantly lower in normal tissues than in KIRC
tissues compared with paired tissue samples, while the FSCN2
exhibited the opposite result.
Relationships between FSCN family
expressions and clinicopathological parameters of KIRC
The present study explored the relationship between
clinical and pathological parameters and the expression of FSCN1-3
based on the TCGA data (https://tcga-data.nci.nih.gov/tcga/) and UALCAN
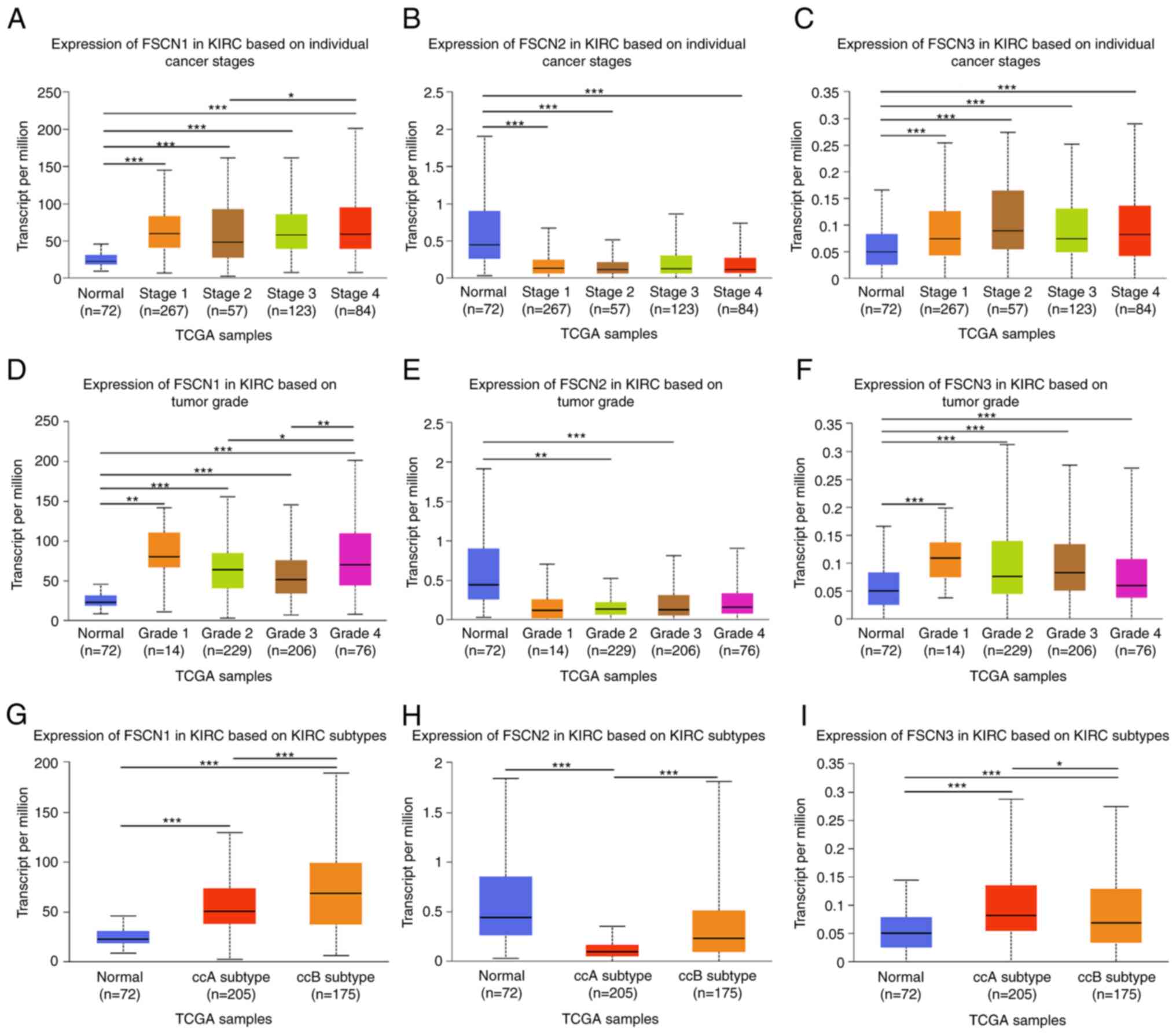
database. As shown in Fig. 2A-C,
with respect to tumor stage, there was a remarkable correlation
between FSCN1/3 mRNA expression level and individual cancer stages.
As cancer stage increased, FSCN1 mRNA expression level increased.
The highest mRNA expression of FSCN1 was found in stage IV.
However, no significant difference was observed between cancer
stages and FSCN2 mRNA expression. As presented in Fig. 2D-F, the mRNA expression of FSCN1 was
markedly associated with tumor grade with the highest mRNA
expression level expressed in grade IV, while mRNA expressions of
FSCN2 and FSCN3 were not associated with tumor grade. As shown in
Fig. 2G-I, the mRNA expression
levels of FSCN1-2 in KIRC good risk (ccA) subtype were
significantly lower compared to the KIRC poor risk (ccB) subtype,
while the expression of FSCN3 showed the opposite result.
Therefore, the results suggested that mRNA expressions of FSCN1-3
were significantly associated with clinicopathological
parameters.
Prognostic value of mRNA expression of
FSCN family members in patients with KIRC
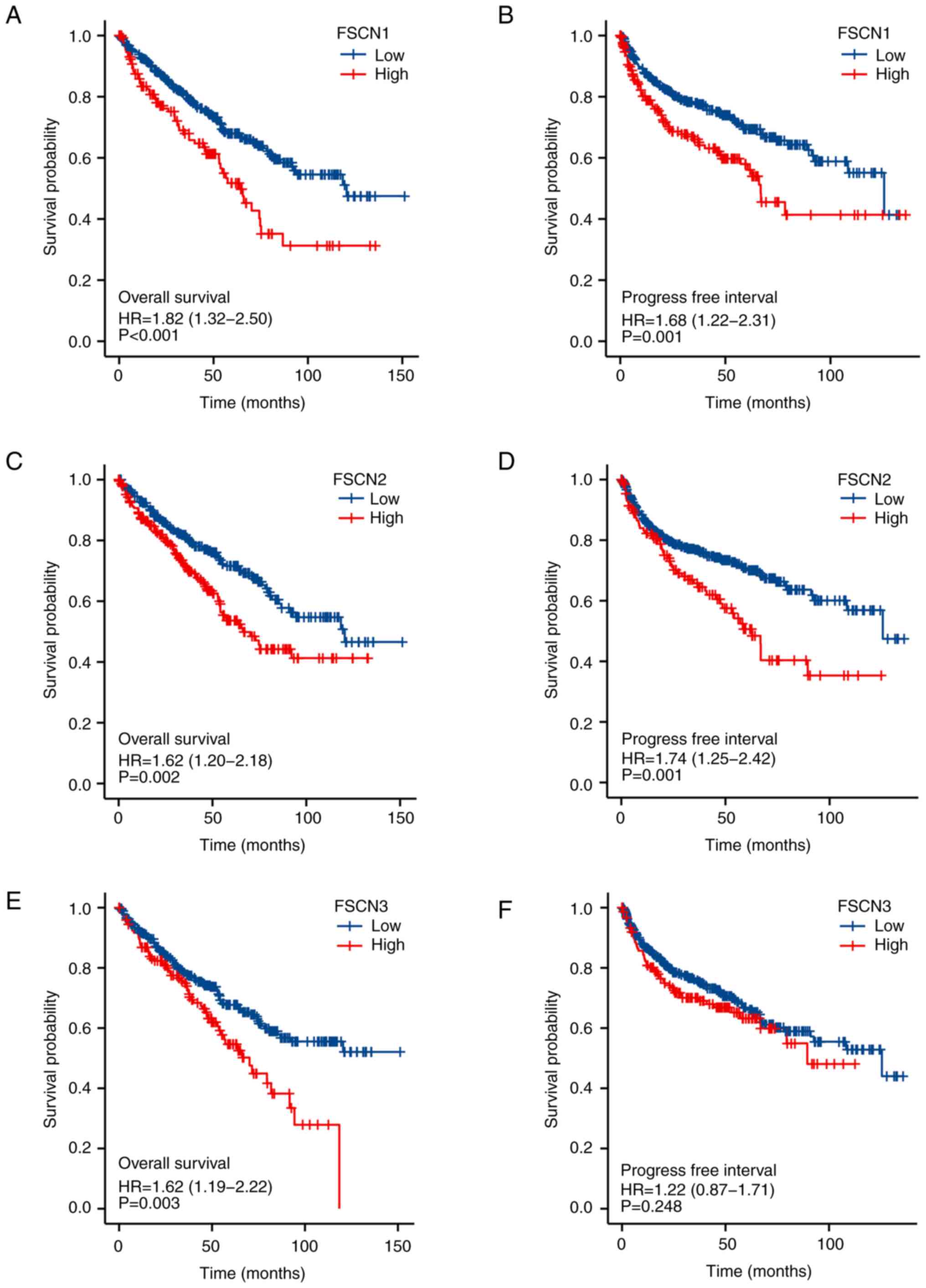
To further explore the prognostic role of FSCN
family members in patients with KIRC, survival analysis was
conducted by R software according to the clinical information in
TCGA database. As shown in Fig.
3A-F, the expression of mRNA of FSCN1-3 family members was
significantly associated with prognosis in patients with KIRC. The
results showed that higher mRNA expressions of FSCN1 (HR=1.82;
95%CI:1.32–2.50 and P<0.001) and FSCN2 (HR=1.74; 95%CI:1.25–2.42
and P=0.001) were associated with poorer OS in patients with KIRC,
whereas the mRNA expression level of FSCN3 (HR=1.22;
95%CI:0.87–1.71 and P=0.248) was not associated with the OS of
patients. Higher mRNA expressions of FSCN1 (HR=1.68;95%CI:1.22–2.31
and P=0.001), FSCN2 (HR=1.62; 95%CI:1.20–2.18 and P=0.002) and
FSCN3 (HR=1.62; 95%CI:1.19–2.22 and P=0.003) were associated with
shorter PFI. These findings indicated mRNA expressions of FSCN1-3
were found to be significantly correlated with the prognosis of
patients with KIRC. Thus, FSCN 1–3 might be useful makers for
predicting the overall survival of patients with KIRC.
Independent prognostic value of mRNA
expression levels of FSCN1-3 in terms of OS in patients with
KIRC
Following the finding that there was a significant
association between FSCN1-3 mRNA levels and OS for patients with
KIRC, the independent prognostic value of mRNA expression of FSCN
family members for patients bearing KIRC was evaluated based on the
TCGA database and prognostic data for Cox survival regression
analysis (25). The univariate Cox
regression analysis showed that high expression of FSCN1 (HR=1.330;
95%CI: 1.108–1.598 and P=0.002), FSCN2 (HR=1.801; 95%CI:
1.259–2.578 and P=0.001), age (HR=1.765; 95%CI: 1.298–2.398 and
P<0.001), pathologic stage (HR=3.946; 95%CI: 2.872–5.423 and
P<0.001) and histologic grade (HR=2.702; 95%CI: 1.918–3.807 and
P<0.001) in the KIRC were significantly correlated with
increased OS. Multivariate analysis showed that FSCN2 (HR=1.659;
95%CI: 1.137–2.422 and P=0.009), age (HR=1.543; 95%CI: 1.132–2.103
and P=0.006), pathologic stage (HR=3.946; 95%CI: 2.206–4.335 and
P<0.001) and histologic grade (HR=1.749; 95%CI: 1.217–2.514 and
P=0.003) were significant prognostic factors for overall survival
(Table SI). Cox regression for OS
analysis revealed that FSCN2, age, pathologic stage and histologic
grade served as independent predictive variables in patients with
KIRC.
Genetic mutations status in FSCN
family members and their associations with OS and progression-free
survival (PFS) of patients with KIRC
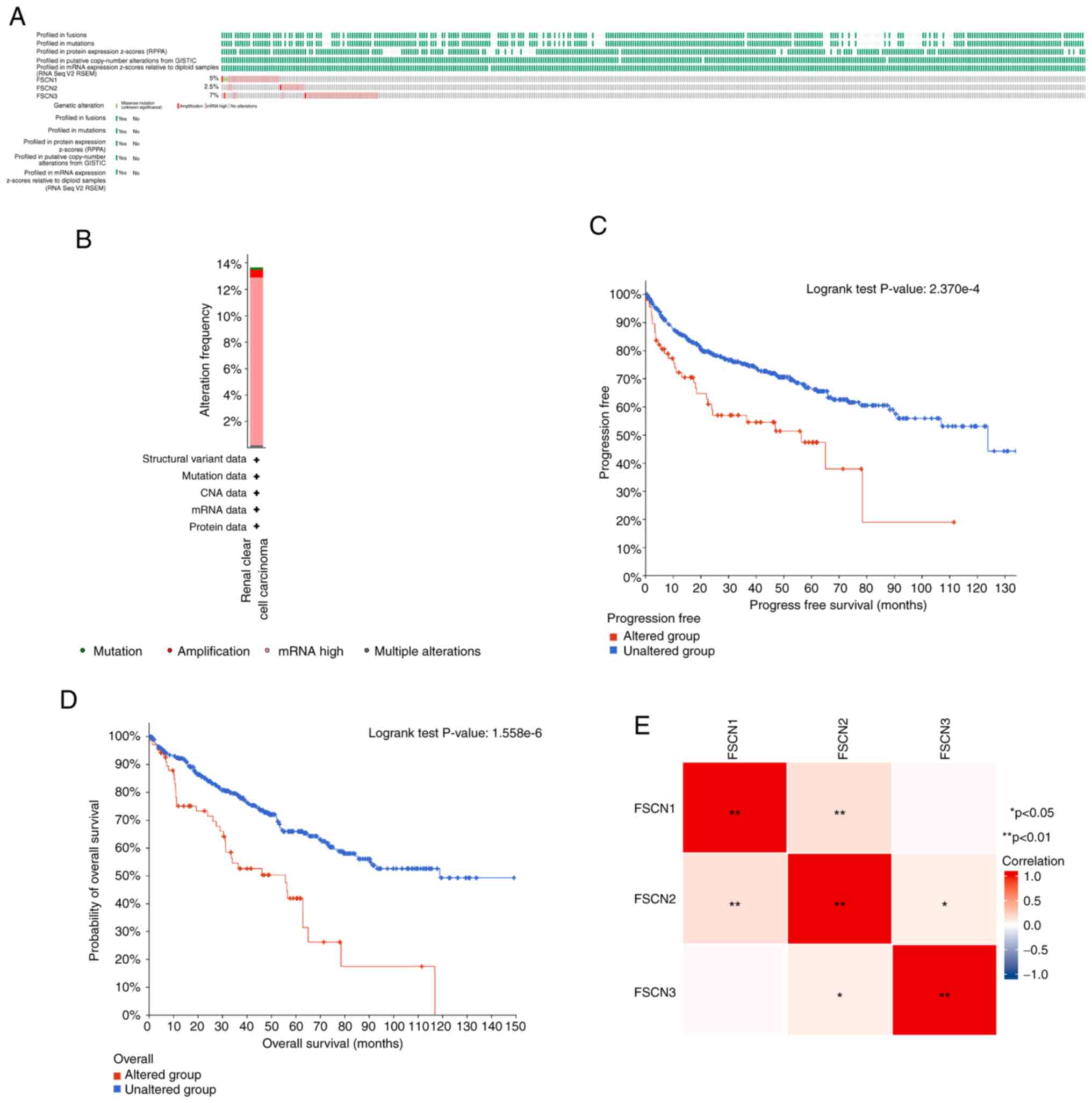
The present study analyzed the genetic alterations
in FSCN family members and their associations with OS and PFS of
patients with KIRC using the cBioPortal online tool to explore the
potential expression pattern of FSCN1-3. To gain further insight
into genetic changes that arise in KIRC, cBioPortal was used to
reanalyze genomic data from 512 sequenced patients with KIRC. As
shown in Fig. 4, the mutation rate
of FSCN3 was the highest, at a percentage of 7% among the FSCN1-3
family. The mutation rate of FSCN1 was 5%, which was twice as high
as the mutation of FSCN2. Kaplan-Meier curve of patients with KIRC
with (altered group) or without mutations (unaltered group) in
FSCN1-3 genes showed significant difference in terms of PFS
(Fig. 4C; P=2.370×10−4)
and PFS (Fig. 4D;
P=1.558×10−6). This result suggested that the poor
prognosis was caused by their mutation. Additionally, the
correlation between FSCN1/2/3 was calculated by analyzing their
mRNA expression. The result showed that FSCN2 had positive
correlations with FSCN1 and FSCN3, while no relationship was found
between FSCN1 and FSCN3 (Fig.
4E).
Predicted functions and pathways of
the alteration in FSCN family and the 50 most frequently altered
adjacent genes in patients with KIRC
After analyzing the genetic alterations in FSCN1/2/3
and the prognostic value of patients with KIRC, the 50 neighbor
genes related to the FSCN1/2/3 mutants were analyzed and an
integrated network was constructed using the STRING database
(https://string-db.org/). Using the cBioPortal
database, the top 50 genes which were co-expressed and associated
with the FSCN1-3 were identified. As shown in Fig. 5A, the actin filament organization
genes including CAPZB, ITGB5, TPM2, ZYX, ARHGEF2 and PPM1F were
significantly associated with FSCN1-3 mutations. GO and KEGG
functional enrichment analyses were performed using the ggplot2 R
package to analyze the functions of FSCN1-3 and 50 neighbor genes
significantly associated with FSCN1-3 (26). As presented in Fig. 5B, biological processes such as GO:
0007015 ‘actin filament organization’, GO: 0051017 ‘actin filament
bundle assembly’, GO:0034329 ‘cell junction assembly’ and
GO:0034330 ‘cell junction organization’ were significantly
modulated by the FSCN1/2/3 mutations in KIRC. Cellular components,
including GO:0005925 ‘focal adhesion’, GO:0005924 ‘cell-substrate
adherens junction’, GO:0030055 ‘cell-substrate junction’,
GO:0030016 ‘myofibril’ and GO:0043292 ‘contractile fiber’ were
significantly related to the FSCN1/2/3 alterations. Additionally,
FSCN family genes mutations significantly affected molecular
functions, such as GO:0003779 ‘actin binding’, GO:0005518 ‘collagen
binding’, GO:0051015 ‘actin filament binding’, GO:0043522 ‘leucine
zipper domain binding’ and GO:0048407 ‘platelet-derived growth
factor binding’. In KEGG analysis, five pathways including has:
04510 ‘Focal adhesion’, has: 04144 ‘Endocytosis’, has: 05410
‘Hypertrophic cardiomyopathy’, has: 05414 ‘Dilated cardiomyopathy’
and has: 04810 ‘Regulation of actin cytoskeleton’ were associated
with the functions of FSCN1-3 mutations in KIRC (Table SII).
Immune infiltrations analysis of the
FSCN1-3 family in KIRC
Correlations between genes and immune infiltrations
were estimated using TIMER. The positive connections existed
between the abundance of CD4+ T cell and the expressions of all
FSCN family members. The expression of FSCN1 showed a positive
correlation with the abundance of CD8+ T cell, while FSCN2 had a
negative correlation. The abundance of macrophage, neutrophil, B
cell and dendritic cell positively showed significant associations
with FSCN1. The expression of FSCN3 showed a positive correlation
with the abundance of CD4+ T cell and neutrophil (Fig. 6A-C). Furthermore, the SCNA of
FSCN1/2/3 were estimated. Results revealed the SCNA of FSCN2
significantly correlated with the infiltration levels of six immune
cells composed of B cells, CD4+ T cells, CD8+ T cells, neutrophils,
macrophages and dendritic cells, while that of FSCN1 was in
significant connections with the infiltrating levels of B and CD4+
T cell. The SCNA of FSCN3 was only significantly associated with
CD4+ T cells (Fig. 6D-F). Together,
FSCN Family members were closely related to the immune infiltration
in patients with KIRC.
Discussion
FSCNs cross-link filamentous actin into tightly
packed parallel bundles and serve a central role in architectural
maintenance and functioning of cell protrusions (8). Growing evidence suggests that FSCNs
serve a critical role not only in tumorigenesis and proliferation
of tumor cells, but also in tumor metastasis (27,28).
However, the association between mRNA expression of distinct FSCNs
family members and prognosis of patients with KIRC remains unclear.
The present study systematically examined the mRNA levels, genetic
alterations, functional enrichment, immune infiltration and
prognostic value of FSCNs.
By considering the combined effect of mutations in
multiple genes within the FSCN gene family, researchers can obtain
a more comprehensive assessment of their effect on prognosis. This
approach enables the capture of synergistic or cumulative effects
resulting from alterations in multiple genes, which may have a
greater influence on disease progression or treatment response
compared to individual gene mutations. Additionally, studying
mutations across the entire family can provide insights into common
disrupted pathways or mechanisms, contributing to a improved
understanding of the underlying biology of the disease. This
knowledge can aid in the identification of potential therapeutic
targets or the development of personalized treatment strategies.
Furthermore, analyzing the correlation between these genes can
offer insights into potential functional redundancy or compensation
within the family. In cases where one family member is mutated,
other members may compensate for its loss of function. By
integrating correlation analysis with mutation analysis,
researchers can identify patterns where mutations in one family
member are associated with changes in the expression or activity of
other family members. Understanding these compensatory mechanisms
provides a more comprehensive view of the functional impact of
mutations within the FSCN family.
FSCN-1 is an actin bundling protein that serves key
functions in cell-cell interactions, adhesion and motility via
regulating the function of filopodial protrusions and
microfilaments (29), which are
involved in the invasion and metastasis of various tumors. It has
been shown that FSCN1 is mainly overexpressed in estrogen
receptor-negative breast tumor tissues and positive FSCN-1
expression is associated with decreased mean tumor-free survival
and overall survival (30).
Furthermore, increased FSCN1 expression in nasopharyngeal carcinoma
is associated with poor prognosis (31). FSCN1 is usually upregulated in a
number of malignant tumors and could be considered as an oncogene
since it promotes tumor cell migration and invasion (32). Among a variety of tumor types, FSCN1
is significantly associated with increased metastatic potential and
more aggressive phenotypes (33–35)
and by inhibiting FSCN1, tumor cells could be prevented from
migrating and metastasizing (36).
Knocking down FSCN1 expression can also have an anti-migration and
anti-invasion effect on ovarian cancer and glioblastoma (37,38). A
study found that knockdown of fascin-1 expression could suppress
cell migration and invasion of non-small cell lung cancer by
regulating the MAPK pathway (39).
Therefore, inhibiting FSCN1 expression might be essential for the
treatment of metastatic cancers. The present study detected that
FSCN1 expressed higher in KIRC tissues compared with normal
tissues. In addition, it was demonstrated that high expression of
FSCN1 was related to shorter OS and PFI in patients with KIRC,
indicating that FSCN1 acted as an oncogenic role in renal cell
carcinoma and promoted the development of renal cell carcinoma. The
results were similar to previous research that concluded that the
increased expression of FSCN1 has been proved to be an adverse
biomarker predicting poor outcomes in esophageal squamous cell
carcinoma (40). The present study
also found that FSCN1 overexpression was significantly related to
advanced individual cancer stage and tumor grade among patients
with KIRC. The expression of FSCN1 was also related to immune cell
infiltration in KIRC, suggesting that FSCN1 might regulate the
immune response to cancer. These findings suggest that FSCN1 might
be a promising prognostic and therapeutic target for patients with
KIRC.
FSCN2, an actin-bundling protein, is a
photoreceptor-specific protein of the fascin family that serves a
significant role in maintaining ear and eye functions (16,41).
Few studies have explored the relationship between FSCN2 and
tumors. In the present study, the survival analyses showed that
high expression of FSCN2 was significantly associated with shorter
PFI and OS in KIRC. However, the expression of FSCN2 was decreased
in KIRC, consistent with the results of the PCR experiment, and
FSCN1 and FSCN2 showed a certain coordinated expression pattern in
the present study. This may be because of their small sample sizes
and ethnic variations leading to inadequate statistical scope.
These findings should be further assessed and confirmed by other
studies. Multivariate analysis was conducted and high expression of
FSCN2 was proved to be an independent positive prognosis indicator
for OS in patients with KIRC. Further efforts are required to
explore the expression of FSCN2 and how FSCN2 affects the patient
survival. The TIMER analysis showed that FSCN2 is positively
correlated with immune infiltration and provided strong evidences
to support the high correlation between CNV and FSCN2 gene
expression, indicating that abnormal expression of FSCN2 might
affect the tumor cell microenvironment and regulate tumor cell
behavior. FSCN2 must be further explored in order to determine how
it affects patient survival.
FSCN3, a newly identified testis-specific
actin-bundling protein, is specifically expressed in elongated
spermatids (17). Little
information is available in the literature regarding the role of
fascin actin-bundling protein 3 in KIRC. In the present study, the
expression level of FSCN3 was significantly increased in KIRC
compared with normal tissues and the results demonstrated that
patients with KIRC with high FSCN3 expression had a shorter OS time
compared with those with low expression. The FSCN3 expression was
positively correlated with the infiltration of immune cells
including neutrophil and T cell CD4 + cell. However, this needs
further study to investigate the FSCN3 gene.
The present study explored the expression and
prognostic value of FSCNs in KIRC by combining public database and
PCR experiments, providing an understanding of the role of FSCNs in
KIRC. However, there were a few limitations to the present study.
First, although enhanced expressions of FSCN1 and FSCN2 closely
related to longer OS and could serve as independent favorable
prognostic factors for OS in KIRC, it is necessary to conduct
further studies with larger sample sizes to validate the findings
of present study and explore the clinical application of FSCNs
members in KIRC. Second, little information is available in the
literature regarding the role of FSCN2 and FSCN3 in tumor.
Additional research is necessary to further explore these potential
mechanisms of FSCN2 and FSCN3.
In conclusion, present study showed that increased
expression levels of FSCN1 and FSCN3 were strongly associated with
shorter OS and that FSCN2 was an independent favorable prognostic
factor for OS in KIRC. The FSCN1 mRNA expression was found to be
significantly associated with clinical cancer stages and histologic
grades in patients with KIRC. The results indicated that FSCN1 and
FSCN2 could be treatment targets for KIRC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Fu Wenda and Dr
Wu Haiyan (pathologists, Department of Pathology, The First
Hospital of Putian City, Putian, China), who independently verified
the pathological diagnosis of tumor tissues.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, PZ and GC designed and directed the project. RC,
BH and MJ performed bioinformatic analysis and the PCR experiment.
BH and MJ confirmed the authenticity of all the raw data. YL
performed the PCR experiment analysis and wrote and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was conducted according to the
ethical principles of the Declaration of Helsinki and was approved
by the Institutional Ethics Committee of First Affiliated Hospital
of Nanchang university (approval no. 202012-110). Written informed
consent was obtained from the patients involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ljungberg B, Albiges L, Abu-Ghanem Y,
Bensalah K, Dabestani S, Fernández-Pello S, Giles RH, Hofmann F,
Hora M, Kuczyk MA, et al: European association of urology
guidelines on renal cell carcinoma: The 2019 update. Eur Urol.
75:799–810. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cairns P: Renal cell carcinoma. Cancer
Biomark. 9:461–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Escudier B, Porta C, Schmidinger M,
Rioux-Leclercq N, Bex A, Khoo V, Grunwald V, Gillessen S and
Horwich A; ESMO Guidelines Committee, : Electronic address:
simpleclinicalguidelines@esmo.org.
Renal cell carcinoma: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up†. Ann Oncol. 30:706–720. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Capitanio U and Montorsi F: Renal cancer.
Lancet. 387:894–906. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bex A, Albiges L, Ljungberg B, Bensalah K,
Dabestani S, Giles RH, Hofmann F, Hora M, Kuczyk MA, Lam TB, et al:
Updated European association of urology guidelines regarding
adjuvant therapy for renal cell carcinoma. Eur Urol. 71:719–722.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhalla S, Chaudhary K, Kumar R, Sehgal M,
Kaur H, Sharma S and Raghava GP: Gene expression-based biomarkers
for discriminating early and late stage of clear cell renal cancer.
Sci Rep. 7:449972017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li A, Dawson JC, Forero-Vargas M, Spence
HJ, Yu X, König I, Anderson K and Machesky LM: The actin-bundling
protein fascin stabilizes actin in invadopodia and potentiates
protrusive invasion. Curr Biol. 20:339–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adams JC: Roles of fascin in cell adhesion
and motility. Curr Opin Cell Biol. 16:590–596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schoumacher M, Goldman RD, Louvard D and
Vignjevic DM: Actin, microtubules, and vimentin intermediate
filaments cooperate for elongation of invadopodia. J Cell Biol.
189:541–556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kureishy N, Sapountzi V, Prag S, Anilkumar
N and Adams JC: Fascins, and their roles in cell structure and
function. Bioessays. 24:350–361. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamashiro S, Yamakita Y, Ono S and
Matsumura F: Fascin, an actin-bundling protein, induces membrane
protrusions and increases cell motility of epithelial cells. Mol
Biol Cell. 9:993–1006. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hayashi Y, Osanai M and Lee GH: Fascin-1
expression correlates with repression of E-cadherin expression in
hepatocellular carcinoma cells and augments their invasiveness in
combination with matrix metalloproteinases. Cancer Sci.
102:1228–1235. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vignjevic D, Schoumacher M, Gavert N,
Janssen KP, Jih G, Laé M, Louvard D, Ben-Ze'ev A and Robine S:
Fascin, a novel target of beta-catenin-TCF signaling, is expressed
at the invasive front of human colon cancer. Cancer Res.
67:6844–6853. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Darnel AD, Behmoaram E, Vollmer RT, Corcos
J, Bijian K, Sircar K, Su J, Jiao J, Alaoui-Jamali MA and Bismar
TA: Fascin regulates prostate cancer cell invasion and is
associated with metastasis and biochemical failure in prostate
cancer. Clin Cancer Res. 15:1376–1383. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shin JB, Longo-Guess CM, Gagnon LH, Saylor
KW, Dumont RA, Spinelli KJ, Pagana JM, Wilmarth PA, David LL,
Gillespie PG and Johnson KR: The R109H variant of fascin-2, a
developmentally regulated actin crosslinker in hair-cell
stereocilia, underlies early-onset hearing loss of DBA/2J mice. J
Neurosci. 30:9683–9694. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu X, Zhao M, Xie Y, Li P, Wang O, Zhou
B, Yang L, Nie Y, Cheng L, Song X, et al: Null mutation of the
Fascin2 gene by TALEN leading to progressive hearing loss and
retinal degeneration in C57BL/6J mice. G3 (Bethesda). 8:3221–3230.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tubb B, Mulholland DJ, Vogl W, Lan ZJ,
Niederberger C, Cooney A and Bryan J: Testis fascin (FSCN3): A
novel paralog of the actin-bundling protein fascin expressed
specifically in the elongate spermatid head. Exp Cell Res.
275:92–109. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin J, Jiang M, Chen R, Zheng P and Chen
G: Comprehensive analysis of the expression, prognostic value and
biological importance of OVO-like proteins in clear cell renal cell
carcinoma. Oncol Lett. 25:1792023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mermel CH, Schumacher SE, Hill B, Meyerson
ML, Beroukhim R and Getz G: GISTIC2.0 facilitates sensitive and
confident localization of the targets of focal somatic copy-number
alteration in human cancers. Genome Biol. 12:R412011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li B, Severson E, Pignon JC, Zhao H, Li T,
Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al: Comprehensive
analyses of tumor immunity: Implications for cancer immunotherapy.
Genome Biol. 17:1742016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim SJ, Choi IJ, Cheong TC, Lee SJ, Lotan
R, Park SH and Chun KH: Galectin-3 increases gastric cancer cell
motility by up-regulating fascin-1 expression. Gastroenterology.
138:1035–1045.e1-e2. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Onodera M, Zen Y, Harada K, Sato Y, Ikeda
H, Itatsu K, Sato H, Ohta T, Asaka M and Nakanuma Y: Fascin is
involved in tumor necrosis factor-alpha-dependent production of
MMP9 in cholangiocarcinoma. Lab Invest. 89:1261–1274. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vignjevic D, Kojima S, Aratyn Y, Danciu O,
Svitkina T and Borisy GG: Role of fascin in filopodial protrusion.
J Cell Biol. 174:863–875. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoder BJ, Tso E, Skacel M, Pettay J, Tarr
S, Budd T, Tubbs RR, Adams JC and Hicks DG: The expression of
fascin, an actin-bundling motility protein, correlates with hormone
receptor-negative breast cancer and a more aggressive clinical
course. Clin Cancer Res. 11:186–192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu D, Chen L, Liao W, Ding Y, Zhang Q, Li
Z and Liu L: Fascin1 expression predicts poor prognosis in patients
with nasopharyngeal carcinoma and correlates with tumor invasion.
Ann Oncol. 21:589–596. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hashimoto Y, Kim DJ and Adams JC: The
roles of fascins in health and disease. J Pathol. 224:289–300.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Machesky LM and Li A: Fascin: Invasive
filopodia promoting metastasis. Commun Integr Biol. 3:263–270.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tan VY, Lewis SJ, Adams JC and Martin RM:
Association of fascin-1 with mortality, disease progression and
metastasis in carcinomas: A systematic review and meta-analysis.
BMC Med. 11:522013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Cho IH, Park JH, Lee MK and Hwang
YS: Fascin is involved in cancer cell invasion and is regulated by
stromal factors. Oncol Rep. 41:465–474. 2019.PubMed/NCBI
|
|
36
|
Han S, Huang J, Liu B, Xing B, Bordeleau
F, Reinhart-King CA, Li W, Zhang JJ and Huang XY: Improving fascin
inhibitors to block tumor cell migration and metastasis. Mol Oncol.
10:966–980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McGuire S, Kara B, Hart PC, Montag A,
Wroblewski K, Fazal S, Huang XY, Lengyel E and Kenny HA: Inhibition
of fascin in cancer and stromal cells blocks ovarian cancer
metastasis. Gynecol Oncol. 153:405–415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Park KS, Yoon SY, Park SH and Hwang JH:
Anti-migration and anti-invasion effects of curcumin via
suppression of fascin expression in glioblastoma cells. Brain Tumor
Res Treat. 7:16–24. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao D, Zhang T, Hou XM and Ling XL:
Knockdown of fascin-1 expression suppresses cell migration and
invasion of non-small cell lung cancer by regulating the MAPK
pathway. Biochem Biophys Res Commun. 497:694–699. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang C, Wang J, Chen Z, Gao Y and He J:
Immunohistochemical prognostic markers of esophageal squamous cell
carcinoma: A systematic review. Chin J Cancer. 36:652017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wada Y, Abe T, Takeshita T, Sato H,
Yanashima K and Tamai M: Mutation of human retinal fascin gene
(FSCN2) causes autosomal dominant retinitis pigmentosa. Invest
Ophthalmol Vis Sci. 42:2395–2400. 2001.PubMed/NCBI
|